Abstract
Fine-scale genetic diversity and contemporary evolution can theoretically influence ecological dynamics in the wild. Such eco-evolutionary effects might be particularly relevant to the persistence of populations facing acute or chronic environmental change. However, experimental data on wild populations is currently lacking to support this notion. One way that ongoing evolution might influence the dynamics of threatened populations is through the role that selection plays in mediating the ‘rescue effect’, the ability of migrants to contribute to the recovery of populations facing local disturbance and decline. Here, we combine experiments with natural catastrophic events to show that ongoing evolution is a major determinant of migrant contributions to population recovery in Trinidadian guppies (Poecilia reticulata). These eco-evolutionary limits on migrant contributions appear to be mediated by the reinforcing effects of natural and sexual selection against migrants, despite the close geographic proximity of migrant sources. These findings show that ongoing adaptive evolution can be a double-edged sword for population persistence, maintaining local fitness at a cost to demographic risk. Our study further serves as a potent reminder that significant evolutionary and eco-evolutionary dynamics might be at play even where the phenotypic status quo is largely maintained generation to generation.
Keywords: adaptation, contemporary evolution, evolution, experimental evolution, genetics – empirical, natural selection and contemporary, population, population dynamics, population ecology, sexual selection
Introduction
Much of conservation biology seeks to understand and enhance the capacity of populations to accommodate chronic disturbance. However, many populations also face catastrophic disturbances that are fleeting but nonetheless overt in their influences on population abundance and odds of persistence (Shaffer 1981; Sousa 1984; Lande 1993; Spiller et al. 1998; Vignieri 2010). While a growing body of theory suggests an important role for genetic variation in determining demographic responses of populations to chronic disturbance (Kinnison and Hairston 2007; Chevin et al. 2010; Hendry et al. 2011), the role of genetic variation in population responses to impermanent catastrophic disturbances has received little attention in theoretical or applied contexts. Such lack of attention might owe in part to a perception that the rapid onset and fleeting nature of such disturbances limits the potential for evolutionary processes to influence population dynamics. In this study, we suggest why this might not always be the case and experimentally assess how genetic variation and ongoing evolution contribute to the recovery of wild populations facing actual catastrophic disturbances in the wild.
A key concept within metapopulation theory (Hanski 1999) is that migrants from productive patches (sources) can sustain other populations in harsh habitat patches where population growth is impaired (sinks) (Pulliam 1988). Within such metapopulations, particular demes might persistently function as sources or sinks, or they might switch between these states owing to catastrophic disturbances. Catastrophic disturbances can range from lasting effects on landscape features and selective conditions (e.g., volcanic eruptions) to more fleeting influences (e.g., occasional floods or droughts). Although often rare, such catastrophic disturbances can have large effects on population dynamics and extinction risk (Shaffer 1981; Lande 1993), and there have been several empirical examples of such effects (reviewed in Sousa 1984; Spiller et al. 1998; Vignieri 2010). Individual survival under such severe and abrupt disturbances may often be dictated by chance, more than adaptive trait variation, providing a distinction from the more subtle disturbances that have often characterized cases of contemporary evolution in the wild (Hendry et al. 2008).
If connected to other populations by individual dispersal, local populations recovering from catastrophic disturbance might receive a critical demographic boost, reducing their risk of extinction (the ‘rescue effect’–Brown and Kodric-Brown 1977). Alternatively, population recovery following disturbance might primarily be the result of demographic contributions from local, surviving individuals (Lindenmayer et al. 2005; Peakall and Lindenmayer 2006; Peery et al. 2010). The relative influence of these two processes depends largely on the fitness of migrants in their new habitat, which might be reduced compared to residents because of local adaptation (Nosil et al. 2005). In this framework, the nature of rescue effects, like many other problems in conservation biology, is not just ecological or evolutionary, but eco-evolutionary (Kinnison and Hairston 2007).
Uncertainty regarding the demographic benefits of migrants is further hinted by theoretical work that variously suggest that migration can impede, prevent, or promote population persistence (Ronce and Kirkpatrick 2001; Kawecki and Holt 2002; Holt et al. 2003; Garant et al. 2007). For example, immediately following a population disturbance, strong selection against maladapted migrants might increase the risk of extinction because of stochasticity associated with small population size, which in the short term (over one generation), might be more important than any genetic processes (outbreeding or genetic bottlenecks) (Lande 1988). However, over the long term, persistent reduction in fitness because of unchecked outbreeding could also lead to population declines and extinction (Ronce and Kirkpatrick 2001). In combination, the greatest risks to population persistence might occur at some intermediate level of migrant maladaptation where they are not so maladapted as to be readily culled by selection, but also not so fit as to greatly reduce demographic risk (Garant et al. 2007). Unfortunately, little experimental data exist on how local adaptation might modify the relative contributions of local and migrant individuals to population recovery in the wild, albeit some studies have variously suggested ways that selection and dispersal might interact to influence population dynamics (Hanski and Saccheri 2006; Duckworth and Badyaev 2007; Moore and Hendry 2009; Van doorslaer et al. 2009). In this study, we present the results of a series of experiments in wild populations of Trinidadian guppies (Poecilia reticulata) that demonstrate the potential for selection on migrants to influence demographic recovery following population collapses resulting from local catastrophic disturbances.
The Trinidadian guppy system
Trinidadian guppies inhabit streams characterized by waterfalls that prevent large predatory fish species from colonizing upstream sites (Endler 1978; Magurran 2005). These waterfalls have two important consequences for our study. First, adjoining guppy populations above versus below these falls show adaptive divergence in response to the contrasting predator regimes (Endler 1995; Magurran 2005). Traits showing adaptive divergence include shape (Hendry et al. 2006), life histories (Reznick and Endler 1982; Gordon et al. 2009), anti-predator behaviors (Magurran et al. 1992; O'Steen et al. 2002), and body coloration (Endler 1978; Millar et al. 2006; Weese et al. 2010). Moreover, these differences are genetically based and evolve on short time scales following experimental translocations between the two predation environments (Endler 1980; Magurran et al. 1992; O'Steen et al. 2002; Gordon et al. 2009). Second, migration and gene flow occur between predation environments, particularly from low-predation (LP) sites above waterfalls into high-predation (HP) sites below waterfalls (Becher and Magurran 2000; Crispo et al. 2006). Thus, within a particular river, the network of Trinidadian guppy populations can be described as an environmentally and phenotypically heterogeneous metapopulation.
Natural guppy populations sometimes experience catastrophic disturbances in the form of very large floods (Grether et al. 2001; Van oosterhout et al. 2007). A series of these floods occurred during the ‘dry season’ (January–March) in 2005 and 2006, reducing the HP population of the Marianne River by several orders of magnitude. For instance, during exhaustive sampling at our focal experimental site, we captured 216 females and 111 males in 2004, but only one female and no males in 2005 and six females and three males in 2006. These same floods did not have a similarly devastating effect on neighboring LP populations (that occur in lower order tributaries) or on the abundance of larger fish predators. After the flooding ended, the depleted populations of HP guppies were therefore likely experiencing higher proportional rates of immigration from the upstream LP habitats, particularly if LP fish were more likely to be distributed over barriers during high water. We here ask how these migrants might influence population recovery. As noted earlier, the answer is not straightforward because although the numerical effect should enhance recovery, strong selection on migrants (Nosil et al. 2005) might reduce this benefit.
We addressed two specific research objectives. First, we quantified selection against migrants by testing for potential differences in both survival and reproductive success between HP and LP guppies. Using equal numbers of both ecotypes, we established experimental populations (in 2 years) at a focal HP site and tested for differential survival using mark-recapture techniques. Based on phenotypic differences presumed to reflect adaptation to predation regimes, we predicted that the LP ecotype would have lower survival compared to the HP ecotype. We also tested for sexual selection on LP males relative to HP males using predator-free enclosures outside of our focal experimental site. Whether this sexual selection would act for or against the LP ecotype was not clear a priori. On the one hand, female guppies commonly prefer to mate with colorful males (Endler and Houde 1995), and so might preferentially mate with the more colorful LP migrants. On the other hand, high mortality rates of migrants and migrant phenotypes could select for positive associative mating by ecotype (Schluter 2005), in which case the LP males might have relatively low mating success with HP females.
Our second objective was to quantify the demographic contributions of local and migrant individuals to population recovery in our focal HP site. To do this, we used population genetic assignment techniques to test for ecotypic differences in the number of offspring contributed to subsequent generations of the experimental populations established at our focal HP site. While LP fish are sure to make an initial numeric addition to the experimental populations, their contribution to population growth (recovery) in subsequent generations will be strongly dependent upon their ability to survive and reproduce in the HP environment. Therefore, we predicted that the demographic contributions of the migrant (LP) guppies would be somewhat less than the local (HP) guppies.
Methods
Study site and mark-recapture techniques
All experiments were conducted in the Marianne River system, which flows from Trinidad's northern mountain range. Within the Marianne River drainage, three source populations were used for our experiments: the HP mainstem source and two LP sources (LP1 and LP2 respectively) (Fig. 1). The HP section of the Marianne River contains several species of potential predatory fishes including several species of goby: Eleotris pisonis, Gobiomorus dormitor, and Dormitator maculatus (Gobiidae); and a river ‘mullet’, Agonostomus monticola (Mugilidae). The LP tributaries of the Marianne River drainage contain less dangerous predators including a killifish (Rivulus hartii) and several species of predatory prawns (Macorbrachium spp.). Additional information describing the location of these tributaries, and their environmental characteristics, can be found in a series of publications describing the color (Millar et al. 2006), shape (Hendry et al. 2006), and population genetic structure (Crispo et al. 2006) of the guppies inhabiting this river.
Figure 1.
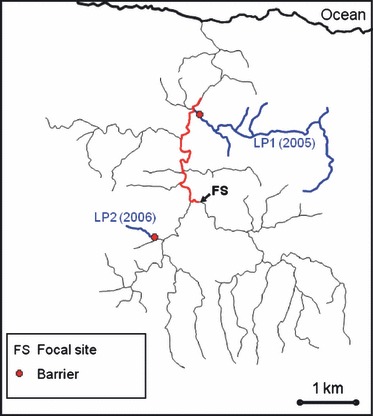
Map of the Marianne River drainage. Our focal site (FS) is where experimental populations were established. LP1 and LP2, shown in blue, indicate the locations of the two low-predation (LP) source populations used in 2005 and 2006, respectively. We have also indicated the location of barriers that are thought to have prevented the colonization of these LP tributaries by predatory fish. Shown in red is the section of the river where we observed that the guppy population had been decimated by floods in 2005 and 2006. We have confirmed the presence of predatory fish throughout the red section. The high-predation guppies introduced into our focal site originated from a series of localized side channels, within the red section (but well below our focal site), where some guppies had resisted the floods. Thus, as none of our guppies originated from the focal site, there is no potential for a home-site advantage.
To study differential survival of HP and LP ecotypes in the HP habitat, we introduced approximately equal numbers of marked guppies from each ecotype into a focal HP site (Fig. 1) and recaptured the fish and their offspring every 2 weeks for approximately 4 months (Table 1) using standard mark-recapture techniques for guppies (Rodd and Reznick 1991; Reznick et al. 1996; Olendorf et al. 2006; Van oosterhout et al. 2007; Gordon et al. 2009; Weese et al. 2010). Two separate experimental introductions were implemented using different LP sources, one in 2005 using LP1 guppies and one in 2006 using LP2 guppies. High-predation fish came almost entirely from mainstem river sections well below our study reach, eliminating the potential for a home-site advantage (Fig. 1). In the second year of our research, we elected to manipulate the source population of the LP migrants rather than the proportion of migrants introduced into the system. Of course, this latter experimental approach would have also revealed valuable insights regarding the role of selection against migrants in mitigating the rescue effect, unfortunately logistical constrains limited us to only two trials of this field experiment.
Table 1.
Genotypes of experimental guppies. Parents and recruits assigning to the high-predation population cluster (HP), or the low-predation population cluster (LP1 or LP2) throughout the duration of both introduction experiments (2005 and 2006). Recapture episodes occurred approximately every 2 weeks
| Year | Genotype | Release | Recap 1 | Recap 2 | Recap 3 | Recap 4 | Recap 5 | Recap 6 | Recap 7 |
|---|---|---|---|---|---|---|---|---|---|
| 2005 | HP | 85 (0) | 62 (1) | 85 (40) | 117 (49) | 135 (54) | 99 (53) | 81 (21) | – |
| LP1 | 83 (0) | 18 (0) | 18 (6) | 12 (3) | 9 (3) | 5 (1) | 10 (3) | – | |
| 2006 | HP | 99 (0) | 72 (0) | 63 (0) | 67 (13) | 79 (24) | 116 (45) | 34 (7) | 28 (4) |
| LP2 | 98 (0) | 55 (0) | 29 (0) | 8 (0) | 9 (3) | 6 (1) | 0 (0) | 0 (0) |
Numbers in parentheses represent the subset of the total number of guppies that were captured during a particular recapture episode (assumed to be offspring of introduced guppies).
Before release, each guppy was individually marked with two sub-cutaneous injections of elastomer dye (Northwest Marine Technology). Using a combination of six different colors and (up to) six different anatomical locations, two sub-cutaneous injections provided 540 individually identifiable marking codes for each sex per year. Each recapture episode occurred over 2 days. On the first day, we sampled through the entire study site until no fish were apparent. We then returned the next day to capture any remaining fish that might have been missed during our first attempt. Our focal site (Fig. 1) was a series of five pools located just downstream from a steep and extensive set of cascades and upstream of another rapid and a small but deep gorge. We anticipated that these ‘barriers’ would discourage guppy emigration out of the site, and guppy immigration into the site. To assess the extent of these two processes, during each recapture episode, we sampled for guppies in the pool immediately above our upstream barrier, and we also sampled all downstream pools within 500 m of the gorge that delimited our focal site. Regarding emigration, it is possible that unrecaptured guppies might have either perished or emigrated out of the areas we sampled. While distinguishing between these options is biologically interesting, from the perspective of local population recovery the ultimate consequence of these two fates is identical – an individual that either emigrates or perishes will fail to make a contribution to population growth. Very few experimental (tagged) guppies were encountered downstream of our focal site (these emigrants were not included in any subsequent analysis), and none were ever encountered upstream of our focal site. Regarding immigration into the focal site, a potentially serious concern is that individuals from nearby naturally recovering HP populations could immigrate into our focal site. This would result in an overestimation of the demographic contribution of the introduced HP individuals to population recovery. However, for two important reasons, we do not anticipate that this would be a serious problem. First, we never encountered any unmarked (or marked) guppies in the pool above our focal site. If immigration from upstream sources into the focal site was common, we should have also observed the presence of unmarked guppies (originating from upstream) in the pool immediately above our focal site (which seemed to contain adequate guppy habitat – but never any guppies). Second, marked guppies that we captured downstream of the focal site never passed the downstream barrier of the focal site to re-enter our experimental pools. Thus, while we did capture unmarked guppies downstream of the focal site (during the later recapture episodes) that could have been either the offspring of our experimental guppies or migrant guppies from adjacent naturally recovering sections of the Marianne River, we doubt this latter category of guppy would strongly influence our results because there is no evidence that guppies swam across the downstream barrier of the focal site (based on the behavior of the marked guppies). The program MARK (White and Burnham 1995) was used to simultaneously estimate recapture and survival probabilities from mark-recapture data. We predicted that HP ecotypes would have higher survival than LP ecotypes, and thus the most likely mark-recapture model would produce ecotype-specific estimates of survival. We performed separate MARK analyses for each sex and year (total of four). The program MARK tests the mark-recapture data for overdispersion (individuals with large gaps in their capture history) using a bootstrapping approach. Our data did not show evidence of overdispersion (P > 0.05); thus, we compared our candidate models using Akaike's Information Criteria (AICc). For each analysis, our suite of candidate models variously included separate parameter estimates (survival and recapture probability) for different recapture episodes and different source populations (ecotypes) (Table 2).
Table 2.
Mark-recapture model selection. Results of four separate MARK (White and Burnham 1995) analyses for each combination of sex and year. For each analysis, rows represent particular candidate models, which each estimate survival (Φ) and recapture (p) probability. Each candidate model variously estimates regime (reg) or recapture-episode (ti) specific parameter values as well as interactions between these effects. Thus, models vary in the number of parameters they estimate (K). The most likely candidate model has the lowest Akiake's Information Criteria score (AICc). ΔAIC is the difference between the AIC for a given model and that for the best model. AIC weights (w) give relative likelihoods of the different models
| Model | AICc | ΔAIC | w | Likelihood | K | Deviance |
|---|---|---|---|---|---|---|
| 2005 Females | ||||||
| {Φ(reg) p(.)} | 426.39 | 0.00 | 0.333 | 1.000 | 3 | 89.17 |
| {Φ(reg*ti) p(.)} | 426.56 | 0.18 | 0.304 | 0.915 | 15 | 62.97 |
| {Φ(reg*ti) p(reg)} | 427.65 | 1.27 | 0.177 | 0.531 | 16 | 61.71 |
| {Φ(reg) p(reg)} | 428.37 | 1.98 | 0.123 | 0.371 | 4 | 89.08 |
| {Φ(reg) p(ti)} | 430.79 | 4.40 | 0.037 | 0.111 | 9 | 80.78 |
| {Φ(reg*ti) p(ti)} | 431.62 | 5.23 | 0.024 | 0.073 | 20 | 56.03 |
| {Φ(ti) p(.)} | 437.24 | 10.85 | 0.001 | 0.004 | 8 | 89.42 |
| {Φ(ti) p(reg)} | 439.42 | 13.04 | 0.000 | 0.002 | 9 | 89.42 |
| {Φ(ti) p(ti)} | 441.88 | 15.50 | 0.000 | 0.000 | 13 | 82.91 |
| {Φ(reg*ti) p(reg*ti)} | 445.68 | 19.29 | 0.000 | 0.000 | 27 | 52.18 |
| 2005 Males | ||||||
| {Φ(reg) p(ti)} | 246.31 | 0.00 | 0.508 | 1.000 | 9 | 32.61 |
| {Φ(reg) p(.)} | 248.36 | 2.05 | 0.182 | 0.358 | 3 | 47.81 |
| {Φ(reg) p(reg)} | 249.17 | 2.86 | 0.122 | 0.239 | 4 | 46.51 |
| {Φ(reg*ti) p(ti)} | 249.39 | 3.09 | 0.109 | 0.214 | 16 | 18.83 |
| {Φ(reg*ti) p(reg*ti)} | 251.49 | 5.19 | 0.038 | 0.075 | 17 | 18.37 |
| {Φ(reg*ti) p(.)} | 252.60 | 6.29 | 0.022 | 0.043 | 11 | 34.26 |
| {Φ(reg*ti) p(reg)} | 252.82 | 6.51 | 0.020 | 0.039 | 12 | 32.11 |
| {Φ(ti) p(reg)} | 269.98 | 23.67 | 0.000 | 0.000 | 9 | 56.28 |
| {Φ(ti) p(ti)} | 274.90 | 28.59 | 0.000 | 0.000 | 13 | 51.78 |
| {Φ(ti) p(.)} | 278.64 | 32.34 | 0.000 | 0.000 | 8 | 67.22 |
| 2006 Females | ||||||
| {Φ(reg) p(ti)} | 578.93 | 0.00 | 0.843 | 1.000 | 10 | 107.39 |
| {Φ(reg*ti) p(reg)} | 583.63 | 4.70 | 0.081 | 0.096 | 16 | 98.93 |
| {Φ(reg*ti) p(.)} | 584.97 | 6.04 | 0.041 | 0.049 | 15 | 102.50 |
| {Φ(reg*ti) p(ti)} | 585.44 | 6.51 | 0.032 | 0.039 | 21 | 89.35 |
| {Φ(reg*ti) p(reg*ti)} | 590.62 | 11.69 | 0.002 | 0.003 | 25 | 85.11 |
| {Φ(ti) p(reg)} | 611.66 | 32.73 | 0.000 | 0.000 | 10 | 140.11 |
| {Φ(reg) p(.)} | 618.00 | 39.07 | 0.000 | 0.000 | 3 | 161.13 |
| {Φ(ti) p(.)} | 618.10 | 39.17 | 0.000 | 0.000 | 9 | 148.69 |
| {Φ(reg) p(reg)} | 618.57 | 39.63 | 0.000 | 0.000 | 4 | 159.64 |
| {Φ(ti) p(ti)} | 619.85 | 40.92 | 0.000 | 0.000 | 15 | 137.38 |
| 2006 Males | ||||||
| {Φ(reg) p(.)} | 401.14 | 0.00 | 0.293 | 1.000 | 3 | 58.71 |
| {Φ(reg) p(ti)} | 401.20 | 0.07 | 0.283 | 0.966 | 7 | 50.33 |
| {Φ(reg) p(reg)} | 402.32 | 1.19 | 0.162 | 0.552 | 4 | 57.82 |
| {Φ(reg*ti) p(reg)} | 402.42 | 1.29 | 0.154 | 0.525 | 12 | 40.49 |
| {Φ(reg*ti) p(.)} | 403.63 | 2.50 | 0.084 | 0.287 | 11 | 43.96 |
| {Φ(reg*ti) p(ti)} | 406.60 | 5.46 | 0.019 | 0.065 | 14 | 40.08 |
| {Φ(reg*ti) p(reg*ti)} | 411.25 | 10.11 | 0.002 | 0.006 | 18 | 35.27 |
| {Φ(ti) p(reg)} | 411.29 | 10.16 | 0.002 | 0.006 | 7 | 60.42 |
| {Φ(ti) p(.)} | 413.26 | 12.12 | 0.001 | 0.002 | 6 | 64.53 |
| {Φ(ti) p(ti)} | 417.25 | 16.11 | 0.000 | 0.000 | 9 | 62.02 |
Enclosure experiment
We performed an enclosure experiment to isolate the effects of sexual selection from viability selection. To do this, we first collected immature guppies from the HP section of the Marianne River (Fig. 1) and maintained females as virgins until they reached maturity. We then constructed a barrier across the mouth of a side channel downstream from our focal site and removed all potential predators and guppies. To test for differences in reproductive success between LP and HP males, we placed virgin HP female guppies into the enclosed side channel along with a mixture of HP and LP males from our source populations (Table 3). Males from our LP1 and LP2 populations were assessed against the same HP source in independent trials. Before release, each fish was marked and provided scale samples for DNA. These fish were left in the enclosure for 2 days, after which guppies were recaptured from the enclosed side channel. A flash flood allowed some guppies to escape from the LP2 versus HP experiment while we were removing the guppies from the enclosure. This reduced our sample of females for this comparison (Table 3), but not males because we had collected scale samples from males (from which we extracted DNA), prior to introducing them into the enclosure. For both experiments, recaptured females were returned to our field station; and after 2 weeks, they were dissected, and four embryos were haphazardly selected for parentage analyses.
Table 3.
Numbers and origins of guppies in enclosure experiment. Numbers of experimental high-predation (HP) females, HP males, and LP males in a predator-free side channel of the Marianne River, and the total number of offspring that were sired by each male ecotype. Sample sizes differ between trials (LP1 versus HP and LP2 versus HP) because a flash flood allowed some guppies to escape from the LP2 versus HP experiment while we were removing the guppies from the enclosure. This reduced our sample of females, but not males as we had collected scale samples from males (from which we extracted DNA), prior to introducing them into the enclosure
| Trial | No. of females | No. of HP males | No. of LP males | HP offspring | LP offspring |
|---|---|---|---|---|---|
| LP1 versus HP | 25 | 12 | 12 | 29 | 15 |
| LP2 versus HP | 8 | 16 | 16 | 14 | 2 |
HP, high predation; LP, low predation.
Mothers, candidate sires, and offspring were genotyped at six microsatellite loci: Pre15, Pre53, Pre8, Pre9, Pre46, and Pre 32. Details of extraction and amplification methods are provided elsewhere (Paterson et al. 2005; Crispo et al. 2006). We assigned paternity using the program Cervus 3.0 (Kalinowski et al. 2007), which uses a likelihood-based approach to estimate the difference in log-likelihood scores between multiple candidate sires. We were conservative in our assignments and only further considered offspring whose father was known with greater than 95% confidence – 44 out of 94 offspring in the LP1 versus HP trial and 16 out of 32 in the LP2 versus HP trial. These data were then analyzed in a general linear model where the dependent variable was the number of confidently assigned offspring sired by individual males, and the independent variables were predation regime, trial (LP1 versus HP; LP2 versus HP), and the interaction term between regime and trial. Our inability to confidently assign parentage to many of our experimental offspring introduces the possibility that one ecotype or the other might be more likely to be identified as a father. While we stress this caveat, we would also like to point out that our results were qualitatively similar (HP males were more successful) in a supporting analysis where we assigned a much larger proportion of offspring to parental ecotype as opposed to individual sires.
Population assignment of wild recruits
DNA was extracted from the scale samples of all guppies initially released in our focal site (Fig. 1), and all individuals were genotyped at 11 microsatellite loci: Pre9, Pre13, Pre15, Pre26, Pre32, Pre38, Pre39, Pre46, Pre53, Pre72, and Pre80 – details of microsatellite amplification are provided elsewhere (Paterson et al. 2005; Crispo et al. 2006). The program STRUCTURE 2.2 (Pritchard et al. 2000) was then used to assign (separately for each year) individuals to either the HP or LP source population. STRUCTURE uses a Bayesian clustering approach to estimate the number of populations in a data set (K), and can probabilistically assign individuals to one of the identified populations or indicate whether an individual has an admixed genotype. We performed analyses that considered values of K between 1 and 15 and used the admixture model, with burn-in and Monte Carlo Markov chain values of 10 000 each. We used the correction of Evanno et al. (2005) to determine the most probable value of K. In each year, these analyses resulted in the identification of a ‘LP’ cluster to which nearly all of the LP guppies assigned with a high probability (see Results). Unmarked guppies sampled during the recapture episodes of our experimental populations were assumed to be the offspring of the originally introduced individuals. For reasons explained earlier, we do not anticipate that unmarked fish immigrating into our focal site would be incorrectly classified as the offspring of our initial experimental fish, although it is possible that a few immigrants invaded our site (likely from HP source populations) and that this might bias our results toward assigning a larger proportion of the subsequent generation to the HP ecotype. Individual offspring were assigned a Q-value that represents the probability that an individual's parents were from one of the HP source clusters (see Results) or the LP source cluster. As all of the female guppies used in this experiment would have entered the site pregnant, most of the recruits into our experimental population should have had either pure LP or pure HP genotypes. Most offspring assigned to the LP genetic cluster with either a very high (Q > 0.9) or very low (Q < 0.1) probability, making it relatively easy to estimate the genetic and demographic contribution of the LP ecotypes to population growth. However, it is possible that hybrids, resulting from in situ copulations, might have been present in the population for our later recapture events (in guppies gestation period is approximately 1 month, and the number of days between parturition and maturity is 40–70 days). Our approach for dealing with potential hybrids was to categorize all individuals with a 50% or greater probability of assignment to the LP cluster as LP fish. Any fish that assigned with the highest probability to one of the HP clusters was classified as a HP offspring. In 2005, four individuals had the highest probability of assignment to the LP cluster, but with a Q-value that was <50%, these individuals were excluded from the calculations of demographic contributions to population growth; no such individuals were detected in 2006. Our criteria for categorizing offspring might have resulted in a few individuals with hybrid genotypes being categorized as either LP or HP offspring; however, such individuals would be relatively few in number and would only have a minor influence on our estimates of demographic contributions for the last couple recapture events. Generally, this analysis allowed us to estimate the genetic and demographic contribution of each ecotype to the subsequent generation of the experimental population.
Results
Differential survival of ecotypes
Our mark-recapture experiment (performed at our focal site) found that LP guppies experienced very high mortality, compared to the HP guppies, when the two were tested together in a novel HP habitat (Tables 1 and 2, Fig. 2). This conclusion is well supported because the most likely candidate models for all four MARK analyses had ecotype-specific estimates of survival, while the least-likely candidate models typically did not (Table 2). All models lacking an ecotype-specific survival estimate have a delta AIC value of at least 10. For the 2005 females and 2006 males, the most likely candidate model estimated an ecotype-specific term for survival, and neither an ecotype-specific nor a recapture-episode-specific term for recapture probability (Table 2). For the 2005 males and 2006 females, the most likely candidate model included an ecotype-specific survival term and a recapture probability term that depended on the recapture episode (Table 2), indicating that our ability to sample all guppies in the focal site differed between recapture episodes. This result is possibly because of variability in environmental conditions (water level or clarity). Consistent with most other guppy mark-recapture studies, females had much higher survival than males (Table 2, Fig. 2).
Figure 2.
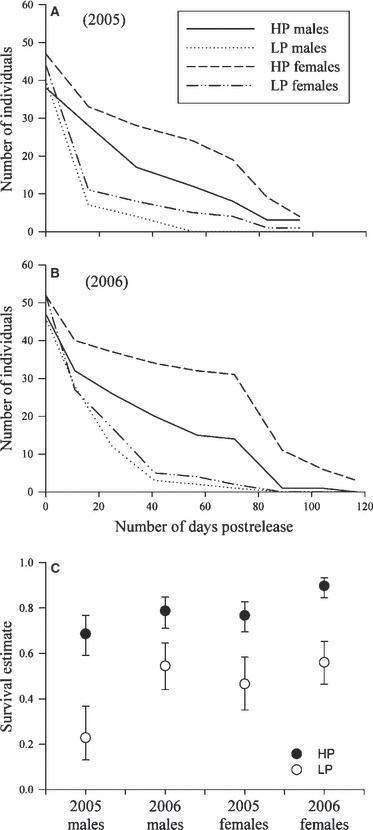
Survival of guppies introduced to our focal site. Numbers of the high- and low-predation guppies originally introduced into our experimental site for 2005 (Fig. 1A) and 2006 (Fig. 1B) plotted against number of days postrelease. Probability of survival over a recapture interval (Ψ) was formally estimated for the experimentally introduced fish using the program MARK (Fig. 1C), errors are 95% confidence intervals.
Differential mating success of ecotypes
In all enclosure experiments, HP males sired more offspring than their LP counterparts (Tables 3 and 4), despite equal numbers of both ecotypes in the enclosures. For the 56 male guppies used in these experiments, reproductive success ranges from 0 to 6 offspring. In the LP1 versus HP trial, there were 29 offspring with HP fathers and 15 offspring with LP fathers. Differences in reproductive success were more dramatic in the LP2 versus HP trial where 14 offspring were sired by HP fathers, whereas only two offspring had LP fathers. Overall, the least-squares mean number of offspring sired by HP males was more than twice the mean number of offspring sired by LP males (2.42:1, P = 0.017) (Table 4). There was also a significant effect of trial in this analysis (Table 4), which is because of the reduced number of females from the LP2 versus HP trial (see Methods).
Table 4.
Results of enclosure experiment. Results of a general linear model that tested for a difference in reproductive success (offspring sired) between high- and low-predation (LP) male guppies from the two separate trials of the enclosure experiment. A total of 56 male guppies, whose reproductive success ranged from 0 to 6, were included in the analysis. From this analysis, the least-squares mean number of offspring sired by high-predation and LP candidate sires was 1.65 and 0.68, respectively
| Factor | DF | F-ratio | P-value |
|---|---|---|---|
| Regime | 1 | 6.1 | 0.017 |
| Trial | 1 | 11.9 | 0.0011 |
| Trial × Regime | 1 | 0.3 | 0.5 |
Differential demographic contributions of ecotypes
In both years, the experimental populations at our focal site initially declined, which was expected because we did not consider offspring as having recruited to the population until they reached maturation (about 40–70 days after birth) (Table 1, Fig. 3). Also in both years, secondary floods (starting approximately 65 days after introduction) caused population declines preceding the end of the experiments (Fig. 3). After these initial declines, population size increased again, and in both years, the majority of these recruits were from the HP ecotype (Figs 3 and 4).
Figure 3.
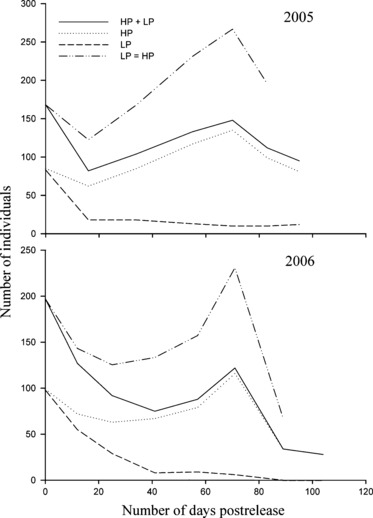
Population size at our focal site. The numbers of guppies (parents and offspring) whose genotypes assign to either the high- (HP) or low-predation (LP) population clusters, and the total number of guppies in the experimental population (HP + LP) plotted against the number of days postrelease. Also included is predicted population size assuming selective equivalence between the high- and low-predation ecotypes (LP = HP). This last line was generated by applying the high-predation (HP) birth rate and death rate to the total population size at the previous recapture episode {Nt = Nt−1−[Nt−1(HP deathrate)] + [Nt−1(HP birthrate)]}.
Figure 4.
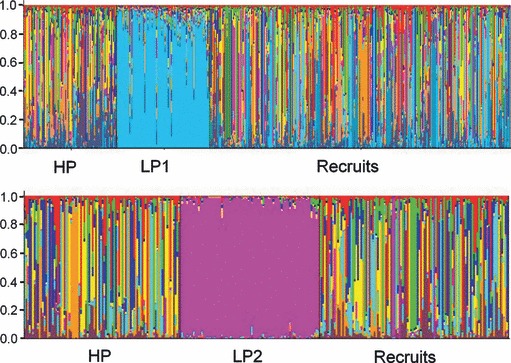
Genetic structure of experimental population. Output of STRUCTURE analyses. In 2005, the most likely number of clusters (K) was 12 (top); in 2006, the most likely number of clusters was 8 (bottom). In both years, these analyses identified a primary low-predation genetic cluster, and multiple high-predation genetic clusters. Each experimental individual (parents and recruits) is represented by a single vertical line. These lines are partitioned into colored segments which represent that individual's estimated membership fraction in a particular genetic cluster (Q-value).
In both years, our STRUCTURE analyses revealed that there was a primary LP genetic cluster to which most of the LP parents assigned with a high Q-value (Table 5, Fig. 4). These analyses also revealed, however, an unexpectedly high number of population clusters within our HP guppies. In 2005, the most likely number of genetic clusters was 12 (including the LP cluster). In 2006, the most likely number of genetic clusters was 8 (including the LP cluster). We collected the HP experimental fish from a few pools within a small area downstream of the focal site. Therefore, we are not certain if these genetic clusters reflect a high degree of biological realism. Indeed, using a similar STRUCTURE analysis, Crispo et al. (2006) studied genetic structure among these same guppy populations (in addition to a much broader geographical sampling) and found the most likely number of clusters to be 7. It is possible that the somewhat unusual findings of the current STRUCTURE analysis are the result of founder effects caused by the recent demographic collapse of the HP populations. This possibility, while potentially interesting in its own right, should not detract from our ability to identify the progeny of the low-predation guppies, as any individual with a LP parent should have a Q-value indicating a high probability of assignment to the LP genetic cluster. In 2005, 218 recruits were assigned to one of the HP genetic clusters, and 16 were assigned to the LP1 genetic cluster (Q-value > 0.50) (Tables 1 and 5; Fig. 4). In 2006, 93 recruits were assigned to the HP population, only four assigned to the LP (LP2) population (Tables 1 and 5; Fig. 4). Thus, although LP ecotypes did contribute to population recovery in a HP environment in both years, the overwhelming majority of recruitment was from the HP ecotype.
Table 5.
Results of STRUCTURE models using the most likely number of population clusters (K = 12 in 2005, and K = 8 in 2006). Proportions of genotypes from each source population, either low predation (LP) (LP1 in 2005 and LP2 in 2006 – see Methods) or high predation (HP). Offspring is the proportions of genotypes from the individuals that recruited into the experimental population (see Methods) within each of the inferred clusters
| Inferred cluster | HP | LP | Offspring |
|---|---|---|---|
| 2005 | |||
| 1 | 0.058 | 0.017 | 0.092 |
| 2 | 0.086 | 0.011 | 0.086 |
| 3 | 0.089 | 0.018 | 0.063 |
| 4 | 0.148 | 0.016 | 0.083 |
| 5 | 0.056 | 0.015 | 0.075 |
| 6 | 0.047 | 0.033 | 0.083 |
| 7 | 0.068 | 0.016 | 0.079 |
| 8 | 0.095 | 0.017 | 0.085 |
| 9 | 0.146 | 0.021 | 0.085 |
| 10 | 0.04 | 0.013 | 0.104 |
| 11 | 0.151 | 0.022 | 0.065 |
| 12 | 0.016 | 0.801 | 0.101 |
| 2006 | |||
| 1 | 0.143 | 0.007 | 0.144 |
| 2 | 0.084 | 0.009 | 0.148 |
| 3 | 0.156 | 0.007 | 0.133 |
| 4 | 0.174 | 0.008 | 0.136 |
| 5 | 0.014 | 0.951 | 0.052 |
| 6 | 0.13 | 0.006 | 0.145 |
| 7 | 0.159 | 0.005 | 0.096 |
| 8 | 0.139 | 0.007 | 0.147 |
To assess how selection on migrants may have influenced the population dynamics of recovery, we must consider how local populations would have responded in the absence of migrants or in the absence of contemporary evolution. In Fig. 3, we plot the observed size of our experimental populations through time, along with the relative numbers of individuals whose genotypes assigned to either the HP or LP (including hybrids) populations. We also present the expected size of the experimental population under a ‘null selection model’– which assumes ecological equivalence between ecotypes (calculated by applying the local HP birth and death rates to the total population size at the previous recapture interval, see Fig. 3). To quantify the demographic benefit of migrants, we can compare the observed population size to the number of individuals with pure HP genotypes. When the experimental population size was maximal, this benefit amounted to 13 recruits (9% of the population) in 2005 and six recruits (5% of the population) in 2006. To estimate the demographic cost of contemporary evolution in the form of selection on migrants, we can compare the observed population size to that estimated under the null selection model. The latter exceed the former by 119 individuals (a 45% cost compared to the null selection model) in 2005 and 108 individuals (a 47% cost) in 2006.
Discussion
We combined natural catastrophes with controlled experiments to assess the combined roles of contemporary evolution and demographic rescue on population recovery following a catastrophic disturbance. A series of massive floods decimated the guppy population in the HP section of the Marianne River. We predicted that population recovery might be accelerated by demographic contributions from neighboring migrant sources into remnant populations. However, we also predicted that, because of local adaptation, the LP ecotype would have higher mortality in the HP environment compared to the local HP ecotype and that selection against migrants would constrain the demographic benefit of any population ‘rescue’. Ultimately, selection against LP guppies was even stronger than we anticipated and thus played a major role in constraining population recovery in our focal HP site. At the same time, such selection also assured that the overwhelming majority of individuals in subsequent generations were offspring of the local ecotype, thus maintaining the long-term fitness of the population.
Differential fitness of HP and LP ecotypes
Consistent with our predictions, HP guppies had much higher survival rates than LP guppies in our focal HP site. This result is unequivocal and applies to both males and females, and both sources of LP guppies (LP1 and LP2). Our head-to-head comparison of ecotype survival is particularly instructive because such assessments quantify the net effects of multifarious selection on comprehensive phenotypes. Differences in survival rates appear to be much stronger than the relatively subtle phenotypic divergence among Marianne River populations in shape (Hendry et al. 2006) and color (Millar et al. 2006; Weese et al. 2010) thought to reflect adaptation to divergent predation regimes. Compared to our findings, studies that have estimated contemporary patterns of selection associated with particular phenotypic traits for guppies have produced more equivocal results. For selection associated with body size (Reznick et al. 1996) and color (Weese et al. 2010), the pattern and strength of selection seems to be similar in both HP and LP sites, inconsistent with predictions distilled from phenotypic differences. Strong survival effects have been noted in another experimental introduction of guppies (Gordon et al. 2009) and in studies of salmon introduced to New Zealand (Kinnison et al. 2008). Taken together, these findings reinforce the idea that many individual traits interact to determine overall adaptation and that assessment based on single characters will often be insufficient.
The ultimate demographic contributions of migrant versus local males to a recovering population will depend not only on viability selection but also on the nature of sexual selection. Thus, using predator-free enclosures, we also tested for relative mating success of migrants relative to residents. Again, the HP ecotype seemed to have higher fitness than the LP ecotype. The average number of offspring per male was nearly three times higher for HP males. Because predators were not present in the enclosures, we suggest that this difference in reproductive success was the result of sexual selection, not viability selection. Because multiple males and females were in each field enclosure, the differences reflect some unknown combination of overt female choice, coercive (i.e., sneak) mating by males, male–male aggression, sperm competition, and female sperm sorting (Magurran 2005). Given the limited number of females used in these enclosure experiments, the lack of replication (of enclosure treatments), and our inability to determine paternity for a large number of experimental offspring, we are somewhat cautious in our conclusion that the HP ecotype have higher reproductive fitness. However, we suggest this small-scale experiment is useful for two reasons: (i) Most previous studies of sexual selection in guppies have used individual fish in a laboratory setting; our use of multiple fish in a field enclosure makes our assessment of reproductive fitness somewhat more realistic, and we encourage future work to implement similar experiments at a larger scale. (ii) While the extent that HP males outperform LP males is questionable, our original intent was to evaluate whether or not the putative attractiveness of LP males could augment gene flow in the face of strong viability selection (see Introduction). Such an effect could have a strong influence on the demographic recovery of the local population over subsequent generations. This does not seem to be the case, even if we accept that the evidence for HP superiority is equivocal.
Thus, owing to strong viability selection and a probable reinforcing effect of sexual selection, LP guppies have lower fitness in a HP environment than do HP guppies, or in other words, there is profound selection against migrants even given the close geographic proximity of migrant sources and evidence that gene flow does occur (Crispo et al. 2006). Lower fitness does not by itself preclude a demographic ‘rescue effect’– that is, these migrants might still have a positive effect on population growth following a disturbance. We therefore specifically quantified the potential rescue effect by monitoring the demographic contributions (offspring recruitment) of each ecotype to our experimental population after the introduction in each year.
Demographic consequences of selection against migrants
We predicted that, because of local adaptation, the demographic contribution of the migrants (LP) would be reduced compared to the contribution of the local (HP) guppies. However, we were surprised by the magnitude of the difference in the demographic contribution made by locals versus migrants. Compared to the expectations of the ‘null selection model’, the observed population size at our focal experimental site was drastically reduced; this comparison is heuristically informative in showing how ongoing contemporary evolution, in the form of selection against migrants, can play a potentially dominant role in the dynamics of wild populations. Such eco-evolutionary dynamics might easily be overlooked in nature, where they could be considered ‘cryptic’ in the sense that they occur in the absence of any apparent change in selective conditions and without overt trait changes generation to generation. Importantly, although HP populations might benefit less from an immediate rescue effect, selection appears to be very effective in limiting genetic loads that might otherwise impair mean local fitness and rates of rebound during subsequent generations or future disturbances (Ronce and Kirkpatrick 2001). Our experiment was not designed to assess potential demographic costs or benefits beyond the F1 generation; however, we anticipate that if selection against migrants was impeded and gene flow allowed for several generations (compromising local adaptation), the HP population might be placed in appreciable risk because of reduction in average fitness. Such effects have been noted in a limited number of studies investigating wild salmon recovery programs that have attempted to use non-native sources for population restoration (McClelland and Naish 2007; Araki et al. 2008). It remains to be seen whether eco-evolutionary effects ultimately place particular populations at higher or lower risk of extinction. Ideally, future work investigating this topic could compare the responses of localized populations that either received or did not receive, an initial demographic contribution from migrants. It would be especially beneficial, under this scenario, to have multiple treatments representing variable levels of population mixing, and to track the genetic and demographic contributions of migrants and locals over multiple generations.
Conservation implications
The metapopulation concept is fundamental to modern conservation biology, including efforts to preserve biodiversity (Damshen et al. 2006) and to predict biological responses to climate change (Loarie et al. 2009). Furthermore, interactions between divergent selection, adaptive divergence and gene flow are fundamental to evolutionary theory (Hendry et al. 2001; Kawecki and Holt 2002; Whitlock 2002). Few empirical studies, however, have specifically linked evolutionary and metapopulation theory to evaluate the eco-evolutionary dynamics associated with selection against migrants (Hanski and Saccheri 2006; Duckworth and Badyaev 2007; Moore and Hendry 2009), much less the role of such dynamics in population recovery from catastrophic population disturbance. Our experimental assessment supports prior theoretical work (Boulding and Hay 2001; Ronce and Kirkpatrick 2001; Kawecki and Holt 2002; Holt et al. 2003; Garant et al. 2007; Kinnison and Hairston 2007) in suggesting important interactions between selection, migration, and demography in nature and places those interactions in a pressing conservation context – population recovery following catastrophe. Whereas prior studies of contemporary evolution in conservation contexts have tended to emphasize modest but persistent disturbance and directional trait change (Visser 2008; Darimont et al. 2009), such conditions are not prerequisite for eco-evolutionary conservation concerns. We have shown that eco-evolutionary dynamics might be a consideration even where disturbance is fleeting, selection patterns persist largely unchanged, net evolution is limited, and populations exchange migrants. The potential for eco-evolutionary dynamics to limit the efficacy of natural rescue effects or human restoration efforts should be considered carefully in light of evidence that humans might be accelerating both the incidence of catastrophic disturbance and the fragmentation of metapopulations into more physically isolated and ecologically divergent populations.
Acknowledgments
Field work was assisted by Sonya Auer, Michael Bailey, Ron Bassar, Craig Blackie, Laura Easty, Cory Gardner, Swanne Gordon, Zaki Jafri, Kevin Lachapelle, Brandon Libby, Nathan Millar, Ian Patterson, David Reznick, and Martin Turcotte. Lynn Anstey assisted with DNA extraction and sequencing. We would like to thank Louis Bernatchez, Robin Waples, and an anonymous reviewer for helpful comments. Funding was provided by the National Science Foundation (MTK and APH), the Maine Agricultural and Forestry Experiment Station (MTK), a doctoral fellowship from the Natural Science and Technology Research Council of Quebec (AKS), and Discovery grants from the Natural Sciences and Engineering Research Council of Canada (APH, PB).
Literature cited
- Araki H, Berejikian BA, Ford MJ, Blouin MS. Fitness of hatchery- reared salmonids in the wild. Evolutionary Applications. 2008;1:342–355. doi: 10.1111/j.1752-4571.2008.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher SA, Magurran AE. Gene flow in Trinidadian guppies. Journal of Fish Biology. 2000;56:241–249. [Google Scholar]
- Boulding EG, Hay T. Genetic and demographic parameters determining population persistence after a discrete change in the environment. Heredity. 2001;86:313–324. doi: 10.1046/j.1365-2540.2001.00829.x. [DOI] [PubMed] [Google Scholar]
- Brown JH, Kodric-Brown A. Turnover rates in insular biogeography: effect of immigration on extinction. Ecology. 1977;58:445–449. [Google Scholar]
- Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biology. 2010;8 doi: 10.1371/journal.pbio.1000357. e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispo E, Bentzen P, Reznick DN, Kinnison MTK, Hendry AP. The relative influence of natural selection and geography on gene flow in guppies. Molecular Ecology. 2006;15:49–62. doi: 10.1111/j.1365-294X.2005.02764.x. [DOI] [PubMed] [Google Scholar]
- Damshen EI, Haddad NM, Orrock JL, Tewksbury JJ, Levey DJ. Corridors increase plant species richness at large scales. Science. 2006;313:1284–1286. doi: 10.1126/science.1130098. [DOI] [PubMed] [Google Scholar]
- Darimont CT, Carlson SM, Kinnison MT, Paquet PC, Reimchen TE, Wilmers CC. Human predators outpace other agents of trait change in the wild. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:952–954. doi: 10.1073/pnas.0809235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth RA, Badyaev AV. Coupling of dispersal and aggression facilitates the rapid range expansion of a passerine bird. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15017–15022. doi: 10.1073/pnas.0706174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA. A predator's view of animal color patterns. Evolutionary Biology. 1978;11:319. 364. [Google Scholar]
- Endler JA. Natural selection on color patterns in Poecilia reticulata. Evolution. 1980;34:76–91. doi: 10.1111/j.1558-5646.1980.tb04790.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Multiple trait coevolution and environmental gradients in guppies. Trends in Ecology & Evolution. 1995;10:22–29. doi: 10.1016/s0169-5347(00)88956-9. [DOI] [PubMed] [Google Scholar]
- Endler JA, Houde AE. Geographic variation in female preferences for male traits in Poecilia reticulata. Evolution. 1995;49:456–468. doi: 10.1111/j.1558-5646.1995.tb02278.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Garant D, Forde SE, Hendry AP. The multifarious effects of dispersal and gene flow on contemporary adaptation. Functional Ecology. 2007;21:434–443. [Google Scholar]
- Gordon SP, Reznick DN, Kinnison MT, Bryant MJ, Weese DJ, Räsänen K, Millar NP, et al. Adaptive changes in life history and survival following a new guppy introduction. American Naturalist. 2009;174:34–45. doi: 10.1086/599300. [DOI] [PubMed] [Google Scholar]
- Grether GF, Millie DF, Bryant MJ, Reznick DN, Mayea W. Rain forest canopy cover, resource availability, and life history evolution in guppies. Ecology. 2001;82:1546–1559. [Google Scholar]
- Hanski I. Metapopulation Ecology. Oxford: Oxford University Press; 1999. [Google Scholar]
- Hanski I, Saccheri I. Molecular-level variation affects population growth in a butterfly metapopulation. Public Library Online System Biology. 2006;4:719–726. doi: 10.1371/journal.pbio.0040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Day T, Taylor EB. Population mixing and the adaptive divergence of quantitative traits in discrete populations: a theoretical framework for empirical tests. Evolution. 2001;55:459–466. doi: 10.1554/0014-3820(2001)055[0459:pmatad]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Kelly ML, Kinnison MT, Reznick DN. Parallel evolution of the sexes? Effects of predation and habitat features on the size and shape of wild guppies. Journal of Evolutionary Biology. 2006;19:741–754. doi: 10.1111/j.1420-9101.2005.01061.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Molecular Ecology. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Kinnison MT, Heino M, Day T, Smith TB, Fitt G, Bergstrom CT, et al. Evolutionary priniciples and their practical application. Evolutionary Applications. 2011;4:159–183. doi: 10.1111/j.1752-4571.2010.00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RD, Gomulkiewicz R, Barfield M. The phenomenology of niche evolution via quantitative traits in a ‘black-hole’ sink. Proceedings of the Royal Society of London Series B. 2003;270:215–224. doi: 10.1098/rspb.2002.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Holt RD. Evolutionary consequences of asymmetric dispersal rates. American Naturalist. 2002;160:333–347. doi: 10.1086/341519. [DOI] [PubMed] [Google Scholar]
- Kinnison MT, Hairston NG., Jr Eco-evolutionary conservation biology: contemporary evolution and the dynamics of persistence. Functional Ecology. 2007;21:444–454. [Google Scholar]
- Kinnison MT, Unwin MJ, Quinn TP. Eco-evolutionary vs. habitat contributions to invasion in salmon: experimental evaluation in the wild. Molecular Ecology. 2008;17:405–414. doi: 10.1111/j.1365-294X.2007.03495.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- Lande R. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. American Naturalist. 1993;142:911–927. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- Lindenmayer DB, Cunningham RB, Peakall R. The recovery of populations of bush rat Rattus fuscipes in forest fragments following major population reduction. Journal of Applied Ecology. 2005;42:649–658. [Google Scholar]
- Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. The velocity of climate change. Nature. 2009;462:1052–1055. doi: 10.1038/nature08649. [DOI] [PubMed] [Google Scholar]
- Magurran AE. Evolutionary Ecology: The Trinidadian Guppy. Oxford: Oxford University Press; 2005. [Google Scholar]
- Magurran AE, Seghers BH, Carvalho GR, Shaw PW. Behavioural consequences of an artificial introduction of guppies, Poecilia reticulata, in N. Trinidad: evidence for the evolution of antipredator behaviour in the wild. Proceedings of the Royal Society of London Series B. 1992;248:117–122. [Google Scholar]
- McClelland EK, Naish KA. What is the fitness outcome of crossing unrelated fish populations? A meta-analysis and an evaluation of future research directions. Conservation Genetics. 2007;8:397–416. [Google Scholar]
- Millar NP, Reznick DN, Kinnison MT, Hendry AP. Disentangling the selective factors that act on male colour in wild guppies. Oikos. 2006;113:1–12. [Google Scholar]
- Moore JS, Hendry AP. Can gene flow have negative demographic consequences? Mixed evidence from stream threespine stickleback. Philosophical Transactions of the Royal Society of London Series B. 2009;364:1533–1542. doi: 10.1098/rstb.2009.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Vines TH, Funk DJ. Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- Olendorf R, Rodd FH, Punzalan D, Houde AE, Hurt C, Reznick DN, Hughes AndKA. Frequency-dependent survival in natural guppy populations. Nature. 2006;441:633–636. doi: 10.1038/nature04646. [DOI] [PubMed] [Google Scholar]
- O'Steen S, Cullum AJ, Bennett AF. Rapid evolution of escape ability in Trinidadian guppies (Poecilia reticulata. Evolution. 2002;56:776–784. doi: 10.1111/j.0014-3820.2002.tb01388.x. [DOI] [PubMed] [Google Scholar]
- Paterson IG, Crispo E, Kinnison MT, Hendry AP, Bentzen P. Characterization of tetranucleotide microsatellite markers in guppy (Poecilia reticulata. Molecular Ecology Notes. 2005;5:269–271. [Google Scholar]
- Peakall R, Lindenmayer D. Genetic insights into population recovery following experimental perturbation in a fragmented landscape. Biological Conservation. 2006;132:520–532. [Google Scholar]
- Peery MZ, Hall LA, Sellas A, Beissinger SR, Moritz C, Bérubé M, Raphael MG, et al. Genetic analyses of historic and modern marbled murrelets suggest decoupling of migration and gene flow after habitat fragmentation. Proceedings of the Royal Society Series B. 2010;277:697–706. doi: 10.1098/rspb.2009.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam HR. Sources, sinks, and population regulation. American Naturalist. 1988;132:652–661. [Google Scholar]
- Reznick DN, Endler JA. The impact of predation on life history evolution in Trinidadian guppies (Poecilia reticulata. Evolution. 1982;36:160–177. doi: 10.1111/j.1558-5646.1982.tb05021.x. [DOI] [PubMed] [Google Scholar]
- Reznick DN, Butler MJ, Todd FH, Ross P. Life-history evolution in guppies (Poecilica reticulata) 6. Differential mortality as a mechanism for natural selection. Evolution. 1996;50:1651–1660. doi: 10.1111/j.1558-5646.1996.tb03937.x. [DOI] [PubMed] [Google Scholar]
- Rodd FH, Reznick DN. Life history evolution in Guppies III. The impact of prawn predation on guppy life histories. Oikos. 1991;62:13–19. [Google Scholar]
- Ronce O, Kirkpatrick M. When sources become sinks: migration meltdown in heterogeneous habitats. Evolution. 2001;55:1520–1531. doi: 10.1111/j.0014-3820.2001.tb00672.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford University Press; 2005. [Google Scholar]
- Shaffer ML. Minimum population sizes for species conservation. BioScience. 1981;31:131–134. [Google Scholar]
- Sousa WP. The role of disturbance in natural communities. Annual Review of Ecology and Systematics. 1984;15:353–391. [Google Scholar]
- Spiller DA, Losos JB, Schoener TW. Impact of a catastrophic hurricane on island populations. Science. 1998;281:695–697. doi: 10.1126/science.281.5377.695. [DOI] [PubMed] [Google Scholar]
- Van doorslaer WV, Vanoverbeke J, Duvivier C, Rousseaux S, Jansen M, Jansen B, Feuchtmayr H, et al. Local adaptation to higher temperatures reduces immigration success of genotypes from a warmer region in the water flea Daphnia. Global Change Biology. 2009;15:3046–3055. [Google Scholar]
- Van oosterhout C, Mohammed RS, Hansen H, Archard GA, McMullan M, Weese DJ, Cable J. Selection by parasites in spate conditions in wild Trinidadian guppies (Poecilia reticulata. International Journal for Parasitology. 2007;37:805–812. doi: 10.1016/j.ijpara.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Vignieri SN. The genetic effects of ecological disturbance: flooding in jumping mice. American Naturalist. 2010;175:126–135. doi: 10.1086/648606. [DOI] [PubMed] [Google Scholar]
- Visser ME. Keeping up with a warming world: assessing the rate of adaptation to climate change. Proceedings of the Royal Society of London Series B. 2008;275:649–659. doi: 10.1098/rspb.2007.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weese DJ, Gordon SP, Hendry AP, Kinnison MT. Spatiotemporal variation in linear natural selection on body color in wild guppies (Poecilia reticulata. Evolution. 2010;64:1802–1815. doi: 10.1111/j.1558-5646.2010.00945.x. [DOI] [PubMed] [Google Scholar]
- White GC, Burnham KP. Program MARK: survival estimation from populations of marked animals. Bird Study. 1995;46:120–138. [Google Scholar]
- Whitlock MC. Selection, load and inbreeding depression in a large metapopulation. Genetics. 2002;160:1191–1202. doi: 10.1093/genetics/160.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]


