Abstract
Klotho (KL) is a putative tumor suppressor gene in breast and pancreatic cancers located at chromosome 13q12. A functional sequence variant of Klotho (KL-VS) was previously reported to modify breast cancer risk in Jewish BRCA1 mutation carriers. The effect of this variant on breast and ovarian cancer risks in non-Jewish BRCA1/BRCA2 mutation carriers has not been reported. The KL-VS variant was genotyped in women of European ancestry carrying a BRCA mutation: 5,741 BRCA1 mutation carriers (2,997 with breast cancer, 705 with ovarian cancer, and 2,039 cancer free women) and 3,339 BRCA2 mutation carriers (1,846 with breast cancer, 207 with ovarian cancer, and 1,286 cancer free women) from 16 centers. Genotyping was accomplished using TaqMan® allelic discrimination or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Data were analyzed within a retrospective cohort approach, stratified by country of origin and Ashkenazi Jewish origin. The per-allele hazard ratio (HR) for breast cancer was 1.02 (95% CI 0.93–1.12, P = 0.66) for BRCA1 mutation carriers and 0.92 (95% CI 0.82–1.04, P = 0.17) for BRCA2 mutation carriers. Results remained unaltered when analysis excluded prevalent breast cancer cases. Similarly, the per-allele HR for ovarian cancer was 1.01 (95% CI 0.84–1.20, P = 0.95) for BRCA1 mutation carriers and 0.9 (95% CI 0.66–1.22, P = 0.45) for BRCA2 mutation carriers. The risk did not change when carriers of the 6174delT mutation were excluded. There was a lack of association of the KL-VS Klotho variant with either breast or ovarian cancer risk in BRCA1 and BRCA2 mutation carriers.
Keywords: Breast cancer, Ovarian cancer-Klotho, BRCA, Modifier gene
Introduction
The Klotho (KL) gene (MIM # 604824) encodes a 1,014 amino acid transmembrane protein [1]. The Klotho protein has pleiotropic activities, including potent inhibition of the insulin receptor (IR) and the IGF-1 receptor (IGF-1R) [2]. This pathway plays a prominent role in breast cancer pathogenesis [3, 4] and specifically in BRCA1–BRCA2-associated breast cancers [5, 6] and epithelial ovarian cancer [7]. These functional activities, taken together with the in vitro evidence of its tumor suppressor activity in breast cancer cell lines and altered expression somatically in breast cancer tissues [8], combined with the chromosomal location of the gene at 13q12, a frequent site of allelic loss in breast and ovarian tumors [9], make KL a putative tumor suppressor gene in breast and ovarian cancer. Since the lifetime risk for developing these tumor types is substantially increased in women who carry germline mutations in BRCA1 or BRCA2 [10, 11], it seemed plausible that KL could affect cancer penetrance in BRCA1 and/or BRCA2 mutation carriers. Indeed, we have previously shown that loss of function sequence variants in the Klotho gene [amino acid substitutions F352V (rs9536314) and C370S (rs9527025) both in linkage disequilibrium—coined KL-VS] that can be evaluated by genotyping the rs9536314 SNP are associated with an altered breast cancer risk in Jewish Ashkenazi BRCA1 mutation carriers [12]. As the penetrance of mutated BRCA1 and BRCA2 alleles may in part be determined by the location and the type of mutation, and by other genetic factors, at times ethnically restricted, and since the mutational spectrum in Ashkenazi Jews in BRCA1 is limited [13, 14], this study aimed to evaluate the putative modifier effect of the KL-VS sequence variants on mutant BRCA1 and BRCA2 allele penetrance in a larger cohort of ethnically diverse mutation carriers.
Methods
Study participants—recruitment and data collection
All study participants were women, aged 18 years or above, who carried a deleterious germline mutation in either BRCA1 or BRCA2. Study participant genotype and phenotype data were submitted from 16 centers participating in the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) [15]. The recruitment strategy and the type of clinical, demographic, and phenotypic data collected from each participant, as well as the means used to ensure that there are no overlaps and no duplicate genotyping, have previously been reported [15, 16]. All study participants took part in research studies at the parent institutions under ethically approved protocols as previously detailed [15, 16].
Genetic analysis
All participating centers genotyped the KL-VS variant (rs9536314) using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) [16], or TaqMan® allelic discrimination with limited sequencing, as previously described [12]. Genotyping quality control procedures were carried out as reported elsewhere (http://www.srl.cam.ac.uk/consortia/cimba/eligibility/eligibility.html).
Statistical analysis
Data were analyzed within a retrospective likelihood approach [17], assuming a proportional hazards model for the effect of the genotype on cancer incidence. For the breast cancer analysis, carriers were censored at the age of the first of the following events: breast cancer diagnosis, ovarian cancer diagnosis, bilateral prophylactic mastectomy, or age at last observation. Only those censored at a breast cancer diagnosis were assumed to be affected. To evaluate the association with ovarian cancer risk, data were analyzed within a competing risks model framework, which evaluates the associations with breast and ovarian cancer risk simultaneously [18]. For this analysis, mutation carriers were followed up to the age of the first breast or ovarian cancer diagnosis and were considered to have developed the corresponding disease. Individuals were censored for breast cancer at the age of bilateral prophylactic mastectomy and for ovarian cancer at the age of bilateral oophorectomy and were assumed to be unaffected for the corresponding disease. The remaining individuals were censored at the age at last observation and were assumed to be unaffected for both diseases. For all models, the effect of each SNP was modeled either as a per-allele hazard ratio (HR) (multiplicative model) or as separate HRs for heterozygotes and homozygotes, and these were estimated on the log scale (i.e., βi). Analyses were carried out using the pedigree analysis software MENDEL [19]. A robust variance-estimation approach was used to allow for the non-independence among related carriers [20]. Analyses were stratified by country of residence and used calendar-year- and cohort-specific cancer incidences for BRCA1 and BRCA2 [21]. Because of the previously reported linkage disequilibrium between the KL-VS variant and BRCA2 6174delT mutation [12], the US American BRCA2 carriers, who contributed the majority of the 6174delT mutation carriers, were further stratified according to whether they carried this particular mutation or not.
Results
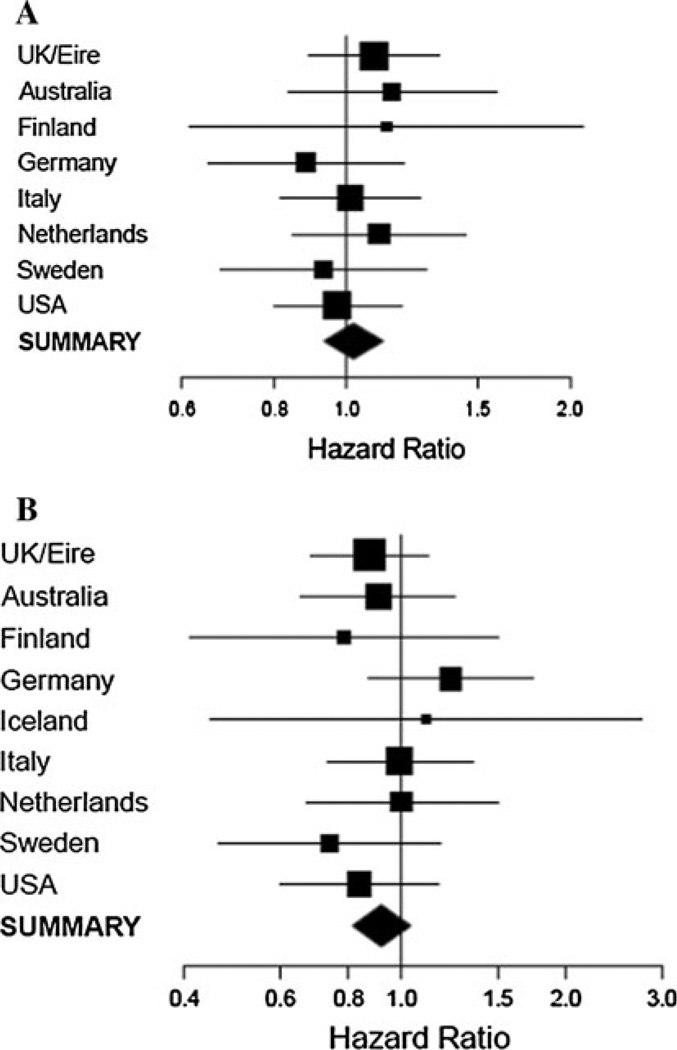
A total of 9,080 mutation carriers (5,741 BRCA1 and 3,339 BRCA2 mutation carriers) from 16 centers were included in the current analysis (Table 1, Supplementary Table 1). In the breast cancer analysis there were 4,870 subjects with breast cancer (3,020 BRCA1 mutation carriers, 1,850 BRCA2 mutation carriers), and 4,210 unaffected carriers (2,721 BRCA1 carriers, 1,489 BRCA2 carriers) (Tables 1, 2). The genotype frequencies and estimated HRs by mutation and disease status for breast and ovarian cancer are shown in Tables 2, 3, and 4. There was no evidence of association between breast cancer risk and the KL-VS genotype for either BRCA1 or BRCA2 mutation carriers (per-allele HR = 1.02, 95% CI 0.93–1.12, P = 0.66 and 0.92, 95% CI 0.82–1.04, P = 0.17 for BRCA1 and BRCA2, respectively). There was no evidence of heterogeneity in the HRs across studies: p-het = 0.91 for BRCA1 and p-het = 0.78 for BRCA2 (Fig. 1a, b). There was also no evidence of association of the Klotho sequence variant with ovarian cancer risk for BRCA1 mutation carriers (per-allele HR = 1.01, 95% CI 0.84–1.20, P = 0.95) or BRCA2 mutation carriers (per-allele HR = 0.90, 95% CI 0.66–1.22, P = 0.49) (Table 4). The results were unchanged when long-term cancer survivors (prevalent cases) were excluded from the analyses, or when carriers of the 6174delT mutation were excluded. Furthermore, the KL-VS variant was not significantly associated with breast cancer risk in an analysis of self-reported Ashkenazi Jewish participants (HR = 1.22, 95% CI 0.91–1.64, P = 0.18 for 374 BRCA1 mutation carriers and HR = 0.97, 95% CI 0.56–1.69, P = 0.93 for 209 BRCA2 mutation carriers).
Table 1.
Summary characteristics for the 9080 eligible BRCA1 and BRCA2 mutation carriers used in the analysis
| Characteristic | BRCA1 | BRCA2 | ||
|---|---|---|---|---|
| Unaffected | Breast Cancer | Unaffected | Breast Cancer | |
| Number | 2,721 | 3,020 | 1,489 | 1,850 |
| Person-years follow-up | 116,795 | 122,809 | 66,559 | 81,486 |
| Median age at censure (IQR) | 42 (33–51) | 39 (34–46) | 43 (35–53) | 43 (37–50) |
| Age at censure, N (%) | ||||
| <30 | 567 (20.8) | 392 (13.0) | 288 (19.3) | 169 (9.1) |
| 30–39 | 707 (26.0) | 1205 (39.9) | 359 (24.1) | 546 (29.5) |
| 40–49 | 727 (26.7) | 968 (32.1) | 391 (26.3) | 678 (36.7) |
| 50–59 | 463 (17.0) | 350 (11.6) | 242 (16.3) | 324 (17.5) |
| 60–69 | 178 (6.5) | 89 (3.0) | 137 (9.2) | 100 (5.4) |
| 70+ | 79 (2.9) | 16 (0.5) | 72 (4.8) | 33 (1.8) |
| Year of birth, N (%) | ||||
| <1920 | 9 (0.3) | 13 (0.4) | 11 (0.7) | 23 (1.2) |
| 1920–1929 | 62 (2.3) | 105 (3.5) | 46 (3.0) | 85 (4.6) |
| 1930–1939 | 196 (7.2) | 279 (9.2) | 115 (7.0) | 214 (11.6) |
| 1940–1949 | 413 (15.2) | 651 (21.6) | 206 (13.8) | 420 (22.7) |
| 1950–1959 | 603 (22.2) | 890 (29.5) | 315 (20.8) | 584 (31.6) |
| 1960–1969 | 755 (27.8) | 794 (26.3) | 429 (27.2) | 458 (23.1) |
| 1970+ | 683 (25.1) | 288 (9.5) | 367 (27.6) | 96 (5.2) |
| Ethnicity, N (%) | ||||
| Ashkenazi Jewish | 172 (6.3) | 202 (6.7) | 105 (7.1) | 104 (5.6) |
| Not Ashkenazi Jewish | 2,549 (93.7) | 2,818 (93.3) | 1,384 (92.9) | 1,746 (94.4) |
IQR interquartile range Carriers of European or Ashkenazi Jewish ancestry only
Table 2.
SNP genotype distribution and association with breast cancer risk. Analysis restricted to mutation carriers of European ancestry
| Mutation | Genotype | Unaffected, N (%) | Affecteda, N (%) | HR | 95% CI | P value |
|---|---|---|---|---|---|---|
| BRCA1 | GG | 1,941 (71.3) | 2,160 (71.5) | 1 | ||
| AG | 701 (25.8) | 775 (25.7) | 1.02 | 0.91–1.14 | ||
| AA | 79 (2.9) | 85 (2.8) | 1.06 | 0.79–1.41 | ||
| 2-df test | 0.90 | |||||
| Per-allele | 1.02 | 0.93–1.12 | 0.66 | |||
| BRCA2 | GG | 909 (61.1) | 1,201 (64.9) | 1 | ||
| AG | 504 (33.9) | 589 (31.8) | 0.96 | 0.83–1.10 | ||
| AA | 76 (5.1) | 60 (3.2) | 0.75 | 0.53–1.05 | ||
| 2-df test | 0.24 | |||||
| Per-allele | 0.92 | 0.82–1.04 | 0.17 |
Breast cancer
Table 3.
Association with breast cancer risk, after excluding prevalent breast cancer cases
| Unaffected, N | Affected, N | HR | 95% CI | P value | |
|---|---|---|---|---|---|
| Excluding prevalent breast cancer cases | |||||
| BRCA1 | 2,721 | 1,513 | 0.99 | 0.89–1.11 | 0.86 |
| BRCA2 | 1,489 | 908 | 0.92 | 0.79–1.06 | 0.26 |
Analysis restricted to mutation carriers of European ancestry
Table 4.
Competing risk analysis
| Unaffected, N (%) |
Breast cancer, N (%) |
Ovarian cancer, N (%) |
Breast cancer | Ovarian cancer | |||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||||
| BRCA1 | |||||||||
| GG | 1,447 (71.0) | 2,142 (71.5) | 512 (72.6) | 1 | 1 | ||||
| AG | 536 (26.3) | 770 (25.7) | 170 (24.1) | 1.01 | 0.90–1.13 | 0.94 | 0.76–1.16 | ||
| AA | 56 (2.8) | 85 (2.8) | 23 (3.3) | 1.12 | 0.83–1.52 | 1.29 | 0.76–2.19 | ||
| 2-df test | 0.76 | 0.51 | |||||||
| Per-allele | 1.03 | 0.93–1.13 | 0.60 | 1.01 | 0.84–1.20 | 0.95 | |||
| BRCA2 | |||||||||
| GG | 787 (61.2) | 1198 (64.9) | 125 (60.4) | 1 | 1 | ||||
| AG | 435 (33.8) | 589 (31.9) | 69 (33.3) | 0.94 | 0.81–1.09 | 0.82 | 0.58–1.14 | ||
| AA | 64 (5.0) | 59 (3.2) | 13 (6.3) | 0.73 | 0.51–1.04 | 0.97 | 0.45–2.08 | ||
| 2-df test | 0.19 | 0.48 | |||||||
| Per-allele | 0.91 | 0.80–1.02 | 0.11 | 0.90 | 0.66–1.22 | 0.49 | |||
Association with breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers. Analysis restricted to mutation carriers of European ancestry
Fig. 1.
a Per-allele stratum-specific HRs for breast cancer risk in BRCA1 carriers and b Per-allele stratum-specific HRs for breast cancer risk in BRCA2 carriers
Discussion
In this study, functional KL gene variants had no discernable effect on either breast or ovarian cancer risk in a large group of ethnically diverse BRCA1 and BRCA2 mutation carriers. These findings are in contrast with our previous report which showed that in Jewish BRCA1 mutation carriers there was an association of the same sequence variant on breast cancer risk [12]. The estimated HRs based on the complete sample as well as the restricted sample of Ashkenazi Jewish mutation carriers was considerably smaller than the relative risk reported previously. Several reasons could account for the lack of replication in this study: the number of self-reported Ashkenazi Jewish individuals in this study was relatively small (BRCA1 n = 374, BRCA2 n = 209). Therefore, if the association is restricted to Ashkenazi Jewish mutation carriers only, the present analysis may be under-powered. Alternatively, given the sample size of this study, the original finding may have been a false positive. It is well established that the mutation location and mutation type affect cancer risk in BRCA1 and BRCA2 mutation carriers [22, 23]. As the mutational spectrum locations and types in the present analysis was more diverse than in the Ashkenazi population, it is also possible that these inconsistencies in the results between this study and the previous one may relate to the effect of the Klotho variant on the specific Ashkenazi mutations based on their location and effect on protein.
Although this particular polymorphism did not show any effect on breast/ovarian cancer risk in BRCA1/BRCA2 mutation carriers, Klotho gene may still modify these risks by another sequence alteration that may be in linkage equilibrium in the Ashkenazi Jewish population or by other variants not or only in weak linkage disequilibrium with the current polymorphism, by epigenetic alterations in gene expression (e.g., methylation, miRNA effects) or in combination and interaction with other polymorphisms that in concert have an overall effect on cancer risk.
In conclusion, the KL-VS sequence variant in the Klotho gene has no major effect on breast or ovarian cancer risk in genetically susceptible, ethnically diverse individuals who carry either a deleterious BRCA1 or a BRCA2 mutation. However, the effect of this gene and its putative involvement in penetrance of mutant BRCA1 or BRCA2 alleles by other genetic or epigenetic mechanisms cannot be ruled out, especially in Ashkenazi Jews.
Supplementary Material
Acknowledgments
This research was supported in part by grants from the Israel Cancer Association to EF for the Israeli consortium of inherited breast cancer; The CIMBA data management and analysis is supported by Cancer Research, UK (C12292/A11174). ACA is a Cancer Research, UK Senior Cancer Research Fellow. GCT and ABS are supported by Fellowships from the Australian National Health and Medical Research Council. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia, and the Cancer Foundation of Western Australia. The kConFab clinical Follow Up Study has been funded by NHMRC, the National Breast Cancer Foundation and Cancer Australia. kConFab general acknowledgements: We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study for their contributions to this resource, and the many families who contribute to kConFab; Georgetown Center (CI) received support from the Familial Cancer Registry and the Tissue Culture Shared Registry at Georgetown University (NIH/NCI Grant P30-CA051008), the Cancer Genetics Network (HHSN261200744000C), and Swing Fore the Cure. Susan L Neuhasen’s support was received from NIH Grant R01CA74415 (to SLN), SLN was partially supported by the Morris and Horowitz Families Endowed Professorship; The MAYO study was supported by NIH Grant CA128978, the Breast Cancer Research Foundation and the Komen Foundation for the Cure; The HEBCS study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, and the Sigrid Juselius Foundation; SWE BRCA collaborators—Per Karlsson, Margareta Nordling, Annika Bergman, and Zakaria Einbeigi, Gothenburg, Sahlgrenska University Hospital; Marie Stenmark-Askmalm and Sigrun Liedgren, Linköping University Hospital; Åke Borg, Niklas Loman, Ha°kan Olsson, Maria Soller, Helena Jernström, Katja Harbst, and Karin Henriksson, Lund University Hospital; Annika Lindblom, Brita Arver, Anna von Wachenfeldt, Annelie Liljegren, Gisela Barbany-Bustinza, and Johanna Rantala, Stockholm, Karolinska University Hospital; Beatrice Melin, Henrik Grönberg, Eva-Lena Stattin, and Monica Emanuelsson, Umea° University Hospital; Hans Ehrencrona, Richard Rosenquist, and Niklas Dahl, Uppsala University Hospital; Epidemiological study of BRCA1 & BRCA2 mutation carriers (EMBRACE): Douglas F. Easton is the PI of the study. EMBRACE Collaborating Centres are: Coordinating Centre, Cambridge: Susan Peock, Debra Frost, Steve D. Ellis, Elena Fineberg, Radka Platte. North of Scotland Regional Genetics Service, Aberdeen: Zosia Miedzybrodzka, Helen Gregory. Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison, Lisa Jeffers. West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Kai-ren Ong, Jonathan Hoffman. South West Regional Genetics Service, Bristol: Alan Donaldson, Margaret James. East Anglian Regional Genetics Service, Cambridge: Marc Tischkowitz, Joan Paterson, Sarah Downing, Amy Taylor. Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark T. Rogers, Emma McCann. St James’s Hospital, Dublin & National Centre for Medical Genetics, Dublin: M. John Kennedy, David Barton. South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous, Sarah Drummond. Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman, Kathryn Hill. West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson, Eshika Haque, Ed Tobias, Alexis Duncan. South East Thames Regional Genetics Service, Guy’s Hospital London: Louise Izatt, Chris Jacobs, Caroline Langman. North West Thames Regional Genetics Service, Harrow: Huw Dorkins. Leicestershire Clinical Genetics Service, Leicester: Julian Barwell. Yorkshire Regional Genetics Service, Leeds: Julian Adlard, Gemma Serra-Feliu. Cheshire & Merseyside Clinical Genetics Service, Liverpool: Ian Ellis, Catherine Houghton. Manchester Regional Genetics Service, Manchester: D. Gareth Evans, Fiona Lalloo, Jane Taylor. North East Thames Regional Genetics Service, NE Thames, London: Lucy Side, Alison Male, Cheryl Berlin. Nottingham Centre for Medical Genetics, Nottingham: Jacqueline Eason, Rebecca Collier. Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber, Irene Jobson. Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durell, Barbara Stayner. The Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins, Elena Castro, Emma Killick, Sue Martin, Gillian Rea, Anjana Kulkarni. North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley. South West Thames Regional Genetics Service, London: Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester, Charlotte Eddy, Vishakha Tripathi, Virginia Attard. Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Donna McBride, Sarah Smalley. EMBRACE is supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo are supported by an NIHR Grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles and Elizabeth Bancroft are supported by Cancer Research UK Grant C5047/A8385; The German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC) GC-HBOC is supported by a Grant of the German Cancer Aid (Grant 109076) and the Centre of Molecular Medicine Cologne, Germany (CMMC); University of Kansas Medical Center (KUMC) would like to thank JoEllen Weaver for her help collecting patient data and samples. A.K.G. was funded by U01CA69631, 5U01CA113916, and the Eileen Stein Jacoby Fund while at FCCC. The author acknowledges support from The University of Kansas Cancer Center and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. is the Chancellors Distinguished Chair in Biomedical Sciences endowed Professor; UPENN funding- Breast Cancer Research Foundation (to KLN), Susan G. Komen for the Cure, MacDonald Women’s Cancer Risk Evaluation Program (to SMD); MSKCC was supported by grants to KO by the Breast Cancer Research Foundation, The Starr Cancer Consortium, The Robert and Kate Niehaus Clinical Cancer Initiative, and the Schreiber Family Research Fund; The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) HEBON Collaborating Centers: Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: F. B. L. Hogervorst, S. Verhoef, M. Verheus, L. J. van‘t Veer, F. E. van Leeuwen, M. A. Rookus, J. P. Knol-Bout; Erasmus Medical Center, Rotterdam, NL: M. Collée, A. M. W. van den Ouweland, A. Jager, M. J. Hooning, M. M. A. Tilanus-Linthorst, C. Seynaeve; Leiden University Medical Center, NL, Leiden: C. J. van Asperen, J. T. Wijnen, M. P. Vreeswijk, R. A. Tollenaar, P. Devilee; Radboud University Nijmegen Medical Center, Nijmegen, NL: M. J. Ligtenberg, N. Hoogerbrugge, C. M. Kets; University Medical Center Utrecht, Utrecht, NL: M. G. Ausems, R. B. van der Luijt; Amsterdam Medical Center, NL: C. M. Aalfs, T. A. van Os; VU University Medical Center, Amsterdam, NL: J. J. P. Gille, Q. Waisfisz, H. E. J. Meijers-Heijboer; University Hospital Maastricht, Maastricht, NL: E. B. Gomez-Garcia, K. E. P. van Roozendaal, Marinus J. Blok, B. Caanen; University Medical Center Groningen University, NL: J. C. Oosterwijk, A. H. van der Hout, M. J. Mourits; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H. F. Vasen. The HEBON study is supported by the Dutch Cancer Society Grants NKI1998-1854, NKI2004-3088, NKI2007-3756 and the ZonMW Grant 91109024.
Appendix
Collaborators
Gisela Barbany Bustinza, Department of Clinical Genetics, Karolinska University Hospital, Stockholm, Sweden; Helena Jernström, Department of Oncology, Lund University, Lund, Sweden; Maria Soller, Department of Clinical Genetics, Lund University Hospital, Lund, Sweden; Marie Stenmark Askmalm, Division of Clinical Genetics, Department of Clinical and Experimental Medicine, Linköping University, Linköping, Sweden; Richard Rosenquist, Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, Uppsala, Sweden; Theo van Os, Department of Clinical Genetics, Academic Medical Center, Amsterdam, The Netherlands; Jacoba P. Knol-Bout, Department of Epidemiology, Netherlands Cancer Institute, Amsterdam, The Netherlands; Hanne E. J. Meijers-Heijboer, Department of Clinical Genetics, VU Medical Center, Amsterdam, The Netherlands; Juul Wijnen, Kara Sarrel, and Mark Robson, Department of Human Genetics & Department of Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands; Kees E. P. van Roozendaal, Department of Clinical Genomics, MUMC, Maastricht, The Netherlands; Caroline M. Kets, Hereditary Cancer Clinic, Radboud University Nijmegen Medical Center, The Netherlands; Ans M. W. van den Ouweland, Department of Clinical Genetics, Family Cancer Clinic, Erasmus University Medical Center, Rotterdam, The Netherlands; Margreet G. E. M. Ausems, Jonathan Beesley, and Amanda B. Spurdle, Department of Medical Genetics, University Medical Center Utrecht, PO Box 85090, 3508 AB Utrecht, The Netherlands; Jan C. Oosterwijk, Department of Genetics, University Medical Center, Groningen University, Groningen, The Netherlands; Debra Frost, Steve D. Ellis, Radka Platte, Elena Fineberg, Centre for Cancer Genetic Epidemiology, Department of Public Health and Primary Care, University of Cambridge, UK; D. Gareth Evans, Fiona Lalloo, Susan M. Domchek, and Timothy R. Rebbeck, Genetic Medicine, Manchester Academic Health Sciences Centre, Central Manchester University Hospitals NHS Foundation Trust, Manchester, UK; Louise Izatt, Clinical Genetics, Guy’s and St. Thomas’ NHS Foundation Trust, London, UK; Ros Eeles and Yuan Chun Ding, Oncogenetics Team, The Institute of Cancer Research and Royal Marsden NHS Foundation Trust, UK; Rosemarie Davidson, Ferguson-Smith Centre for Clinical Genetics, Yorkhill Hospitals, Glasgow, UK; Diana Eccles, University of Southampton Faculty of Medicine, Southampton University Hospitals NHS Trust, Southampton UK; Trevor Cole and Bella Kaufman, West Midlands Regional Genetics Service, Birmingham Women’s Hospital Healthcare NHS Trust, Edgbaston, Birmingham, UK; Jackie Cook and Bella Kaufman, Sheffield Clinical Genetics Service, Sheffield Children’s Hospital, Sheffield, UK; Marc Tichkowitz, Department of Medical Genetics, University of Cambridge, UK; Fiona Douglas, Institute of Genetic Medicine, Centre for Life, Newcastle Upon Tyne Hospitals NHS Trust, Newcastle upon Tyne, UK; Carole Brewer, Department of Clinical Genetics, Royal Devon & Exeter Hospital, Exeter, UK; Shirley Hodgson, Medical Genetics Unit, St George’s, University of London, UK; Catherine Houghton, Cheshire & Merseyside Clinical Genetics Service, Liverpool Women’s NHS Foundation Trust, Liverpool, UK; Joseph Vijai, Clinical Cancer Genetics Laboratory, Memorial Sloane Kettering Cancer Center, New York, NY; Alfons Meindl, Department of Gynaecology and Obstetrics, Division of Tumor Genetics, Klinikum rechts der Isar, Technical University Munich, Germany; Ina Ruehl, Department of Gynaecology and Obstetrics, Ludwig-Maximillians University Munich, Germany; Norbert Arnold, Department of Gynaecology and Obstetrics, University Hospital of Schleswig-Holstein, Campus Kiel, Christian-Albrechts University Kiel, Germany; Raymonda Varon-Mateeva, Institute of Human Genetics, Campus Virchov Klinikum, Charite Berlin, Germany; Dorothea Gadzicki, Institute of Cell and Molecular Pathology, Hannover Medical School, Hannover, Germany; Sabine Preisler-Adams, Institute of Human Genetics, University of Münster, Münster, Germany; Dieter Niederacher, Department of Gynaecology and Obstetrics, University Hospital Düsseldorf, Heinrich-Heine University Duüsseldorf, Germany; Karin Kast, Department of Gynaecology and Obstetrics, University Hospital Carl Gustav Carus, Technical University Dresden, Germany; Andrea Gehrig, Centre of Familial Breast and Ovarian Cancer, Department of Medical Genetics, Institute of Human Genetics, University Würzburg, Germany; Christian Sutter, Institute of Human Genetics, Division of Molecular Genetics, University Heidelberg, Germany; Helmut Deissler, Department of Gynaecology and Obstetrics, University Hospital Ulm, Germany; Kristiina Aittomäki, Department of Clinical Genetics, Helsinki University Central Hospital, Meilahdentie 2, 00290 Helsinki, Finland; Xianshu Wang, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota 55905, USA; Noralane M. Lindor, Department of Medical Genetics, Mayo Clinic, Rochester, Minnesota 55905, USA; Siranoush Manoukian, Unit of Medical Genetics, Department of Preventive and Predicted Medicine, Fondazione IRCCS Istituto Nazionale Tumori (INT), Milan, Italy; Bernard Peissel, Unit of Medical Genetics, Department of Preventive and Predicted Medicine, Fondazione IRCCS Istituto Nazionale Tumori (INT), Milan, Italy; Bernardo Bonanni, Division of Cancer Prevention and Genetics, Istituto Europeo di Oncologia, Milan, Italy; Loris Bernard, Department of Experimental Oncology, Istituto Europeo di Oncologia., Milan, Italy and Consortium for Genomics Technology (Cogentech), Milan, Italy; Alessandra Viel, Unit of Experimental Oncology 1, Centro di Riferimento Oncologico, IRCCS, Aviano (PN), Italy; Barbara Pasini, Department of Genetics, Biology and Biochemistry, University of Turin, Turin, Italy; Laura Ottini, Department of Molecular Medicine, “Sapienza” University of Rome, Rome, Italy.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-011-1938-8) contains supplementary material, which is available to authorized users.
Conflict of interest All authors declare that they have no conflict of interest.
Contributor Information
Yael Laitman, The Susanne Levy Gertner Oncogenetics Unit 1, The Danek Gertner Institute of Human Genetics, Chaim Sheba Medical Center, Tel-Hashomer 52621, Israel.
Karoline B. Kuchenbaecker, Department of Public Health & Primary Care, CIMBA Coordinating Center, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge CB1 8RN, UK
Johanna Rantala, Department of Clinical Genetics, Karolinska University Hospital, Stockholm, Sweden.
Frans Hogervorst, Family Cancer Clinic, Netherlands Cancer Institute, Amsterdam, The Netherlands.
Susan Peock, Department of Public Health and Primary Care, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge, UK.
Andrew K. Godwin, Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, Kansas City, KS 66160, USA
Adalgeir Arason, Department of Pathology, Landspitali University Hospital, Reykjavík, Iceland; Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
Tomas Kirchhoff, Department of Environmental Medicine, NYU Cancer Institute, New York University School of Medicine, New York, NY, USA.
Kenneth Offit, Memorial Sloan-Kettering Cancer Center, New York, NY, USA.
Claudine Isaacs, Georgetown University, 3800 Reservoir Road, NW, Washington, DC 20007, USA.
Rita K. Schmutzler, Department of Gynaecology and Obstetrics, Centre of Familial Breast and Ovarian Cancer and Centre for Integrated Oncology (CIO), University Hospital of Cologne, Cologne, Germany
Barbara Wappenschmidt, Department of Gynaecology and Obstetrics, Centre of Familial Breast and Ovarian Cancer and Centre for Integrated Oncology (CIO), University Hospital of Cologne, Cologne, Germany.
Heli Nevanlinna, Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Central Hospital, Haartmaninkatu 8, 00290 Helsinki, Finland.
Xiaoqing Chen, Queensland Institute of Medical Research, 300 Herston Rd, Herston, QLD 4006, Australia.
Georgia Chenevix-Trench, Queensland Institute of Medical Research, 300 Herston Rd, Herston, QLD 4006, Australia.
Sue Healey, Queensland Institute of Medical Research, 300 Herston Rd, Herston, QLD 4006, Australia.
Fergus Couch, Departments of Laboratory Medicine and Pathology, and Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA.
Paolo Peterlongo, Unit of Molecular Bases of Genetic Risk and Genetic Testing, Department of Preventive and Predicted Medicine, Fondazione IRCCS Istituto Nazionale Tumori (INT), Milan, Italy; IFOM, Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy.
Paolo Radice, Unit of Molecular Bases of Genetic Risk and Genetic Testing, Department of Preventive and Predicted Medicine, Fondazione IRCCS Istituto Nazionale Tumori (INT), Milan, Italy; IFOM, Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy.
Katherine L. Nathanson, Departments of Medicine and Biostatistics and Epidemiology, Abramson Cancer Center, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA
Maria Adelaide Caligo, University Hospital of Pisa, Pisa, Italy.
Susan L. Neuhausen, Department of Population Sciences, Beckman Research Institute of City of Hope, Duarte, CA 91010, USA
Patricia Ganz, UCLA Schools of Public Health & Medicine, Division of Cancer Prevention & Control Research, Jonsson Comprehensive Cancer Center at UCLA, Los Angeles, CA 90095-6900, USA.
Olga M. Sinilnikova, Unité Mixte de Génétique Constitutionnelle Des Cancers Fréquents, Centre Hospitalier Universitaire de Lyon/Centre Léon Bérard, and Equipe Labellisée LIGUE 2008, UMR5201 CNRS, Centre Léon Bérard, Université de Lyon, Lyon, France
Lesley McGuffog, Department of Public Health & Primary Care, CIMBA Coordinating Center, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge CB1 8RN, UK.
Douglas F. Easton, Department of Public Health & Primary Care, CIMBA Coordinating Center, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge CB1 8RN, UK
Antonis C. Antoniou, Department of Public Health & Primary Care, CIMBA Coordinating Center, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge CB1 8RN, UK
Ido Wolf, The Oncology Institute, Chaim Sheba Medical Center, Tel-Hashomer 52621, Israel; The Sackler School of Medicine, Tel-Aviv University, Tel-Aviv, Israel.
Eitan Friedman, Email: feitan@post.tau.ac.il, eitan.friedman@sheba.health.gov.il, eitan211@netvision.net.il, The Susanne Levy Gertner Oncogenetics Unit 1, The Danek Gertner Institute of Human Genetics, Chaim Sheba Medical Center, Tel-Hashomer 52621, Israel; The Sackler School of Medicine, Tel-Aviv University, Tel-Aviv, Israel.
References
- 1.Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, Nabeshima Y-i. Identification of the human Klotho gene and its two transcripts encoding membrane and secreted Klotho protein. Biochem Biophys Res Commun. 1998;242:626–630. doi: 10.1006/bbrc.1997.8019. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M. Regulation of fibroblast growth factor-23 signaling by Klotho. J Biol Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf I, Sadetzki S, Catane R, Karasik A, Kaufman B. Diabetes mellitus and breast Cancer. Lancet Oncol. 2005;6:103–111. doi: 10.1016/S1470-2045(05)01736-5. [DOI] [PubMed] [Google Scholar]
- 4.Yee D. Targeting insulin-like growth factor pathways. BR J Cancer. 2006;94:465–468. doi: 10.1038/sj.bjc.6602963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudelist G, Wagner T, Rosner M, Fink-Retter A, Gschwantler-Kaulich D, Czerwenka K, Kroiss R, Tea M, Pischinger K, Köstler WJ, Attems J, Mueller R, Blaukopf C, Kubista E, Hengstschläger M, Singer CF. Intratumoral IGF-I protein expression is selectively upregulated in breast Cancer patients with BRCA1/2 mutations. Endocr Relat Cancer. 2007;14:1053–1062. doi: 10.1677/ERC-06-0075. [DOI] [PubMed] [Google Scholar]
- 6.Maor S, Yosepovich A, Papa MZ, Yarden RI, Mayer D, Friedman E, Werner H. Elevated insulin-like growth factor-I receptor (IGF-IR) levels in primary breast tumors associated with BRCA1 mutations. Cancer Lett. 2007;257:236–243. doi: 10.1016/j.canlet.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Lu L, Katsaros D, Wiley A, de la Longrais IAR, Puopolo M, Yu H. Klotho expression in epithelial ovarian Cancer and its association with insulin-like growth factors and disease progression. Cancer Invest. 2008;26:185–192. doi: 10.1080/07357900701638343. [DOI] [PubMed] [Google Scholar]
- 8.Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, Kuro-o M, Karlan B, Kaufman B, Koeffler HP, Rubinek T. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast Cancer. Oncogene. 2008;27:7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 9.van deRLooij M, Papp J, Sztán M, Pulay T, Elfadil I, Besznyak I, Tóth J, Devilee P, Oláh E Allelic imbalance and micro-satellite instability in BRCA1 associated breast and ovarian tumors. Int J Oncol. 2001;18(4):775–780. [PubMed] [Google Scholar]
- 10.Levy-Lahad E, Friedman E. Cancer risks among BRCA1 and BRCA2 mutation carriers. BR J Cancer. 2007;96:11–15. doi: 10.1038/sj.bjc.6603535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begg CB, Haile RW, Borg A, Malone KE, Concannon P, Thomas DC, Langholz B, Bernstein L, Olsen JH, Lynch CF, Anton-Culver H, Capanu M, Liang X, Hummer AJ, Sima C, Bernstein JL. Variation of breast Cancer risk among BRCA1/2 carriers. J Am Med Assoc. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf I, Laitman Y, Rubinek T, Abramovitz L, Novikov I, Beeri R, Kuro-O M, Koeffler HP, Catane R, Freedman LS, Levy-Lahad E, Karlan BY, Friedman E, Kaufman B. Functional variant of KLOTHO: a breast Cancer risk modifier among BRCA1 mutation carriers of Ashkenazi origin. Oncogene. 2010;29(1):26–33. doi: 10.1038/onc.2009.301. [DOI] [PubMed] [Google Scholar]
- 13.Roa BB, Boyd AA, Volcik K, Richards CS. Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat Genet. 1996;14(2):185–187. doi: 10.1038/ng1096-185. [DOI] [PubMed] [Google Scholar]
- 14.Laitman Y, Borsthein RT, Stoppa-Lyonnet D, Dagan E, Castera L, Goislard M, Gershoni-Baruch R, Goldberg H, Kaufman B, Ben-Baruch N, Zidan J, Maray T, Soussan-Gutman L, Friedman E. Germline mutations in BRCA1 and BRCA2 genes in ethnically diverse high risk families in Israel. Breast Cancer Res Treat. 2011;127(2):489–495. doi: 10.1007/s10549-010-1217-0. [DOI] [PubMed] [Google Scholar]
- 15.Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE CIMBA. An international initiative to identify genetic modifiers of Cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9(2):104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antoniou AC, Sinilnikova OM, Simard J, Léoné M, Dumont M, Neuhausen SL, Struewing JP, Stoppa-Lyonnet D, Barjhoux L, Hughes DJ, Coupier I, Belotti M, Lasset C, Bonadona V, Bignon YJ, Rebbeck TR, Wagner T, Lynch HT, Domchek SM, Nathanson KL, Garber JE, Weitzel J, Narod SA, Tomlinson G, Olopade OI, Godwin A, Isaacs C, Jakubowska A, Lubinski J, Gronwald J, Górski B, Byrski T, Huzarski T, Peock S, Cook M, Baynes C, Murray A, Rogers M, Daly PA, Dorkins H, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, Niederacher D, Deissler H, Spurdle AB, Chen X, Waddell N, Cloonan N, Kirchhoff T, Offit K, Friedman E, Kaufmann B, Laitman Y, Galore G, Rennert G, Lejbkowicz F, Raskin L, Andrulis IL, Ilyushik E, Ozcelik H, Devilee P, Vreeswijk MP, Greene MH, Prindiville SA, Osorio A, Benitez J, Zikan M, Szabo CI, Kilpivaara CI, Nevanlinna H, Hamann U, Durocher F, Arason A, Couch FJ, Easton DF, Chenevix-Trench G Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers Study (GEMO); Epidemiological Study of BRCA1 and BRCA2 Mutation Carriers (EMBRACE); German Consortium for Hereditary Breast and Ovarian Cancer (GCHBOC); Kathleen Cuningham Consortium for Research into Familial Breast Cancer (kConFab); Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) RAD51 135G’C modifies breast Cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antoniou AC, Easton DF. Models of genetic susceptibility to breast Cancer. Oncogene. 2006;25:5898–5905. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 18.Barnes D, Lee A, Easton D, Antoniou A EMBRACE Investigators, kConFab Investigators. Evaluation of the association methods for analysing modifiers of disease risk in carriers of high risk mutations. Genet Epidemiol. 2011 doi: 10.1002/gepi.21620. (in press) [DOI] [PubMed] [Google Scholar]
- 19.Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 20.Boos DD. On generalised score tests. Am Stat. 1992;46:327–333. [Google Scholar]
- 21.Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, Risch HA, Eyfjord JE, Hopper JL, Southey MC, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tryggvadottir L, Syrjakoski K, Kallioniemi OP, Eerola H, Nevanlinna H, Pharoah PD, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian Cancers: updates and extensions. BR J Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson D, Easton D Breast Cancer Linkage Consortium. Variation in Cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet. 2001;68(2):410–419. doi: 10.1086/318181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ware MD, DeSilva D, Sinilnikova OM, Stoppa-Lyonnet D, Tavtigian SV, Mazoyer S. Does nonsense-mediated mRNA decay explain the ovarian Cancer cluster region of the BRCA2 gene? Oncogene. 2006;25(2):323–328. doi: 10.1038/sj.onc.1209033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.