Abstract
Sporadic outbreaks of human H3N2 influenza A virus (IAV) infections in swine populations have been reported in Asia, Europe and North America since 1970. In South America, serological surveys in pigs indicate that IAVs of the H3 and H1 subtypes are currently in circulation; however, neither virus isolation nor characterization has been reported. In November 2008, an outbreak of respiratory disease in pigs consistent with swine influenza virus (SIV) infection was detected in Argentina. The current study describes the clinical epidemiology, pathology, and molecular and biological characteristics of the virus. Phylogenetic analysis revealed that the virus isolate shared nucleotide identities of 96–98 % with H3N2 IAVs that circulated in humans from 2000 to 2003. Antigenically, sera from experimentally inoculated animals cross-reacted mainly with non-contemporary human-origin H3N2 influenza viruses. In an experimental infection in a commercial swine breed, the virus was of low virulence but was transmitted efficiently to contact pigs and caused severe disease when an infected animal acquired a secondary bacterial infection. This is the first report of a wholly human H3N2 IAV associated with clinical disease in pigs in South America. These studies highlight the importance of two-way transmission of IAVs and SIVs between pigs and humans, and call for enhanced influenza surveillance in the pig population worldwide.
Introduction
Influenza A viruses (IAVs) of the H1N1, H3N2 and H1N2 subtypes are endemic in commercial pig populations worldwide (Brown, 2000; Olsen et al., 2006). These subtypes are represented by multiple lineages that differ antigenically and genetically as result of antigenic drift and reassortment (Brown, 2000; Olsen et al., 2006). Outbreaks in pigs, produced by transmission in toto of H3N2 human influenza viruses, were reported in Asia, Europe and North America as early as 1970 (Karasin et al., 2000; Song et al., 2003; de Jong et al., 2007; Yu et al., 2008). In 1984 in Europe, a double reassortant containing the surface genes of the human-like swine H3N2 virus and the internal genes of the swine H1N1 virus of avian origin was introduced into the pig population, causing clinical signs and eventually replacing the previously circulating human-origin A/Hong Kong/68-like H3N2 strain (Van Reeth et al., 2008). From 1977 to 1995, most of the H3N2 virus strains isolated from swine in Europe, Hong Kong and Canada were closely related to each other and to the human strains (de Jong et al., 2007). Since the late 1990s, however, the complexity of swine influenza viruses (SIVs) in commercial pig settings has increased dramatically. In China, for instance, wholly human, double (human and avian) and triple (human, swine and avian) reassortant H3N2 viruses have emerged in pigs (Yu et al., 2008). In the USA, H3N2 SIVs emerged between 1997 and 1998 as double and triple reassortant viruses containing gene segments of avian (PB2 and PA subunits of the viral polymerase), human [PB1, haemagglutinin (HA) and neuraminidase (NA) proteins], and swine origin [nucleoprotein (NP), matrix (M) protein and non-structural (NS) protein], which eventually led to the establishment of the triple reassortant SIVs in pigs in North America (Zhou et al., 2000). Furthermore, epidemiological evidence suggests that the constellation of internal genes of the triple reassortant viruses, known as the triple reassortant internal genes cassette or TRIG, possesses a biological advantage and predominates in pigs in North America. In fact, since 1998, multiple introductions of H1N1 and H3N2 human influenza viruses have occurred in pigs in North America, always associated with a TRIG cassette. More importantly, the TRIG cassette was associated with the emergence of the 2009 H1N1 pandemic virus, a triple reassortant virus whose genome contains genes derived from avian (PB2 and PA), human (PB1), North American swine (HA, NP and NS) and Eurasian swine (NA and M) influenza lineages (Donaldson et al., 2009). Like H1N1 SIVs, the North American triple reassortant H3N2 swine influenza virus shows both genetic and antigenic diversity. Specifically, triple reassortant H3N2 swine influenza viruses are divided into three distinct phylogenetic clusters (I–III) based on the HA gene. Consistent with their phylogeny, these genetic clusters are antigenically diverse, and most isolates can be placed into one of three serogroups. However, there are several isolates from genetic cluster III that do not cross-react with serogroup 3 antisera, which suggests that constant antigenic drift occurs among these viruses (Richt et al., 2003; Gramer et al., 2007).
Despite this wealth of knowledge, there are still major gaps in our understanding of SIVs, including their ecology, as well as the environmental, anthropological and molecular factors that contribute to two-way transmission between swine and humans. In South America, studies related to influenza virus activity in domestic animals, including pigs, are scarce. Limited serological surveys in Venezuela, Colombia and Brazil have revealed that IAV H3N2 and H1N1 subtypes are common (Perfumo, 2010). However, as neither virus isolation nor sequencing analysis has been reported, the biological and molecular characteristics of these viruses remain unknown. In Argentina, a retrospective serological study performed in 13 farms showed that human and/or swine IAVs that are antigenically closely related to human strains have been present in pigs since at least 2002 (Piñeyro et al., 2010). Interestingly, despite the positive serological response, clinical disease was absent in these farms and no viruses were obtained. In this report, however, we provide evidence of a respiratory disease outbreak in Argentina caused by a wholly human H3N2 IAV in nursery barns of an intensively managed pig farm. We describe the clinical outcome, pathology and molecular features of this isolate. Finally, the pathogenicity and transmissibility of the virus were evaluated experimentally in swine.
Results and Discussion
Herd history
A respiratory disease outbreak affected a 6000-sow herd in Buenos Aires province in November 2008. The farm had its own source of replacement gilts. Pigs were moved all-in/all-out from the farrowing barns (site 1) to the nursery (site 2) at the weaning age of 20 days. Each of the nine nursery barns was filled in a week, with a mean of 3000 pigs per barn. The pig population at risk was 27 000. Respiratory clinical signs consistent with SIV infection were observed initially at the nursery. The clinical disease cycle persisted for 8 weeks, spreading at the rate of one barn per week. In each barn, clinical signs included cough, dyspnoea, sneezing, oculonasal exudates, lethargy, anorexia and fever. The disease affected initially ~2 % of 40-day-old pigs. Morbidity increased rapidly, reaching up to 40 % in 48–55-day-old pigs, and subsequently resolved before these animals were moved to site 3 (at 80 days old). A comparison of productive and health parameters (mean daily weight gain, feed intake and mortality) of eight barns before and during the outbreak showed that they did not differ significantly. However, the body weight of the pigs examined post-mortem was below the target weight for their age (mean of 12.0 kg bodyweight in 49-day-old pigs).
Pathology, immunohistochemistry and serology of pigs from the affected farm
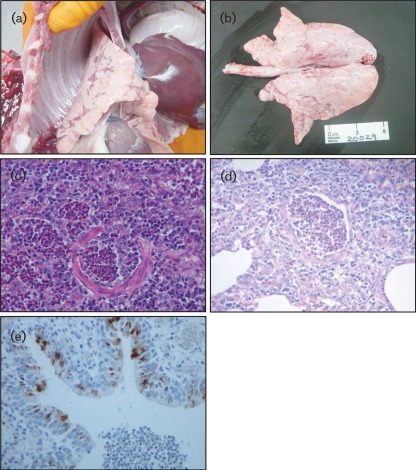
Nine pigs that were examined post-mortem on the farm displayed cranioventral plum-coloured consolidated areas in their lungs (mean lung percentage 16.6 %). In two of them, distinctive scattered dark red foci of lobular consolidation (with a chessboard-like distribution) were observed (Fig. 1a, b). Gross examination led to a presumptive diagnosis of viral pneumonia in seven pigs and bacterial pneumonia in two pigs. In five of the seven pigs with viral pneumonia, the most remarkable microscopic findings were severe necrotizing bronchiolitis and bronchointerstitial pneumonia, two of them with alveolar collapse and the rest presenting exudative inflammation of alveoli. Small and medium-sized bronchioles were plugged with neutrophil cellular debris and proteinaceous exudates (Fig. 1c, d). The affected airways were denuded or lined with flat regenerative epithelia. At the lobular level, the adjacent alveolar parenchyma was collapsed and/or the alveolar lumen filled with macrophages, neutrophils and denuded epithelial cells. Immunohistochemistry showed a distinctive positive reaction for influenza NP antigen only in the bronchial and bronchiolar epithelia, with minimal inflammatory changes and a few mononuclear cells in the alveoli. Both nuclei and cytoplasm were reactive (Fig. 1e). Severely affected bronchioles were negative for NP antigen. Taken together, these findings are consistent with SIV infection, although we cannot rule out the possibility that other factors may have contributed to the severity of the disease and pathology under field settings.
Fig. 1.
(a, b) Cranioventral consolidation. Note the distinctive chessboard-like distribution over the lung in (b). (c, d) Histopathology of the case shown in (a). Medium-sized bronchioles were lined with immature flat epithelial cells (c), whilst small bronchioles showed necrosis and sloughed-off epithelium (d). The lumen was filled with detached necrotic epithelial cells and neutrophils. (e) Immunohistochemical staining showed IAV antigen on the epithelial cells in bronchioles that showed slight inflammatory changes and a few scattered mononuclear cells within the alveoli. Magnification, ×40 (c–e).
A non-contemporary wholly human H3N2 virus isolated from pig samples
Nasal swabs and lung tissue samples from five pigs with a presumptive diagnosis of viral pneumonia were subjected to RNA extraction and real-time RT-PCR analysis using primers for the influenza virus M gene. All the samples were PCR positive. Lung homogenates and nasal swabs from PCR-positive samples were used independently to inoculate Madin–Darby canine kidney (MDCK) cells in order to perform virus isolation. After two passages, one influenza virus was isolated from the lung homogenate samples, which was subsequently PCR amplified and sequenced. Full genome sequencing of A/swine/Argentina/CIP051-A2/2008 (H3N2) (referred to hereafter as A2/08) revealed nucleotide identities of 96–98 % with human H3N2 influenza viruses that circulated in Oceania and North America from 2000 to 2003 (Table 1). Unfortunately, due to the lack of virus sequence information from Argentina during this time period, it was not possible to infer retrospectively the precise origin of the A2/08 isolate.
Table 1. Full-length sequences of the PB2, PB1, PA, HA, NP, NA, M and NS genes analysed by nucleotide blast analysis and the most closely related IAV gene for each respective segment.
| Segment | Identity (%) | blast hit | Subtype | GenBank accession no. |
| PB2 | 98 | A/Wellington/34/2001 | H3N2 | CY012319.1 |
| 96 | A/New York/418/2002 | H3N2 | CY003207.1 | |
| 96 | A/Auckland/604/2001 | H3N2 | CY023073.1 | |
| PB1 | 97 | A/New York/80/2001 | H3N2 | CY000575.1 |
| 97 | A/Western Australia/15/2001 | H3N2 | CY013222.1 | |
| PA | 98 | A/New York/75/2002 | H3N2 | CY001306.1 |
| 98 | A/Auckland/602/2001 | H3N2 | CY022530.1 | |
| HA | 97 | A/Hong Kong/CUHK28038/2000 | H3N2 | EU856960.1 |
| 97 | A/Auckland/603/2001 | H3N2 | CY023058.1 | |
| NP | 96 | A/Auckland/610/2002 | H3N2 | CY025053.1 |
| 96 | A/Western Australia/45/2003 | H3N2 | CY015839.1 | |
| 96 | A/New York/408/2002 | H3N2 | CY003130.1 | |
| NA | 98 | A/New York/114/2002 | H3N2 | CY000539.1 |
| 98 | A/New York/85/2001 | H3N2 | CY000387.1 | |
| 98 | A/Auckland/614/2002 | H3N2 | CY022207.1 | |
| M | 97 | A/New York/85/2001 | H3N2 | CY000386.1 |
| 96 | A/New York/420/2002 | H3N2 | CY003778.1 | |
| NS | 96 | A/Western Australia/29/2002 | H3N2 | CY015736.1 |
| 96 | A/New York/86/2002 | H3N2 | CY000285.1 |
Interestingly, sequence analysis of the PB1 gene revealed that A2/08 did not encode a full-length PB1–F2 protein due to the presence of two stop codons in the PB1–F2 ORF (at codons 35 and 58). Additionally, A2/08 had a trinucleotide deletion at coding positions 643–645, which created an amino acid deletion at position 49 of NS2/NEP and at position 205 of NS1.
A2/08 replicates to high titres in the lungs but exhibits low virulence for swine
In order to evaluate the pathogenicity of the A2/08 virus under experimental conditions, two independent experiments were carried out in commercial pigs that were serologically negative for IAV. In both studies, pigs were inoculated intratracheally with 108 TCID50 A2/08 per pig and monitored daily for clinical signs of disease. At 5 days post-infection (p.i.), animals were necropsied to measure virus replication in the lungs and to evaluate both macroscopic and microscopic pathology. There was no increase in body temperature after infection in either study, and only mild clinical signs of disease such as anorexia, lethargy and sneezing were observed (Table 2), suggesting that the virus is of low virulence for pigs under experimental conditions.
Table 2. Clinical signs in pigs infected with A2/08 .
| Clinical sign | No. of pigs showing clinical signs/total no. pigs | ||
| A2/08 infected pigs* | A2/08 contact pigs† | PBS control | |
| Fever | 0/8 | 1/3 | 0/2 |
| Lethargy | 8/8 | 3/3 | 0/2 |
| Anorexia | 8/8 | 3/3 | 0/2 |
| Nasal discharge | 0/8 | 1/3 | 0/2 |
| Sneezing | 8/8 | 3/3 | 0/2 |
| Cough | 0/8 | 1/3 | 0/2 |
| Conjunctivitis | 0/8 | 0/3 | 0/2 |
The infected group depicts results from two independent studies using a total of eight pigs (n = 5 and n = 3, respectively).
One contact animal developed severe disease and was euthanized by 9 days post-contact (p.c.).
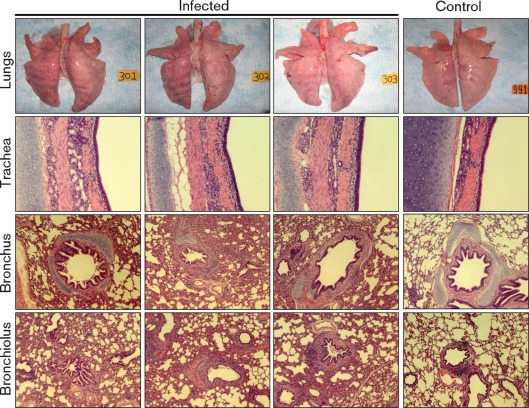
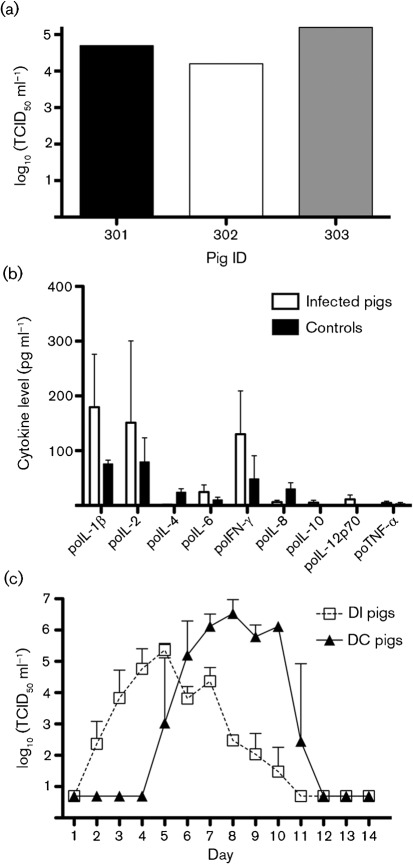
Pathological analysis showed rib imprints characteristic of viral interstitial pneumonia, and the texture of consolidated lung was firmer than normal (Fig. 2). In one of these pigs, a small focus of lung consolidation in the middle right lobe was observed, whilst the caudal lung was unaffected. Histopathologically, the A2/08 virus induced necrotizing bronchitis and bronchiolitis, characterized by sloughing of the epithelial lining of the airway and the presence of sloughed epithelial cells and mixed leukocytes in the lumen (Fig. 2). There were no significant microscopic changes in the trachea (Fig. 2). The microscopic examination of pneumonia mirrored the changes found in the field cases but the changes were less severe, suggesting that other factors might have contributed to the presentation of the disease at the farm. Virus titres in bronchoalveolar lavage fluid (BALF) ranged from 104 to 105 TCID50 ml−1, suggesting that A2/08 replicates efficiently in the lower respiratory tract of pigs (Fig. 3a). Control pigs remained healthy throughout the study, and had no significant lung pathology or detectable virus in BALF. To characterize further the pathogenicity of A2/08 in pigs, we measured the levels of nine cytokines in porcine BALF. A2/08-infected pigs exhibited higher-than-average levels of the pro-inflammatory cytokines interleukin (IL)-1β, IL-6 and gamma interferon (IFN-γ) compared with the control group; however, due to the small number of animals in each group and the variability among them, the group means were not statistically significant (at P<0.05 compared with PBS-inoculated groups by Student’s t-test; Fig. 3b). Interestingly, the pulmonary levels of the T helper 2 cytokine IL-4 was downregulated (P = 0.007) in the directly infected (DI) group compared with the uninfected controls (Fig. 3b). Although other investigations have been unable to detect significant changes in IL-4 expression in either macaques (Kobasa et al., 2007) or pigs (Kim et al., 2008) during influenza virus infection, the role of IL-4 during the course of H3N2 influenza infections in the pig warrants further investigation.
Fig. 2.
Gross and microscopic pathology of pigs directly infected with A2/08. Groups of pigs were directly inoculated intratracheally with 108 TCID50 A2/08 per pig and the lungs were collected at 5 days p.i. to evaluate lung pathology. Tissues were processed routinely for histopathology and stained with haematoxylin and eosin (H&E). Macroscopic examination of the lungs with pneumonia showed distinct rib imprints on the surface of the lungs (upper row). Microscopic examination of the trachea, bronchi and bronchioli are shown in the lower three rows. Each column represents an individual animal. The lung of animal 303 shows dark red area of lung consolidation in the middle right lobe typical of swine influenza pneumonia. Representative pictures from a control pig inoculated with PBS are show on the right.
Fig. 3.
(a) Replication of A2/08 in swine lungs. BALF was collected from DI pigs at 5 days p.i. and viral titres were determined as TCID50 on MDCK cells. Values are shown as the log10 TCID50 ml−1 for each infected pig. The pig ID corresponds to that shown in Fig. 2. (b) Pulmonary cytokine response induced by A2/08 in pigs. The levels of nine porcine (po) cytokines in BALF samples were determined by multiplex sandwich ELISA. TNF-α, Tumour necrosis factor alpha. (c) Virus shedding of A2/08. Groups of pigs were directly inoculated intratracheally with 108 TCID50 A2/08 per pig and three DC pigs were co-housed with the DI pigs on 1 day p.i. Nasal swabs were collected daily and titrated as TCID50 on MDCK cells. Values in (b) and (c) are shown as mean±sem.
A2/08 is highly transmissible among pigs and can predispose animals to fatal bacterial pneumonia
In order to determine the extent of shedding and the transmissibility of A2/08 virus in pigs, three naïve pigs were co-housed [direct contact (DC) group] with DI pigs at 1 day p.i. and followed for 14 days. A2/08 was shed in large amounts in both the DI and the DC groups (Fig. 3c). However, the kinetics and magnitude of virus excretion differed between the two groups. In the DI group, virus shedding began at 2 days p.i. and reached the highest titre at 5 days p.i.; thereafter, virus titres declined progressively and disappeared by 11 days p.i. A2/08 was found to be highly transmissible among pigs, as all contact animals became infected as early as 4 days p.c., equivalent to 5 days p.i. In this group, virus shedding peaked at 8 days p.c. and then gradually decreased, with no virus being detected by 12 days p.c. Although the DI group was inoculated with a high virus dose (108 TCID50), nasal virus titres were higher in the DC group than in the DI group (Fig. 3c). Interestingly, one contact pig developed severe disease and had to be euthanized at 9 days p.c. At necropsy, its lungs showed multifocal distribution of small abscesses (1–2 mm) in all lobes, compatible with secondary bacterial infection (data not shown). Together, these results indicated that the A2/08 virus is transmitted very efficiently to contact pigs and that it can potentially trigger fatal secondary bacterial pneumonia.
A2/08 antiserum reacts with non-contemporary H3N2 influenza viruses
Haemagglutination inhibition (HI) antibodies induced by a single infection with A2/08 failed to cross-react with circulating human H3N2 virus (A/Brisbane/59/07), with the 1968 H3N2 pandemic strain (A/Hong Kong/1/68), with a typical avian H3N2 strain (A/duck/Hong Kong/3/75) or with a more recent avian triple reassortant H3N2 (A/turkey/Ohio/313053/04). However, antisera from infected pigs induced cross-reactive antibodies to non-contemporary North American human (A/Memphis/14/98) and swine (A/swine/Wisconsin/14094/99) H3N2 influenza viruses. HI titres were highest against the homologous A2/08 strain [geometric mean titre (GMT) of 453], followed by A/Memphis/14/98 (GMT of 226) and A/swine/Wisconsin/14094/99 (GMT of 57) (Table 3). Together, these results suggested that A2/08 is antigenically related to non-circulating H3N2 strains, which was consistent with the phylogenetic analysis.
Table 3. Antigenic profile of A2/08 virus.
M/98, A/Memphis/14/98 (H3N2); sw/99, A/swine/Wisconsin/14094/99 (H3N2); ty/04, A/turkey/Ohio/313053/04 (H3N2); BR/07, A/Brisbane/59/07 (H3N2); HK/68, A/Hong Kong/1/68 (H3N2); Dk/75, A/duck/Hong Kong/3/75. Positive HI titres are indicated in bold.
| Pig ID | Days p.i. | Virus | ||||||
| A2/08 | M/98 | sw/99 | ty/04 | BR/07 | HK/68 | Dk/75 | ||
| Pig 28 | 0 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| Pig 29 | 0 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| Pig 30 | 0 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| Pig 32 | 0 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| Pig 34 | 0 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| Pig 28 | 14 | 320 | 160 | 40 | <10 | <10 | <10 | <10 |
| Pig 29 | 14 | 640 | 320 | 80 | <10 | <10 | <10 | <10 |
| Pig 30 | 14 | 1280 | 320 | 80 | <10 | <10 | <10 | <10 |
| Pig 32 | 14 | 320 | 160 | 40 | <10 | <10 | <10 | <10 |
| Pig 34 | 14 | 80 | 40 | <10 | <10 | <10 | <10 | <10 |
Our report is consistent with previous observations of infection of pigs with human H3N2 influenza viruses in the field (Brown, 2000). In North America, only two wholly human influenza viruses have been identified in pigs, although these viruses did not appear to transmit efficiently in pigs and have not been maintained in the pig population (Karasin et al., 2000). Although experimental infection with the A2/08 virus resulted in efficient transmission in pigs, there is no evidence that the virus is maintained in commercial pigs in Argentina, as there is not enough surveillance data to determine the spatial distribution of this strain in swine herds. Unlike outbreaks of H3N2 TRIG viruses that cause high levels of abortion in pregnant sows and increased sow mortality, the outbreak associated with the A2/08 virus was not accompanied by reproductive disorders or an increase in mortality. Our experimental inoculation study in pigs was consistent with previous reports that depict mild pulmonary changes in pigs infected with wholly human H3N2 virus (Landolt et al., 2003). In our study, microscopic lung lesions were observed in pigs inoculated with the A2/08 virus, although they were less severe than those observed in field cases. However, viral antigen was detected in the respiratory tract, consistent with active virus replication and viral pneumonia. In addition, the isolation of non-contemporary human H3N2 virus was not unexpected, given that a human H3N2 virus derived from early human viruses has been isolated in Heilongjiang province, PR China (Yu et al., 2007).
In summary, we have reported the first outbreak of a wholly human H3N2 subtype in Argentina (and South America). A presumptive diagnosis was performed by clinical, epidemiological and histopathological analysis, including immunohistochemical studies. Confirmatory diagnosis was performed by virus isolation and molecular characterization. Experimental reproduction of infection showed that the virus was transmitted efficiently between pigs, and that the inoculated pigs had characteristic lesions of influenza virus, suggesting that this virus is completely adapted to swine and has the potential to be maintained in the swine population. The data generated in this study support the notion of the two-way transmission of IAVs between pigs and humans, as demonstrated by the 1918 H1N1 and 2009 H1N1 pandemic viruses. Our study has important implications, indicating that increased influenza virus surveillance is highly desirable in commercial swine operations worldwide.
Methods
Pathology and immunohistochemistry.
Nine pigs (45–50 days old) with severe respiratory clinical signs were selected and submitted for post-mortem studies. Tissue samples of the nose, trachea, lungs, regional lymph nodes, tonsils, spleen, liver and kidney were fixed in 10 % buffered formalin, embedded in paraffin and stained with haematoxylin and eosin (H&E). Immunohistochemistry was performed on tissues of suspected cases using an anti-NP antibody. Briefly, selected paraffin sections of lungs with microscopic lesions were examined. Samples were first treated with anti-NP influenza A mAb, isotype MsIgG2a (Chemicon International), diluted 1 : 500, incubated at 4 °C overnight and then probed with biotin-conjugated anti-mouse rabbit IgG and goat antiserum at room temperature for 10 min. The colour reaction was performed by incubation with diaminobenzidine for 1–2 min, followed by counterstaining with H&E and examination under a light microscope. Positive- and negative-control samples were processed simultaneously.
Virus identification.
Bronchial swabs and lung tissue samples were collected from the pigs described above. Viral RNA was extracted from swab suspensions and tissue samples using a QIAamp Viral RNA Mini kit (Qiagen) and used for real-time RT-PCR to detect the M gene of IAV (Spackman et al., 2002). PCR was performed in an ABI Prism 7500 SDS apparatus (Applied Biosystems).
Virus isolation.
Samples that were positive by real-time RT-PCR were processed for virus isolation in MDCK cells. Briefly, 200 µl of a 10 % suspension of lung homogenate or nasal swab suspension in PBS was used for viral isolation. The suspensions were clarified by centrifugation at 3000 g for 10 min at 4 °C, and the supernatants were inoculated onto MDCK cells in accordance with standardized protocols described by the Office International des Epizooties (OIE, 2000). Virus growth was determined by examining the cytopathic effect and by real-time RT-PCR.
Genetic analysis and phylogenetic characterization.
One virus, named A/swine/Argentina/CIP051-A2/2008 (H3N2) (A2/08), was obtained from MDCK cells. The viral genome was amplified by RT-PCR as described previously (Hoffmann et al., 2001). Sequencing was performed using a BigDye Terminator kit (Applied Biosystems) on an ABI 3730 sequencer (Applied Biosystems). Genomic information was derived from overlapping sequences covered by forward and reverse primers. At least two independent RT-PCRs were carried out for each gene and used for sequencing. Sequences were assembled, edited and analysed using Lasergene 8.1 (DNASTAR). Nucleotide blast analysis (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was used initially to identify the most closely related IAV gene for each respective segment.
Animal studies.
Animal studies were carried out according to protocol R-08-16, ‘Transmissibility of influenza A viruses in swine’, approved by the Institutional Animal Care and Use Committee of the University of Maryland, MD, USA. Five-week-old Yorkshire pigs were obtained from a commercial farm and shown to be negative for antibodies against IAVs by a competitive NP ELISA. Pigs were housed in a Biosafety Level 2 facility for the duration of the study. Food and water were provided ad libitum. Pigs were acclimatized for 5 days prior to the beginning of the study. In the first experiment and in order to evaluate the pathogenicity and transmissibility of the virus isolate, animals were randomly assigned to three groups: DI (n = 5), DC (n = 3) and PBS controls (n = 2), to evaluate the pathogenicity and transmissibility of the virus isolate. Clinical examinations including rectal temperature were recorded daily. Pigs were sedated by intramuscular injection of 6 mg tiletamine-zolazepam (Telazol; Pfizer Animal Health) kg−1 and 2.2 mg xylazine kg−1. The DI group was inoculated intratracheally with 108 TCID50 A2/08 per pig. On 1 day p.i., the room was thoroughly disinfected and the DC animals were co-housed with the inoculated animals. The PBS-control animals were housed in a separate isolation room and inoculated with sterile PBS. After inoculation, the pigs were assessed daily for clinical manifestations of disease, and nasal swabs were collected daily from each pig. Swabs were placed in 1 ml brain–heart infusion medium containing antibiotics (10 000 IU penicillin ml−1, 5000 µg streptomycin ml−1, 1000 µg gentamicin sulfate ml−1, 700 µg kanamycin sulfate ml−1 and 10 µg amphotericin B ml−1; Sigma) and stored at −80 °C until use for virus titration. At 5 days p.i., two pigs from the DI group and the two PBS-control pigs were euthanized with 1 ml Beuthanasia-D (Intervet/Schering-Plough) per 4.5 kg. Macroscopic lung lesions were graded on the basis of the percentage of surface area of each lung lobe exhibiting gross consolidation. Each lung was lavaged with 50 ml PBS to obtain BALF and the viral titre was determined as TCID50. Lung tissue samples were collected for histopathology in 10 % buffered formalin, embedded in paraffin and stained with H&E. The remaining pigs were euthanized at 14 days p.i. An independent pathogenicity study was conducted in which three pigs were inoculated intratracheally with 108 TCID50 A2/08 per pig. Following inoculation, rectal temperature and clinical signs of disease were monitored daily until termination of the experiment at 5 days p.i. when the animals were euthanized humanely and necropsy was performed.
Cytokine levels in porcine BALF.
The levels of nine porcine cytokines (IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12 and TNF-α) in BALF samples were determined by multiplex sandwich ELISA (Aushon Biosystems).
Titration of virus stocks and virus present in biological samples.
Viral stocks and virus present in animal samples (nasal swabs or BALF) were titrated on MDCK cells and the TCID50 ml−1 was determined as described previously (Reed & Muench, 1938). Briefly, samples were serially diluted tenfold in OptiMEM I medium (Gibco) containing antibiotics, as described above, and 1 µg TPCK-trypsin (Sigma) ml−1. Next, 200 µl of the inoculum was overlaid onto confluent monolayers of MDCK cells seeded in 96-well plates. The cells with sample were incubated for 3 days and the end-point viral titre was determined by a haemagglutination assay on the supernatant using turkey red blood cells.
Cross-reactivity of A2/08 antiserum against H3N2 influenza viruses.
Serum samples obtained at 0 and 14 days p.i. from the DI group were treated with receptor-destroying enzyme (Accurate Chemical and Scientific) to remove non-specific receptors. Antibody titres were evaluated by an HI assay, as described previously (Webster et al., 2005). HI assays were performed using homologous and heterologous viruses. Heterologous viruses comprised the following strains: A/Memphis/14/98, A/swine/Wisconsin/14094/99, A/turkey/Ohio/313053/04, A/Brisbane/59/07, A/Hong Kong/1/68 and A/duck/Hong Kong/3/75. HI titres of 1 : 40 or higher were considered positive. Values were expressed as GMT.
Acknowledgements
The authors would like to thank Yonas Araya for assistance with animal studies, Theresa Wolter Marth and Andrea Ferrero for coordination of animal studies and Andrea Puebla for assistance with sequencing. This work was partially supported with grants from ANPCyT OC/AR PICT 2005-33987; by UNLP V184; by the NIAID, Center for Research on Influenza Pathogenesis (CRIP) through University of Maryland contract no. HHSN266200700010C; by Proyecto BiotecSur Cadena Carne Aviar 1; and by Proyecto Específico INTA Exoticas y Emergentes (AESA201731).
References
- Brown I. H. (2000). The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 74, 29–46 10.1016/S0378-1135(00)00164-4 [DOI] [PubMed] [Google Scholar]
- de Jong J. C., Smith D. J., Lapedes A. S., Donatelli I., Campitelli L., Barigazzi G., Van Reeth K., Jones T. C., Rimmelzwaan G. F., et al. & other authors (2007). Antigenic and genetic evolution of swine influenza A (H3N2) viruses in Europe. J Virol 81, 4315–4322 10.1128/JVI.02458-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson L. J., Rutter P. D., Ellis B. M., Greaves F. E., Mytton O. T., Pebody R. G., Yardley I. E. (2009). Mortality from pandemic A/H1N1 2009 influenza in England: public health surveillance study. BMJ 339, b5213 10.1136/bmj.b5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramer M. R., Lee J. H., Choi Y. K., Goyal S. M., Joo H. S. (2007). Serologic and genetic characterization of North American H3N2 swine influenza A viruses. Can J Vet Res 71, 201–206 [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R. (2001). Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146, 2275–2289 10.1007/s007050170002 [DOI] [PubMed] [Google Scholar]
- Karasin A. I., Schutten M. M., Cooper L. A., Smith C. B., Subbarao K., Anderson G. A., Carman S., Olsen C. W. (2000). Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: evidence for wholly human and reassortant virus genotypes. Virus Res 68, 71–85 10.1016/S0168-1702(00)00154-4 [DOI] [PubMed] [Google Scholar]
- Kim H. M., Lee Y.-W., Lee K.-J., Kim H. S., Cho S. W., van Rooijen N., Guan Y., Seo S. H. (2008). Alveolar macrophages are indispensable for controlling influenza viruses in lungs of pigs. J Virol 82, 4265–4274 10.1128/JVI.02602-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D., Jones S. M., Shinya K., Kash J. C., Copps J., Ebihara H., Hatta Y., Kim J. H., Halfmann P., et al. & other authors (2007). Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323 10.1038/nature05495 [DOI] [PubMed] [Google Scholar]
- Landolt G. A., Karasin A. I., Phillips L., Olsen C. W. (2003). Comparison of the pathogenesis of two genetically different H3N2 influenza A viruses in pigs. J Clin Microbiol 41, 1936–1941 10.1128/JCM.41.5.1936-1941.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE (2000). Highly pathogenic avian influenza. In Manual of Standards for Diagnostic Tests and Vaccines, 4th edn, pp. 212–220 Paris: Office International des Epizooties [Google Scholar]
- Olsen C. W., Brown I. H., Easterday B. C., Van Reeth K. (2006). Swine influenza. In Diseases of Swine, pp. 469–482 Edited by Straw B., Zimmmerman J. J., D'Alleire S., Taylor D. J. Iowa, USA: Blackwell Publishing [Google Scholar]
- Perfumo C. J. (2010). Current situation of swine influenza in South America. In Proceedings of III Fórum Internacional de Suinocultura, pp. 223–233. Curitiba, Paraná, Brazil [Google Scholar]
- Piñeyro P. E., Baumeister E., Cappuccio J. A., Machuca M. A., Quiroga M. A., Tedoroff T., Perfumo C. J. (2010). [Seroprevalence of the swine influenza virus in fattening pigs in Argentina in the 2002 season: evaluation by hemagglutination-inhibition and ELISA tests]. Rev Argent Microbiol 42, 98–101 (in Spanish). [DOI] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty per cent endpoints. Am J Hyg 27, 493–497 [Google Scholar]
- Richt J. A., Lager K. M., Janke B. H., Woods R. D., Webster R. G., Webby R. J. (2003). Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol 41, 3198–3205 10.1128/JCM.41.7.3198-3205.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D. S., Lee J. Y., Oh J. S., Lyoo K. S., Yoon K. J., Park Y. H., Park B. K. (2003). Isolation of H3N2 swine influenza virus in South Korea. J Vet Diagn Invest 15, 30–34 10.1177/104063870301500107 [DOI] [PubMed] [Google Scholar]
- Spackman E., Senne D. A., Myers T. J., Bulaga L. L., Garber L. P., Perdue M. L., Lohman K., Daum L. T., Suarez D. L. (2002). Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40, 3256–3260 10.1128/JCM.40.9.3256-3260.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K., Brown I. H., Dürrwald R., Foni E., Labarque G., Lenihan P., Maldonado J., Markowska-Daniel I., Pensaert M., et al. & other authors (2008). Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002–2003. Influenza Other Respir Viruses 2, 99–105 10.1111/j.1750-2659.2008.00043.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R., Cox N., Stohr K.(2005).WHO Manual on Animal Influenza Diagnosis and Surveillance. http://www.who.int/vaccine_research/diseases/influenza/WHO_manual_on_animal-diagnosis_and_surveillance_2002_5.pdf
- Yu H., Zhang G.-H., Hua R.-H., Zhang Q., Liu T.-Q., Liao M., Tong G.-Z. (2007). Isolation and genetic analysis of human origin H1N1 and H3N2 influenza viruses from pigs in China. Biochem Biophys Res Commun 356, 91–96 10.1016/j.bbrc.2007.02.096 [DOI] [PubMed] [Google Scholar]
- Yu H., Hua R.-H., Zhang Q., Liu T.-Q., Liu H.-L., Li G.-X., Tong G.-Z. (2008). Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol 46, 1067–1075 10.1128/JCM.01257-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N. N., Senne D. A., Landgraf J. S., Swenson S. L., Erickson G., Rossow K., Liu L., Yoon K.-J., Krauss S., Webster R. G. (2000). Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet Microbiol 74, 47–58 10.1016/S0378-1135(00)00165-6 [DOI] [PubMed] [Google Scholar]