Abstract
A biologically contained influenza A virus that stably expresses a foreign gene can be effectively traced, used to generate a novel multivalent vaccine and have its replication easily assessed, all while satisfying safety concerns regarding pathogenicity or reversion. This study generated a PB2-knockout (PB2-KO) influenza virus that harboured the GFP reporter gene in the coding region of its PB2 viral RNA (vRNA). Replication of the PB2-KO virus was restricted to a cell line stably expressing the PB2 protein. The GFP gene-encoding PB2 vRNA was stably incorporated into progeny viruses during replication in PB2-expressing cells. The GFP gene was expressed in virus-infected cells with no evidence of recombination between the recombinant PB2 vRNA and the PB2 protein mRNA. Furthermore, other reporter genes and the haemagglutinin and neuraminidase genes of different virus strains were accommodated by the PB2-KO virus. Finally, the PB2-KO virus was used to establish an improved assay to screen neutralizing antibodies against influenza viruses by using reporter gene expression as an indicator of virus infection rather than by observing cytopathic effect. These results indicate that the PB2-KO virus has the potential to be a valuable tool for basic and applied influenza virus research.
Introduction
Influenza A viruses cause epidemics annually characterized by a contagious respiratory illness, mild to severe fever and, in some instances, death (Palese & Shaw, 2007). Currently available therapeutic and prophylactic interventions include two types of vaccine (inactivated and live) and two classes of antivirals [M2 ion-channel blockers, such as amantadine and rimantadine, and neuraminidase (NA) inhibitors, such as oseltamivir and zanamivir; Davies et al., 1964; Hayden, 2001]. Nevertheless, seasonal influenza is a contagious disease with one of the highest impacts on public health epidemiology. Furthermore, during the 2009–2010 influenza season, a novel influenza A virus strain, the 2009 H1N1 pandemic virus, emerged and spread worldwide, causing the first influenza pandemic in 40 years, with a considerable impact on global health and economics (http://www.cdc.gov/flu/about/disease/index.htm). In the USA alone, an estimated 61 million H1N1 cases, including 274 000 hospitalizations and 12 470 deaths, were reported (http://www.cdc.gov/flu/about/season/index.htm). To accelerate the development of novel countermeasures against influenza virus infection, biological innovation is imperative.
The biological manipulation of virus machinery has unleashed immense potential in the field of medical research. Genetic engineering of viruses through recombinant DNA-based technologies allows the exploitation of the genetic parasitism of a virus, while disarming its pathogenic power. Viruses can be rendered replication incompetent and non-pathogenic, or manipulated to introduce and express a foreign gene in a receptive host.
The stable expression of a foreign gene in a replication-incompetent influenza virus allows effective tracking of the manipulated virus. In pursuit of a biologically contained foreign-gene-expressing virus with extensive applications in the field of virology, we focused on the PB2 protein, an influenza virus polymerase subunit that forms part of the trimeric viral RNA-dependent RNA polymerase that is essential for virus replication. We demonstrated previously that the partial coding sequences of the 3′ and 5′ ends of the PB2 viral RNA (vRNA) confer its more critical role in efficient infectious virion generation relative to the other vRNAs in the vRNA hierarchy (Muramoto et al., 2006). This finding suggests that a PB2-knock out (PB2-KO) influenza virus harbouring a reporter gene flanked by the coding and non-coding sequences of the PB2 vRNA would replicate only in PB2-expressing cells while stably expressing the reporter gene.
Here, we established a cell line that stably expressed PB2 protein and used it to characterize a PB2-KO virus that possessed the GFP gene. We also investigated the potential for various virus strain-derived haemagglutinin (HA) and NA genes, as well as other reporter genes, to be accommodated by the PB2-KO virus. Finally, we used the PB2-KO virus as a platform to screen neutralizing antibodies against pandemic viruses from 2009.
Results
Characterization of the PR8/PB2–GFP virus
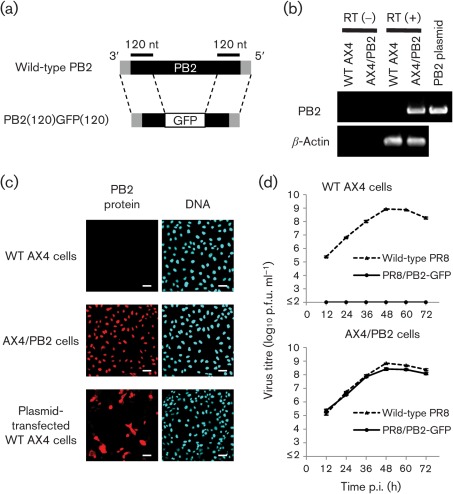
To establish a cell line that stably expressed PB2 protein, we transduced AX4 cells, which are human 2,6-sialyltransferase-overexpressing Madin–Darby canine kidney (MDCK) cells that allow better replication of clinical influenza isolates compared with wild-type MDCK cells (Hatakeyama et al., 2005), with a retroviral vector that possessed the cDNA of the A/Puerto Rico/8/34 (H1N1, PR8) PB2 protein followed by an internal ribosome entry site sequence derived from encephalomyocarditis virus and the blasticidin resistance gene. We designated the blasticidin-resistant cell clone AX4/PB2. To confirm the expression of mRNA for the PB2 protein in AX4/PB2 cells, total RNA was extracted from AX4/PB2 and wild-type AX4 cells and subjected to RT-PCR with PB2-specific primers. PB2 mRNA was detected in AX4/PB2 cells but not in wild-type AX4 cells (Fig. 1b). To further validate expression of the PB2 protein in AX4/PB2 cells, immunofluorescence staining of AX4/PB2 cells was performed using a PB2-specific mAb. Fluorescent signals were detected in the nuclei (the normal intracellular location of PB2) of AX4/PB2 cells and in both the nucleus and cytoplasm (abnormal intracellular location of PB2, probably due to transient overexpression of PB2) of some of the PB2 protein expression plasmid-transfected AX4 cells (which served as a positive control), whereas no signal was detected in wild-type AX4 cells (Fig. 1c). These results indicated that the AX4/PB2 cells stably expressed the PB2 protein.
Fig. 1.
Characterization of PR8/PB2–GFP virus. (a) Schematic diagram of wild-type PB2 and PB2(120)GFP(120) vRNAs. PB2(120)GFP(120) vRNA possesses the 3′ non-coding region, 120 nt of the coding sequence of PB2 vRNA, the GFP gene and 120 nt of the 3′ and 5′ non-coding regions of PB2 vRNA. The non-coding region and coding regions of PB2 vRNA are represented by shaded and filled bars, respectively. (b) PB2 gene expression in AX4/PB2 cells. RNA was extracted from wild-type AX4 and AX4/PB2 cells. RT-PCR was performed using an oligo(dT) primer followed by cDNA synthesis and PCR with PB2-specific or canine β-actin-specific primers. (c) PB2 protein expression in AX4/PB2 cells. Cells were reacted with anti-PB2 antibody (clone 18/1; left panels) and the nuclei stained with Hoechst 33342 (right panels). Bars, 50 µm. (d) Growth kinetics of PR8/PB2–GFP monitored over 72 h. Wild-type AX4 and AX4/PB2 cells were infected with wild-type PR8 or PR8/PB2–GFP virus at an m.o.i. of 0.001. Supernatants collected at the indicated time points were assayed for infectious virus in plaque assays in AX4/PB2 cells.
To investigate whether PB2-expressing cells supported PB2-KO virus replication, a PR8-based PB2-KO virus possessing PB2(120)GFP(120) vRNA (Fig. 1a), designated PR8/PB2–GFP (Table 1), was generated by and used to infect AX4/PB2 and wild-type AX4 cells (Fig. 1d). Although no infectious virus was detected in wild-type AX4 cells, replication of PR8/PB2–GFP virus in AX4/PB2 cells was comparable to that of wild-type PR8 (Fig. 1d). These results indicated that the replication of PB2-KO virus was restricted to PB2-expressing cells.
Table 1. Viruses generated in this study.
na, Not applicable.
| Virus | Origin of:* | |||
| HA gene | NA gene | PB2 gene | Remaining genes | |
| Wild-type PR8 | PR8 | PR8 | PR8 | PR8 |
| PR8/PB2–GFP | PR8 | PR8 | PB2(120)GFP(120) | |
| PR8ΔPB2 | PR8 | PR8 | na | |
| WSN/PB2–GFP | WSN | WSN | PB2(120)GFP(120) | |
| CA04/PB2–GFP | CA04 | CA04 | PB2(120)GFP(120) | |
| VN1203/PB2–GFP | VN1203 | VN1203 | PB2(120)GFP(120) | |
| PR8/PB2–Rluc | PR8 | PR8 | PB2(120)Rluc(120) | |
| PR8/PB2–Fluc | PR8 | PR8 | PB2(120)Fluc(120) | |
| CA04/PB2–Rluc | CA04 | CA04 | PB2(120)Rluc(120) | |
PR8, A/Puerto Rico/8/34 (H1N1); WSN, A/WSN/33 (H1N1); CA04, A/California/04/09 (H1N1); VN1203, A/Vietnam/1203/04 [H5N1; the various basic amino acid residues in the HA cleavage site (RERRRKKR↓G) were replaced with a non-virulent-type cleavage sequence (RETR↓G)]. PB2(120)GFP(120), PB2(120)Rluc(120) and PB2(120)Fluc(120) are the GFP, firefly luciferase and Renilla luciferase genes, respectively, flanked by the 3′ and 5′ non-coding sequences and 120 nt of the 3′ and 5′ coding sequences of the PB2 gene.
The stability of the reporter gene in PB2-KO virus was ascertained by serial passaging of PR8/PB2–GFP virus in AX4/PB2 cells. Most of the plaques formed by the passaged viruses expressed the fluorescent protein, which was clearly visible and quantifiable under a fluorescent microscope. However, to count the number of GFP-positive and -negative plaques by eye, the plaques were subjected to staining with an anti-GFP mAb by means of an immunostaining assay. Under these conditions, 80–90 % of the plaque-forming viruses expressed GFP after five serial passages (Table 2). PB2-KO virus failed to form plaques in wild-type cells, even after five serial passages in AX4/PB2 cells, indicating that reversion of PB2-KO virus to a replication-competent genotype by recombination between the PB2–GFP vRNA and the cell-expressed PB2 mRNA was unlikely. We also attempted to rescue a PB2 gene-deficient virus possessing seven vRNA segments (PR8ΔPB2, Table 1); however, neither cytopathic effect (CPE) nor nucleoprotein (NP) expression was observed in AX4/PB2 or wild-type AX4 cells inoculated with the transfectant supernatant for PR8ΔPB2 (data not shown). These results highlighted the importance of the PB2 vRNA for efficient generation of infectious virions (Muramoto et al., 2006) and suggested that the GFP-negative plaques were not formed by the PB2 gene-deficient virus but by viruses possessing a mutation(s) or an internal deletion of the GFP gene ORF in the PB2 gene.
Table 2. Genetic stability of PB2-KO virus.
| Experiment no. | No. GFP-positive plaques* from viruses in: | ||||
| Passage 1 | Passage 2 | Passage 3 | Passage 4 | Passage 5 | |
| 1 | 10 | 9 | 9 | 10 | 8 |
| 2 | 10 | 10 | 9 | 10 | 9 |
| 3 | 10 | 9 | 9 | 10 | 9 |
The respective virus supernatants were subjected to standard virus plaque assays in confluent AX4/PB2 cells. Ten plaques were marked per well, which were then subjected to an immunodetection assay using an anti-GFP antibody to detect GFP-expressing virus plaques. The number of plaques that stained brown is presented. The results of three independent experiments are shown.
Functional expression of different HA and NA genes in PB2-KO virus
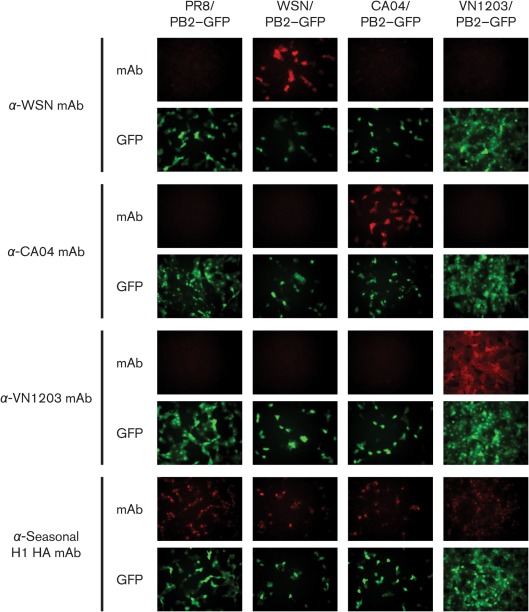
Two surface glycoproteins on influenza A virions, HA and NA, are the main protective antigens (Wright et al., 2007). In particular, HA mediates cell attachment; therefore, an antibody against HA is crucial for virus neutralization. We tested whether the relevant glycoproteins of our PR8 virus-based PB2-KO virus could be replaced with those of other virus strains. To this end, we generated GFP-encoding PB2-KO viruses by substituting PR8 HA and NA vRNAs with those derived from the laboratory H1N1 strain A/WSN/33 (WSN/PB2–GFP), the 2009 pandemic H1N1 strain A/California/04/2009 (CA04/PB2–GFP) and the highly pathogenic H5N1 strain A/Vietnam/1203/2004 (VN1203/PB2–GFP) (Table 1). AX4/PB2 cells were infected with these viruses and subjected to an immunofluorescence assay with various anti-HA mAbs. In the GFP-positive virus-infected cells, virus strain-specific HA expression was detected (Fig. 2). Intriguingly, an anti-seasonal H1 HA mAb IVC102 reacted with all viruses tested, including H5 subtype VN1203/PB2–GFP, probably because this antibody recognized an epitope conserved among these viruses. We also confirmed, by sequencing, that the corresponding NA vRNAs were incorporated into virions (data not shown). These results indicated that the HA and NA genes of other influenza viruses can also be accommodated in the PB2-KO virus and hence expressed in virus-infected cells.
Fig. 2.
Accommodation of various HA genes in PB2-KO virus and HA expression in PB2-KO virus-infected cells. AX4/PB2 cells were infected with PR8/PB2–GFP, WSN/PB2–GFP, CA04/PB2–GFP or VN1203/PB2–GFP. At 16 h p.i., the cells were stained with mAbs specific for WSN, CA04, VN1203 or seasonal H1 HA protein, respectively. Expression of HA and GFP was examined by fluorescence microscopy.
Expression of alternative reporter genes in PB2-KO virus
The activity of the luciferase reporter gene is readily quantifiable and therefore its incorporation into PB2-KO virus should allow the measurement of virus replication based on luciferase activity. To test this possibility, we generated PB2-KO viruses possessing either the firefly (PR8/PB2-Fluc) or Renilla (PR8/PB2-Rluc) luciferase gene in the PB2 vRNA (Table 1). AX4/PB2 and wild-type AX4 cells were infected with these viruses at various m.o.i. and subjected to a luciferase assay at 8 h post-infection (p.i.). In virus-infected AX4/PB2 cells, Fluc and Rluc activities were detected in a dose-dependent manner; viruses infected at an m.o.i. of 0.1 and 0.001 were adequate for detecting significant Fluc and Rluc activities, respectively (Fig. 3a). By contrast, to detect significant GFP intensity in virus-infected cells, we needed to infect PR8/PB2–GFP at an m.o.i. of 1 or higher (Fig. 3b). These results indicated that the Fluc and Rluc genes can be accommodated in PB2-KO virus and represent a more quantitative indicator for virus replication than the GFP gene. Wild-type AX4 cells infected with PR8/PB2–Fluc and PR8/PB2–Rluc also exhibited detectable Fluc and Rluc activities, respectively, at an m.o.i. of >1 for PR8/PB2–Fluc and >0.01 for PR8/PB2–Rluc, although the activity of both reporter genes was more than tenfold lower than that detected in AX4/PB2 cells (Fig. 3a). As the PB2 protein was not provided in trans to the wild-type AX4 cells, the expression of these reporter genes suggested that viral ribonucleoproteins (i.e. PB2, PB1, PA and NP) derived from incoming viruses mediated the transcription of the PB2 vRNA of PB2-KO virus at a significantly high level in wild-type AX4 cells.
Fig. 3.
Accommodation of various reporter genes in PB2-KO virus. (a) Luciferase activity in PB2-KO virus-infected cells. Wild-type AX4 and AX4/PB2 cells were infected with PR8/PB2–Fluc or PR8/PB2–Rluc at the indicated m.o.i. At 8 h p.i., Fluc and Rluc activities in the cells were measured using a dual-luciferase reporter assay system. (b) GFP intensity in PB2-KO virus-infected cells. AX4/PB2 cells were infected with PR8/PB2–GFP at the indicated m.o.i. At 8 h p.i., GFP intensity was measured using a microplate reader. In (a) and (b), the results from virus-infected cells were compared with those from uninfected cells (indicated by ‘0’) and P values were calculated using Student’s t-test. *, P<0.05. RFU, Relative fluorescent units.
PB2-KO virus-based microneutralization
Biologically contained, reporter gene-expressing influenza viruses have the potential to supersede conventional virus replication evaluation systems in part because of the ability to quantify growth via plate reader assays. The neutralization activity of antisera is typically determined using conventional microneutralization assays (Itoh et al., 2009; Kobasa et al., 2004), which allow the detection of neutralizing antibodies based on the presence or absence of virus infection-induced CPE or virus antigens, as detected by ELISA. To use PB2-KO virus to detect neutralizing antibodies against 2009 pandemic viruses, we generated a PB2-KO virus that possessed the Rluc gene-encoding PB2 vRNA and A/California/04/2009-derived HA and NA vRNAs (CA04/PB2–Rluc; Table 1). CA04/PB2–Rluc exhibited slightly attenuated growth kinetics in wild-type AX4/PB2 cells compared with those of wild-type CA04, although we did not find a statistically significant difference by Student’s t-test (Fig. 4a). CA04/PB2–Rluc (100 p.f.u.) was mixed with serially diluted antiserum samples collected from CA04-infected ferrets and incubated at 37 °C for 1 h. Serum from seasonal H1N1 influenza virus A/Iowa/01/2006-infected and uninfected ferrets served as a negative control. The virus–serum mixtures were used to inoculate AX4/PB2 cells. At 24 h p.i., Rluc activity in cells was measured using a Renilla luciferase assay system. To compare the detection sensitivity, the same antiserum samples were also tested for neutralization activity using a CPE-based conventional microneutralization assay with wild-type CA04 and wild-type AX4 cells. In the PB2-KO virus-based assay, 1 : 1280- and 1 : 640-diluted ferret sera induced a significant decrease in Rluc activity in virus-infected cells (Fig. 4b). By contrast, the neutralizing titres of the same ferret serum samples as determined in the conventional microneutralization assay were 160 and 80 (data not shown). In addition, neutralization activity against CA04 was detected even in serum from an H1N1 A/Iowa/01/2006-infected ferret (up to 1 : 160 dilution) by the PB2-KO virus-based assay (Fig. 4b) and using the conventional assay (up to 1 : 20 dilution). Importantly, cross-reactivity of this ferret serum against CA04 was not observed using a haemagglutination inhibition assay. These results indicated that the PB2-KO virus coupled with PB2-expressing cells offers a neutralizing antibody detection method that is more sensitive than the conventional microneutralization and haemagglutination inhibition assays.
Fig. 4.
PB2-KO virus-based microneutralization assay. (a) Growth kinetics of CA04/PB2–Rluc monitored over 72 h. Wild-type AX4 and AX4/PB2 cells were infected with wild-type CA04 or CA04/PB2–Rluc virus at an m.o.i. of 0.001. Supernatants collected at the indicated time points were assayed for infectious virus by plaque assay in AX4/PB2 cells. (b) AX4/PB2 cells were infected with 100 p.f.u. CA04/PB2–Rluc that had been pre-mixed with serially diluted ferret serum samples in triplicate wells. Rluc activity in cells was measured using a Renilla luciferase assay system at 24 h p.i. The results from virus-infected cells were compared with those from cells that were infected with serum-untreated (−) virus. P values were calculated using Student’s t-test. *, P<0.05. RLU, Relative light units.
Discussion
Here, we demonstrated that PB2-KO influenza viruses are replication incompetent in wild-type cells but undergo numerous replication cycles in PB2 protein-expressing cells (Fig. 1d). In addition, reporter genes, such as luciferase genes, which are readily quantifiable with high sensitivity, flanked by the PB2 vRNA packaging signals, were stably maintained in progeny viruses (Table 1) and expressed in virus-infected cells (Figs 2 and 3). We also confirmed that different virus strain-derived HA and NA genes were accommodated by PB2-KO viruses (Fig. 2). These results indicated that PB2-KO viruses have a broad potential use throughout the field of influenza virology.
As a practical application, a PB2-KO virus-based microneutralization assay was developed and used to detect neutralizing antibodies against the 2009 pandemic virus (Fig. 4). This PB2-KO virus-based assay proved to be more sensitive than the conventional microneutralization assay in terms of neutralizing antibody detection. This robust, plate reader-based assay should be a powerful tool for the large-scale screening of neutralizing antibodies, such as a serological survey or the establishment of mAbs.
The use of replication-incompetent PB2-KO viruses as a screening platform (Fig. 1c, d) may enable the detection of neutralizing antibodies against highly pathogenic viruses such as H5N1 and 1918 strains, which normally must be handled in Biosafety Level 3 facilities and under Biosafety Level 2 containment, although an additional layer of biosafety (e.g. modification of the amino acid sequence of the HA cleavage site) would be required. Kong et al. (2006) previously developed a neutralizing antibody screening system based on influenza HA-pseudotyped lentiviruses, which also allows the detection of neutralizing antibodies against the Biosafety Level 3 agents. However, these lentiviruses do not express influenza virus NA, which, along with HA, has the potential to induce neutralizing antibodies (Nayak et al., 2010); therefore, our PB2-KO virus-based assay should reflect the neutralizing antibody titres more accurately. Although cells that stably express reporter gene-encoding influenza vRNA have also been shown to allow the sensitive detection of neutralizing antibodies (Hossain et al., 2010; Li et al., 2009), infectious viruses are required for these recombinant cell-based assays.
Another potential application of the PB2-KO virus is its use as a novel influenza vaccine, which we believe is feasible for the following reasons. Firstly, PB2-KO virus generated high titres (>108 p.f.u. ml−1) in the AX4/PB2 cell line (Fig. 1d); secondly, the fact that HA and NA proteins could be expressed (Fig. 2) demonstrated that PB2-KO virus is customizable to encode desired antigens; thirdly, the vRNA transcription that occurred in PB2-KO virus-infected cells (Fig. 3a) may stimulate cellular innate immunity by producing dsRNA; and lastly, the stable maintenance of a foreign gene inserted in the PB2 vRNA (Table 2) could serve as a carrier of an additional antigen, enabling the engineering of PB2-KO as a safe multivalent vaccine. In a preliminary study, PR8/PB2–GFP protected mice against lethal challenge with wild-type PR8 (S. T. Victor, M. Ozawa, and Y. Kawaoka, unpublished data).
To date, several recombinant influenza viruses that lack a particular viral protein have been shown to replicate comparably to wild-type virus in cell culture when the missing protein is provided in trans. M2-lacking influenza virus efficiently replicates in M2-expressing cells and has demonstrated potential as a live attenuated vaccine (Watanabe et al., 2009). A distinct advantage of the PB2-KO virus over its M2-lacking counterpart is that the former is replication incompetent in normal cells and thus safer. Furthermore, it remains unknown whether a foreign gene encoded in the M2 protein-coding region can be incorporated into progeny viruses and expressed in virus-infected cells.
Martínez-Sobrido et al. (2010) developed an improved screening assay for the rapid detection of neutralizing antibodies by using influenza virus possessing the GFP gene flanked by the HA vRNA packaging signals. Although this HA-KO virus underwent numerous replication cycles only in cells that expressed the HA protein, the stability of reporter genes in this HA-KO virus was not tested in the study. In fact, we generated a WSN-based HA-KO virus possessing the GFP gene flanked by the HA vRNA packaging signals (9 and 80 nt of the 3′ and 5′ coding sequences, respectively, along with the non-coding sequences at both ends) by reverse genetics and found that only 54.2 % (13/24 plaques) of the plaques formed by our HA-KO virus that was amplified in WSN HA-expressing MDCK cells expressed GFP. Moreover, an HA vRNA-deficient virus possessing seven vRNA segments underwent numerous rounds of replication in HA-expressing MDCK cells (data not shown), in contrast to the PB2 vRNA-deficient PR8ΔPB2 virus (see Results). These results suggest that the reporter gene-encoding HA vRNA in HA-KO virus could easily be dropped during replication in HA-expressing cells. A replication-competent virus that possesses the GFP gene in its NA vRNA has also been used to detect neutralizing antibodies (Rimmelzwaan et al., 2011). An in trans bacterial sialidase improved the restricted replication of this NA-KO virus and allowed reasonable virus titre recovery; however, the reporter gene stability of the NA-KO virus remains uncertain. By contrast, the stability of reporter genes in the PB2-KO virus (Table 2) guarantees the reliability of its application assays.
More recently, GFP gene-possessing replication-competent influenza viruses have been generated using recombinant NS1 (Manicassamy et al., 2010) or NA (Li et al., 2010) genes. Although these viruses have potential as research tools, their replicability raises biosafety issues, which are not a concern with the PB2-KO virus. Overall, the fact that the PB2-KO virus produced in this study stably expresses a foreign gene and is replication incompetent makes it ideal in terms of reliability and biosafety.
In conclusion, we generated a biologically contained foreign-gene-expressing influenza virus by replacing the viral PB2 gene with reporter genes. Replication of the virus was restricted to cells that expressed the PB2 protein in trans. The reporter gene was stably inherited in progeny viruses during replication in PB2-expressing cells. Various HA, NA and reporter genes were accommodated in the PB2-KO virus. This virus therefore shows promise in terms of its numerous applications for basic and applied studies of influenza virus.
Methods
Cells.
293T human embryonic kidney cells (a derivative of the 293 line into which the gene for simian virus 40 T antigen was inserted; DuBridge et al., 1987) were maintained in Dulbecco’s modified Eagle medium (Lonza) supplemented with 10 % FCS (Invitrogen). MDCK cells were maintained in minimum essential medium (MEM; Invitrogen) supplemented with 5 % newborn calf serum (NCS; Sigma). AX4 cells, derived from MDCK cells and transfected with the cDNA of human 2,6-sialyltransferase (Hatakeyama et al., 2005), were maintained in 5 % NCS/MEM with 2 µg puromycin (Sigma) ml−1. AX4/PB2 cells [AX4 cells stably expressing the PB2 protein derived from A/Puerto Rico/8/34 (H1N1, PR8), established by transduction with a retroviral vector; see Results] were maintained in 5 % NCS/MEM with 2 µg puromycin ml−1 and 10 µg blasticidin (InvivoGen) ml−1. All cells were maintained at 37 °C in 5 % CO2.
Reverse genetics and virus propagation.
Reverse genetics was performed with plasmids containing the cDNAs of the PR8 viral genes between the human RNA polymerase I promoter and the mouse RNA polymerase I terminator (referred to as PolI plasmids) and eukaryotic protein expression plasmids (NP, PA, PB1 and PB2) under the control of the chicken β-actin promoter (Niwa et al., 1991), as described previously (Neumann et al., 1999). Briefly, the wild-type PR8 virus was engineered using eight previously produced wild-type constructs derived from PR8 (Horimoto et al., 2007), whereas the PB2-KO mutant comprised pPolIPB2(120)GFP(120) (Fig. 1a; Muramoto et al., 2006) and the remaining seven segmental PolI plasmids. The pPolIPB2(120)GFP(120) plasmid contains the A/WSN/33 (H1N1, WSN)-derived 3′ PB2 non-coding region, 120 nt that correspond to the PB2 coding sequence at the 3′ end of the vRNA followed by the GFP coding sequence, 120 nt that correspond to the PB2 coding sequence at the 5′ end of the vRNA and finally the 5′ PB2 non-coding region (Muramoto et al., 2006). Similarly, pPolIPB2(120)Fluc(120) and pPolIPB2(120)Rluc(120) were constructed to generate PB2-KO viruses possessing the firefly luciferase (Fluc) or Renilla luciferase (Rluc) genes, respectively. The eight PolI plasmids and protein expression plasmids were mixed with the transfection reagent TransIT-293 (Mirus), incubated at room temperature for 15 min and added to 106 293T cells cultured in Opti-MEM I (Invitrogen). At 48 h post-transfection, the supernatant containing wild-type PR8 or PB2-KO virus was harvested and propagated in 10-day-old embryonated chicken eggs or AX4/PB2 cells, respectively. Wild-type CA04 was also generated using reverse genetics, as described previously (Yamada et al., 2010), and propagated in MDCK cells. The propagated viruses were titrated by plaque assay in MDCK cells to determine the number of p.f.u. of virus.
Immunofluorescence staining of the PB2 protein.
Confluent AX4 and AX4/PB2 cells seeded in 35 mm glass-bottomed dishes (Asahi Techno Glass) were fixed in PBS containing 4 % paraformaldehyde (Wako Pure Chemical Industries) and permeabilized with 0.1 % Triton X-100. Cells were incubated with an anti-PB2 antibody (clone 18/1; Hatta et al., 2000) and further incubated with an Alexa Fluor 594-labelled anti-mouse secondary antibody (Invitrogen) in Hoescht 33342 (Invitrogen). Samples were observed under a confocal laser microscope (LSM510META; Carl Zeiss).
RT-PCR.
To detect PB2 mRNA in AX4/PB2 cells, total RNA was extracted using an RNeasy RNA Extraction kit (Qiagen). Viral RNAs were isolated from virions using a QIAmp Viral RNA Mini kit (Qiagen). Reverse transcription and cDNA synthesis were performed with an oligo(dT) primer and SuperScript III reverse transcriptase (Invitrogen). Samples in which the reverse transcription step was omitted were prepared as negative controls. The synthesized cDNA was amplified by PCR with Phusion PCR polymerase (Finnzymes) and PB2-specific primers. RT-PCR conditions, including primer sequences, are available on request.
Growth kinetics and virus titration.
To determine virus growth rates, triplicate wells of confluent AX4 or AX4/PB2 cells were infected at an m.o.i. of 0.001. After 1 h of virus adsorption, cells were washed in MEM containing 0.3 % BSA and overlaid with MEM containing l-(tosylamido-2-phenyl) ethyl chloromethyl ketone-treated trypsin (0.5 µg ml−1). Supernatants were collected every 12 or 24 h for 3 days and assayed for infectious virus in plaque assays in AX4/PB2 cells.
Immunostaining.
To assess the stability of the GFP reporter gene incorporation in the PB2-KO virus, AX4/PB2 cells were infected with various PB2-KO virus dilutions (from undiluted to 10−10). The supernatant from the second-to-last well in which a CPE was observed was harvested and diluted for subsequent infections. Supernatants from five rounds of virus passaging were subjected to a standard virus plaque assay. Once the number of plaques formed was counted, the agar was removed and wells containing plaques were fixed with 100 % methanol for 30 min. Wells were then washed with PBS and incubated with an anti-GFP mAb (clone GFP-20; Sigma) at room temperature for 1 h. Immunohistochemical staining was performed using a biotinylated anti-mouse antibody according to the Vectastain Elite ABC kit instructions (Vector Laboratories). GFP-positive plaques were visualized using Fast 3,3′-diaminobenzidine tablets (Sigma), and the number of GFP-positive plaques was calculated as a percentage of the total number of plaques that formed in the respective wells.
Immunofluorescent staining of the HA protein.
GFP-encoding PB2-KO virus possessing HA and NA vRNAs derived from PR8, WSN, A/California/04/09 (CA04) or A/Vietnam/1203/04 (VN1203) was generated by reverse genetics, as described above, and propagated in AX4/PB2 cells. The various basic amino acid residues in the VN1203 HA cleavage site were replaced with a non-virulent-type cleavage sequence. Confluent AX4/PB2 cells were infected with these viruses at an m.o.i. of 0.2–1. At 16 h p.i., cells were fixed with 4 % paraformaldehyde in PBS and permeabilized with 0.1 % Triton X-100. Cells were then incubated with an anti-WSN HA antibody (clone WS 3-54; prepared in house), an anti-CA04 HA antibody (clone IT-096; eENZYME), an anti-H5 HA antibody (clone 8A3; Rockland Immunochemicals) and an anti-seasonal H1 HA antibody (clone IVC102; Abcam) and then incubated with an Alexa Fluor 594-labelled anti-mouse secondary antibody. Samples were observed under a fluorescence microscope.
Luciferase assay.
Cells infected with PB2-KO virus encoding the Fluc or Rluc gene were subjected to a luciferase assay using a Dual-Luciferase Reporter Assay System (Promega) at 8 h p.i. according to the manufacturer’s instructions. Fluc and Rluc activities were measured with a microplate reader (Infinite M1000; Tecan).
Microneutralization assay.
Sera were collected from two ferrets infected with 106 p.f.u. wild-type CA04 at 3 weeks p.i. and from two uninfected ferrets. Twofold serial dilutions of receptor-destroying enzyme (Denka Seiken)-treated ferret sera were mixed with 100 p.f.u. wild-type CA04 or Rluc-encoding PB2-KO virus possessing CA04-derived HA and NA vRNAs (CA04/PB2–Rluc). After incubation at 37 °C for 1 h, wild-type AX4 or AX4/PB2 cells were inoculated with the wild-type virus–serum or PB2-KO virus–serum mixtures, respectively, in triplicate wells and incubated for 3 days or 24 h for the wild-type and PB2-KO viruses, respectively. The neutralization activity of the ferret sera was determined on the basis of the CPE observed by microscopy or the Rluc activity as measured using the Renilla Luciferase Assay System (Promega) for the wild-type or PB2-KO virus, respectively.
Acknowledgements
We thank Ashley Luka and Lisa Burley for technical assistance. We also thank Susan Watson for editing the manuscript. This work was supported by ERATO (Japan Science and Technology Agency), by a grant-in-aid for Specially Promoted Research from the Ministries of Education, Culture, Sports, Science, and Technology, by grants-in-aid from Health, Labor, and Welfare of Japan, by a Contract Research Fund for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, by National Institute of Allergy and Infectious Disease Public Health Service research grants, and by the Bill & Melinda Gates Foundation (grant #OPPGH5383).
References
- Davies W. L., Grunert R. R., Haff R. F., McGahen J. W., Neumayer E. M., Paulshock M., Watts J. C., Wood T. R., Hermann E. C., Hoffmann C. E. (1964). Antiviral activity of 1-adamantanamine (amantadine). Science 144, 862–863 10.1126/science.144.3620.862 [DOI] [PubMed] [Google Scholar]
- DuBridge R. B., Tang P., Hsia H. C., Leong P.-M., Miller J. H., Calos M. P. (1987). Analysis of mutation in human cells by using an Epstein–Barr virus shuttle system. Mol Cell Biol 7, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S., Sakai-Tagawa Y., Kiso M., Goto H., Kawakami C., Mitamura K., Sugaya N., Suzuki Y., Kawaoka Y. (2005). Enhanced expression of an α2,6-linked sialic acid on MDCK cells improves isolation of human influenza viruses and evaluation of their sensitivity to a neuraminidase inhibitor. J Clin Microbiol 43, 4139–4146 10.1128/JCM.43.8.4139-4146.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M., Asano Y., Masunaga K., Ito T., Okazaki K., Toyoda T., Kawaoka Y., Ishihama A., Kida H. (2000). Mapping of functional domains on the influenza A virus RNA polymerase PB2 molecule using monoclonal antibodies. Arch Virol 145, 1947–1961 10.1007/s007050070068 [DOI] [PubMed] [Google Scholar]
- Hayden F. G. (2001). Perspectives on antiviral use during pandemic influenza. Philos Trans R Soc Lond B Biol Sci 356, 1877–1884 10.1098/rstb.2001.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T., Murakami S., Muramoto Y., Yamada S., Fujii K., Kiso M., Iwatsuki-Horimoto K., Kino Y., Kawaoka Y. (2007). Enhanced growth of seed viruses for H5N1 influenza vaccines. Virology 366, 23–27 10.1016/j.virol.2007.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. J., Perez S., Guo Z., Chen L.-M., Donis R. O. (2010). Establishment and characterization of a Madin–Darby canine kidney reporter cell line for influenza A virus assays. J Clin Microbiol 48, 2515–2523 10.1128/JCM.02286-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Shinya K., Kiso M., Watanabe T., Sakoda Y., Hatta M., Muramoto Y., Tamura D., Sakai-Tagawa Y., et al. & other authors (2009). In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460, 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D., Takada A., Shinya K., Hatta M., Halfmann P., Theriault S., Suzuki H., Nishimura H., Mitamura K., et al. & other authors (2004). Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431, 703–707 10.1038/nature02951 [DOI] [PubMed] [Google Scholar]
- Kong W.-P., Hood C., Yang Z.-Y., Wei C.-J., Xu L., García-Sastre A., Tumpey T. M., Nabel G. J. (2006). Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc Natl Acad Sci U S A 103, 15987–15991 10.1073/pnas.0607564103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Larrimer A., Curtiss T., Kim J., Jones A., Baird-Tomlinson H., Pekosz A., Olivo P. D. (2009). Influenza virus assays based on virus-inducible reporter cell lines. Influenza Other Respir Viruses 3, 241–251 10.1111/j.1750-2659.2009.00095.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Feng L., Pan W., Dong Z., Li C., Sun C., Chen L. (2010). Generation of replication-competent recombinant influenza A viruses carrying a reporter gene harbored in the neuraminidase segment. J Virol 84, 12075–12081 10.1128/JVI.00046-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., García-Sastre A. (2010). Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107, 11531–11536 10.1073/pnas.0914994107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Sobrido L., Cadagan R., Steel J., Basler C. F., Palese P., Moran T. M., García-Sastre A. (2010). Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J Virol 84, 2157–2163 10.1128/JVI.01433-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto Y., Takada A., Fujii K., Noda T., Iwatsuki-Horimoto K., Watanabe S., Horimoto T., Kida H., Kawaoka Y. (2006). Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol 80, 2318–2325 10.1128/JVI.80.5.2318-2325.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak B., Kumar S., DiNapoli J. M., Paldurai A., Perez D. R., Collins P. L., Samal S. K. (2010). Contributions of the avian influenza virus HA, NA, and M2 surface proteins to the induction of neutralizing antibodies and protective immunity. J Virol 84, 2408–2420 10.1128/JVI.02135-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Watanabe T., Ito H., Watanabe S., Goto H., Gao P., Hughes M., Perez D. R., Donis R., et al. & other authors (1999). Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96, 9345–9350 10.1073/pnas.96.16.9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- Palese P., Shaw M. L. (2007). Orthomyxoviridae: the viruses and their replication. In Fields Virology, 5th edn, pp. 1647–1689 Edited by Knipe D. M., Howley P. M. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Rimmelzwaan G. F., Verburgh R. J., Nieuwkoop N. J., Bestebroer T. M., Fouchier R. A., Osterhaus A. D. (2011). Use of GFP-expressing influenza viruses for the detection of influenza virus A/H5N1 neutralizing antibodies. Vaccine 29, 3424–3430 10.1016/j.vaccine.2011.02.082 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Watanabe T., Kawaoka Y. (2009). Influenza A virus lacking M2 protein as a live attenuated vaccine. J Virol 83, 5947–5950 10.1128/JVI.00450-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P. F., Neumann G., Kawaoka Y. (2007). Orthomyxoviruses. In Fields Virology, 5th edn, pp. 1691–1740 Edited by Knipe D. M., Howley P. M. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Yamada S., Hatta M., Staker B. L., Watanabe S., Imai M., Shinya K., Sakai-Tagawa Y., Ito M., Ozawa M., et al. & other authors (2010). Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog 6, e1001034 10.1371/journal.ppat.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]