Abstract
The somatic recombination of lymphocyte antigen receptor loci is integral to lymphocyte differentiation and adaptive immunity. Here we review the relationship of this highly choreographed process with the zinc finger protein CTCF and with cohesin, a protein complex best known for its essential functions in post-replicative DNA repair and chromosome segregation during the cell cycle. At lymphocyte antigen receptor loci, CTCF and cohesin shape long-range interactions and contribute to V(D)J recombination by facilitating lineage- and developmental stage-specific transcription and accessibility.
Keywords: Cohesin, CTCF, transcription, recombination
V(D)J recombination
B- and T-lymphocytes are unusual among the cells in our bodies in that each cell has a receptor for antigen. These receptors are highly variable and their specificity characterises each B- and T-cell. This diversity is key to the workings of the adaptive immune system, which protects us from infections: B cell receptors are secreted as immunoglobulins (Ig) that bind soluble antigens and T cell receptors (TCRs) scan the surface of cells for foreign material.
The gene segments that encode the clone-specific portion of B- and T-cell receptors reside in lymphocyte antigen receptor loci, which comprise multiple copies of variable (V), diversity (D) and joining (J) gene segments arranged over large genomic areas. The diversity of antigen receptors in B- and T-lymphocytes is generated by a somatic rearrangement process that involves the cutting and pasting of gene segments [1]. This process is often referred to as V(D)J recombination because it V, D and J gene segments - the D in V(D)J is bracketed because D segments are only present in the TCR loci Tcrb and Tcrd and the Immunoglobulin (Ig) heavy chain locus Igh, but not in Tcra, Tcrg, or the Ig light chain loci Igk and Igl.
Rearrangement proceeds in a stereotypic, developmentally controlled order where D to J rearrangements occur before V to DJ rearrangements, Igh loci rearrange before Igk and Igl and light chain loci, and Tcrb before Tcra. Importantly, complete (V to (D)J or V to J) rearrangement of the Ig and TCR loci is restricted to B and T cells, respectively. The lineage- and developmental stage-specificity of V(D)J recombination is achieved by controlling the accessibility of lymphocyte receptor loci - and of the appropriate V, D and J domains within these loci - to the recombination machinery. This accessibility is mediated at least in part by transcription of unrearranged or partially rearranged loci, also known as germline transcription [1].
V(D)J recombination is essential for the generation of functional B- and T-cells and hence for adaptive immunity, yet it poses a number of formidable challenges.
First, V(D)J recombination requires the formation of DNA double strand breaks (DSB) by the RAG recombinase. Since the process takes place in lymphoid progenitors, which have virtually unlimited proliferative potential, this generates a potentially explosive mix and these breaks must be repaired swiftly and in a controlled fashion to avoid illegitimate genomic rearrangements that could result in neoplastic transformation. As an important safeguard, the RAG recombinase must have hold of recombination signal sequences at both sites that are to be recombined before it can proceed with initiating a DSB [1]. The challenge is how to satisfy this requirement for ‘synapsis’ between sites that are widely separated in the linear DNA sequence.
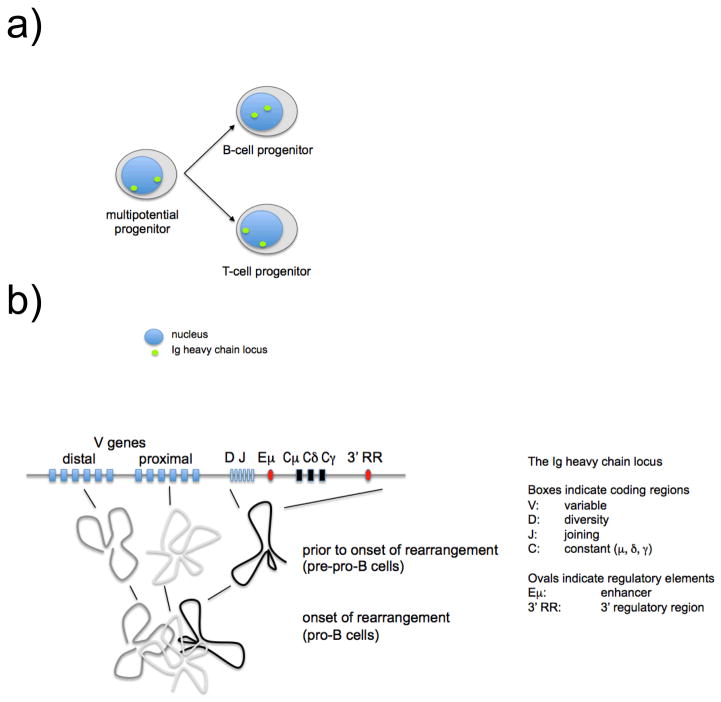
Second, for proper coordination with lymphocyte differentiation, the rearrangement of lymphocyte receptor loci must be lineage- and developmental stage-specific. However, RAG recombinases are common to B- and T-cell progenitors, and other factors involved in V(D)J recombination are ubiquitous, for example those that mediate non-homologous end joining (NHEJ). Specificity comes from the regulation of locus transcription and the control of chromatin accessibility, perhaps supported by locus positioning in specific compartments of the nucleus [2, 3] and by factors that shape chromosomal domains in three-dimensional (3D) nuclear space [4, 5] (Figure 1).
Figure 1. Developmentally regulated positioning and compaction of the Ig heavy chain locus.
a) The position of loci within the nucleus often correlates with their expression: transcriptionally active regions of the genome are often located towards the centre of the nucleus [30]. Igh loci are positioned at the periphery of the nucleus in multi-lineage progenitors and remain there in T cell progenitors. Prior to their rearrangement in B cell progenitors, however, Igh loci are re-positioned towards the center of the nucleus (based on [3]).
b) A two-stage model of the developmental regulation of Igh locus conformation where the folding of individual domains (step 1) occurs prior to onset of rearrangement to be followed by the approximation of the domains (step 2) at the onset of rearrangement [4, 5].
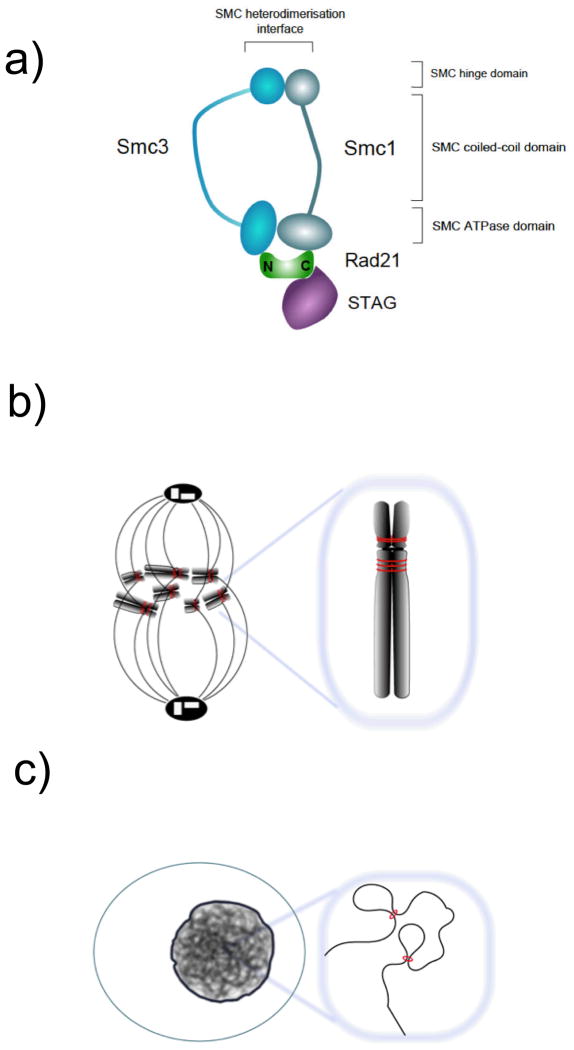
Recent work has drawn attention to the potential role in V(D)J recombination of cohesin, a protein complex that is best known for its essential role in post-replicative DSB repair and faithful chromosome segregation [6] (figure 2a, b) and has also been linked to the regulation of gene expression. Cohesin is recruited to specific binding sites on chromosome arms by components that mediate between tissue-specific transcription factors and the transcriptional machinery [7, 8, 9] and by the mammalian insulator protein CTCF [10, 11]. In turn, cohesin appears to modulate transcription at active genes, to contribute to CTCF's ability to 'insulate' promoters from other gene regulatory elements such as enhancers, and to form 'boundaries' between active and inactive chromosomal domains [10, 12]. In addition, cohesin forms long-range interactions between its binding sites [13, 14] and in this way may 'shape' chromosomal domains in nuclear space (Figure 2c).
Figure 2. Composition and function of the cohesin complex.
a) The composition of the cohesin complex.
b) The cohesin complex holds sister chromatids together from the time of DNA replication in S-phase until mitosis and forms long-range interactions between its binding sites in interphase. These binding sites are defined by the positioning of cohesin loading factors at active promoters and enhancers [7–9], by the mammalian insulator protein CTCF [10, 11], or both [8].
It is easy to imagine at least three possible ways in which cohesin might contribute to the regulation of lymphocyte receptor loci and the control of V(D)J recombination.
The first and perhaps most obvious possibility is that cohesin might position and shape the vast landscape of lymphocyte receptor loci into domains that are manageable for the recombination machinery, possibly allowing recombination signal sequences that are widely separated in the linear DNA sequence to undergo synapsis for recombination [4, 5, 15]. Second, cohesin might facilitate lineage- and developmental stage-specific transcription and accessibility of lymphocyte receptor loci to the recombination machinery [16, 17]. Third, cohesin might block premature or lineage-inappropriate transcription and accessibility of lymphocyte receptor loci to the recombination machinery by imposing boundaries and transcriptional insulation at CTCF binding sites [12, 18, 19].
Current evidence supports the second and third ideas: cohesin- and CTCF-dependent dependent transcription [16, 17] and insulation [18] have clearly been shown to influence V(D)J recombination. However, while CTCF and cohesin may shape the conformation of lymphocyte receptor loci [16, 17, 18, 20], convincing data that cohesin or CTCF facilitate V(D)J recombination by promoting synapsis between distant coding elements of the Ig or TCR loci are eagerly awaited.
To explore how lineage- and developmental-stage specificity of V(D)J recombination is achieved, we take a closer look at the contributions made by CTCF and by cohesin.
Shaping the landscape of lymphocyte receptor loci
Considering the enormous size of lymphocyte receptor loci, it has been argued that there must be mechanisms that shape these loci into a manageable 3D conformation prior to rearrangement [4]. Such mechanisms would ensure not only that DSB introduced by the Rag recombinase can be repaired properly and without unscheduled chromosomal rearrangements, but also provide equal opportunity for rearrangement of proximal and distal V genes. 3D-fluorescence in situ hybridisation (FISH) studies indeed suggested that lymphocyte receptor loci compact prior to rearrangement [21, 22]. Subsequent detailed studies defined the relative positioning of multiple points within the Igh locus by triangulation [4]. As a result of developmentally regulated changes in the conformation of the locus, the relative distances between gene segments (for example between proximal or distal V gene segments and D gene segments) in 3D space appeared to be independent of their separation along the linear sequence of DNA [4].
Genetic evidence indicates that the transcription factors YY1, Pax5 and Ikaros are required both for the compaction of the Igh locus and the rearrangement of distal V genes [21, 23, 24]. It remains unclear, however, how these factors regulate locus compaction, and whether the documented rearrangement defects result directly from changes in locus compaction, or indirectly from other defects in the absence of these factors.
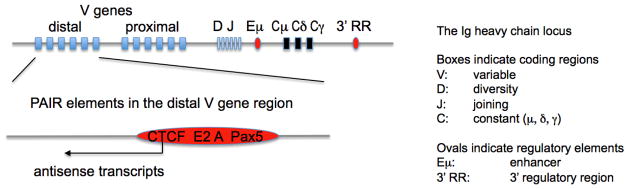
Lymphocyte antigen receptor loci contain numerous binding sites for CTCF and cohesin [16, 17, 20, 25, 26]. For example, the distal V gene cluster of the Igh locus contains a number of regulatory elements termed PAIR [25] that bind E2A, CTCF and cohesin throughout B cell development, recruit Pax-5 specifically in pro-B cells, and mediate developmentally regulated antisense transcription (figure 3). Their contribution to Igh locus conformation and rearrangement remains to be explored. In some instances, CTCF and cohesin binding is developmentally regulated [25, 26]. Consistent with their known ability to regulate long-range interactions (references from introduction), CTCF (and probably cohesin) contribute to shaping the conformation of the Ig loci [16, 18, 20] as explained below.
Figure 3. A new class of regulatory elements within the distal V gene regions of the Ig heavy chain locus.
PAIR elements bind E2A, CTCF and cohesin throughout B cell development, recruit Pax-5 specifically in pro-B cells, and mediate developmentally regulated antisense transcription [25].
Sen and colleagues [5] used FISH to show that the radial repositioning of the Igh locus towards the centre of the nucleus and its compaction depended on the Eμ enhancer and used a combination of chromosome conformation capture and ChIP to study locus conformation at high resolution. They provided evidence for two levels of chromosomal compaction. The first was described as folding into several domains, one of which required the Eμ enhancer. The Eμ-dependent domain was at the 3' end of the locus and extended from DFL16.1 to the 3' regulatory region, encompassing the DH and JH gene segments and constant region exons. Eμ-dependence of this domain is intrinsic to its definition, since Eμ is an interaction partner. Each of these domains appears to consist of several loops that, with the notable exception of Eμ itself, have CTCF binding sites positioned at their bases. Consistent with previous reports that the Igh locus fails to contract in YY1-deficient pro-B cells [23] the authors found YY1 at the base of loops within the 3' Eμ-dependent domain. In a second step, domains at 5' and 3' ends of the locus are spatially juxtaposed in an Eμ-dependent fashion, although the roles of CTCF, cohesin and YY1 in defining the conformation of the Igh locus conformation were not tested directly. However, studies in CTCF-deficient pro-B cells found that CTCF was not absolutely required for rearrangement of proximal and distal VH gene segments [16].
Genetic engineering experiments have demonstrated that physical proximity in the linear DNA sequence can facilitate V(D)J recombination [27]. However, there is as yet no direct evidence that the positioning or the conformation of antigen receptor loci directly affect V(D)J recombination by promoting synapsis between distant coding elements.
Regulating lymphocyte receptor locus transcription and accessibility
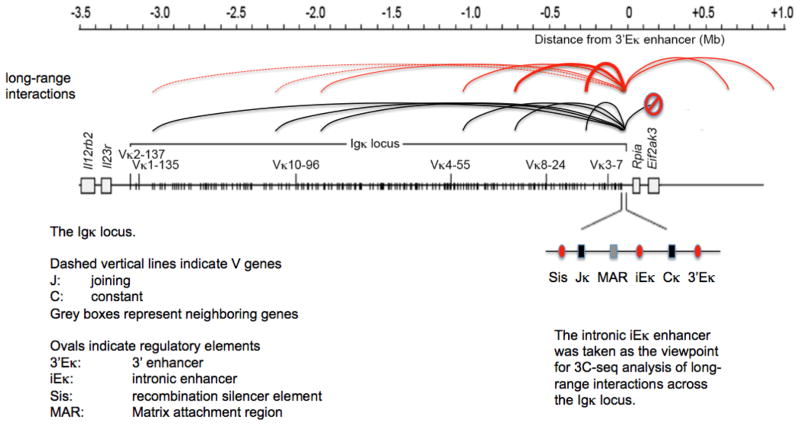
Experiments in which the gene encoding CTCF was deleted in developing B cells showed increased germline transcription of the proximal Vκ region and reduced germline transcription over the Jκ region in pre-B cells [16]. The rearrangement of Igk V gene segments was skewed in the absence of CTCF, such that proximal Vκ gene segments were over-used. Over 50% of all Vκ segments that were rearranged in CTCF-deficient pre-B cells were located in the most proximal 200 kb region, which was not used at all in control cells. This aberrant rearrangement was accompanied by preferential long-range interactions of Igk enhancers with proximal, but not distal Vκ gene segments [16] (Figure 4). Although the precise mechanisms that underlie this phenotype remain unclear, one explanation would be that CTCF controls Vκ rearrangement by shielding proximal Vκ segments from the effects of Eκ enhancers, which could be seen as a variation on the insulator theme discussed below. A similar rearrangement defect with increased proximal and reduced distal Vκ usage was reported in pre-B cells lacking a 3.7kb genomic region including the Sis regulatory element (Figure 4), which is located between Vκ and Jκ and among other factors is bound by CTCF [28].
Figure 4. Loss of CTCF affects long-range interactions and rearrangement at the Igκ light chain locus [16].
The intronic iEκ enhancer was taken as the viewpoint for 3C-seq analysis of long-range interactions across the Igκ locus. In control cells (black lines), the iEκ enhancer interacted more or less equally with multiple Vκ genes across the Igκ locus. Interactions of iEκ were restricted to the Igκ locus. In CTCF-deficient cells, by contrast, the iEκ enhancer interacted preferentially with enhancer-proximal Vκ genes, while enhancer-distal Vκ genes were neglected (the thickness of the lines indicates the relative strength of each interaction). Interactions of iEκ were no longer restricted to the Igk locus.
This pattern of long-range interactions was reflected in the pattern of transcription across the Igk locus and in the range of Vκ gene rearrangements, which were severely restricted in CTCF deficient cells.
The deletion of CTCF severely shortened the life-span of B cell progenitors. This has the potential to complicate the interpretation of altered long-range interactions, germ line transcription and rearrangement in these experiments.
The rearrangement of the Tcra locus in CD4+ CD8+ double positive (DP) thymocytes was investigated by conditional deletion of the cohesin subunit Rad21 using a Cd4Cre transgene [17]. In this experimental system cohesin levels diminished just as T cell precursors exited the cell cycle to undergo sequential Tcra rearrangements. As a result, Rad21-deficient thymocytes were present in normal numbers and had a normal life-span. By coordinating the loss of cohesin protein with the developmentally regulated cell cycle arrest in DP thymocytes, this study separated the essential cell cycle functions of cohesin (which are not required in resting cells) from cohesin effects on Tcra rearrangement [17]. Hence, the effects of cohesin depletion in this experimental setting can be interpreted with greater confidence.
Cohesin binding at Tcra marked the major enhancer and promoter for germ line transcription of the Jα elements, Eα and TEA (figure 5). Additional cohesin sites were found at many V gene promoters and at the boundary between Tcra and the neighbouring house keeping gene (figure 5). Jα germ line transcription was reduced in cohesin deficient thymocytes, a consequence of reduced long-range interactions between Eα and TEA. Reduced germ line transcription was associated with a decrease in H3K4me3, a post-translational histone modification that is of significance for V(D)J rearrangement since the recombinase component RAG2 directly binds to H3K4me3. In this manner, cohesin deletion caused reduced recruitment of RAG to Jα segments. Tcra rearrangement occurs in several sequential steps, beginning with the most proximal Jα and Vα segments. These so-called primary rearrangements are later replaced with secondary rearrangements to progressively more distal Jα and Vα elements. Because the loss of cohesin protein occurred after the time of primary rearrangements in this system [17], cohesin deletion mainly impaired secondary rearrangements involving distal Jα elements.
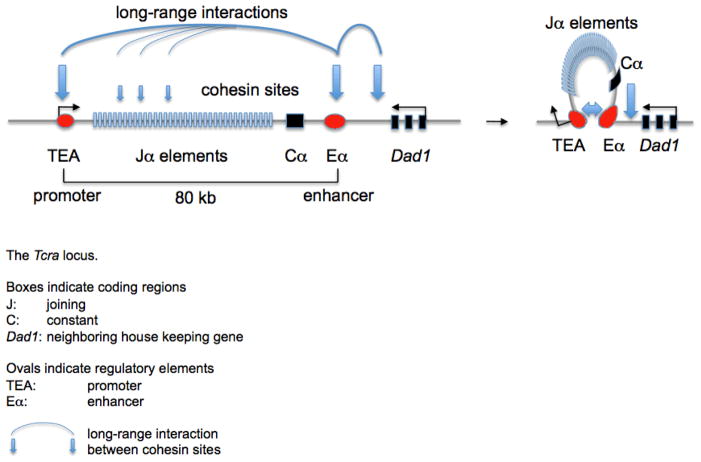
Figure 5.
Cohesin facilitates enhancer-promoter interactions, transcription, and rearrangement of the Tcra locus. The Eα enhancer and TEA promoter are 80 kb apart in the linear sequence of DNA (left). Cohesin-facilitated contacts between these regulatory elements (right) promote germline transcription, histone modification, recruitment of the recombination machinery and, ultimately, somatic recombination of Tcra [17].
Taken together, these data demonstrate that cohesin and CTCF control the rearrangement of lymphocyte antigen receptor genes by mechanisms that involve the regulation of transcription by long-range interactions.
Boundaries are important
In the mouse Igh locus, a region of approximately 100 kb separates the most proximal V gene segments (VH81X, also known as VH7183.a2.3) from the nearest gene segment (called DFL16.1). Within this ‘intergenic’ region lies a 4.1kb intergenic control region’, IGCR1, which contains two CTCF binding elements, CBE1 and CBE2 (Figure 6).
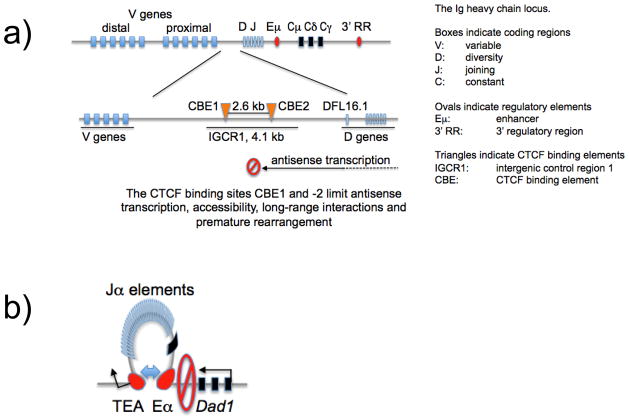
Figure 6. CTCF and cohesin demarcate functional boundaries in and around lymphocyte receptor gene loci.
a) CTCF binding sites limit antisense transcription, accessibility, long-range interactions and premature rearrangement of Igh [18].
b) CTCF and cohesin cooperate to form a boundary at Tcra. A CTCF- and cohesin-binding site with known insulator function is located between Tcra and the neighbouring house keeping gene Dad1. Cohesin deletion results in increased transcription of Dad1, suggesting that cohesin is required for the formation of a boundary that limits Eα enhancer activity to the Tcra locus [17].
Antisense transcription from the D towards the V gene region is important for the step-wise, developmentally regulated accessibility of the locus [29] and the CTCF binding sites in the IGCR1 region have been linked to attenuating these transcripts [19]. A recent study tested the role of the two closely spaced CTCF sites upstream of DFL16.1, first by deleting the IGCR1 region and second, and most convincingly, by mutating the CTCF binding elements CBE1 and CBE2 [18]. As a result, the rearrangement of the proximal VH7183 and VHQ52 families was increased, whereas the rearrangement frequency of the distal VHJ558 family was reduced. Remarkably, VH to DJH rearrangement was no longer ordered, subject to allelic exclusion, or restricted to the B cell lineage: VH81X V gene rearrangements could occur prior to DJ rearrangement, and even in developing T cells [18]. These defects were traced back to defective germ line transcription, accessibility and long-range interactions [18]. These findings provide strong evidence for the notion that the two deleted CTCF binding sites form boundaries that are required to regulate transcription and locus accessibility so that V to DJ rearrangement is restricted to the appropriate developmental stages and cell lineage.
Suggestions of reduced boundary functions were also apparent in the studies discussed earlier that deleted CTCF in developing B cells and cohesin in thymocytes. In addition to V(D)J recombination, cohesin depletion impaired the functional separation between Tcra and the neighbouring house keeping gene Dad1 [17]. Cohesin deletion increased Dad1 transcription at the expense of Cα, suggesting that cohesin was required for the functional integrity pf a previously characterised boundary at a CTCF binding site between the Tcra enhancer Eα and Dad1 (figure 6b). In CTCF-deficient pre-B cells, long-range interactions of the enhancer elements iEκ and 3'Eκ were no longer restricted to the Igk locus as in control cells, but reached beyond the boundaries of the Igk domain [16] (indicated by a ‘stop’ sign in figure 4). The increased transcription and recombination of proximal Vκ gene segments observed in CTCF-deficient pre-B cells [16] could be interpreted along similar lines: CTCF might ‘shield’ proximal Vκ segments from the effects of the nearby Eκ enhancers.
Concluding remarks
In order to generate a diverse repertoire, V(D)J recombination must occur over large genomic distances and DNA-FISH studies have demonstrated that rearrangement is preceded by the developmentally regulated compaction of lymphocyte receptor loci [21, 22]. Compaction may promote the juxtaposition of distal elements to facilitate V(D)J recombination and reduce the risk of illegitimate chromosomal rearrangements and the ability of cohesin and CTCF to form long-range interactions [13, 14] marks them as candidate factors that could bring distant gene elements together prior to RAG-mediated rearrangement. Indeed, numerous CTCF and cohesin binding sites have been mapped and appear to form long-range interactions within lymphocyte receptor loci [16–18, 20, 24]. CTCF is positioned at the base of several loops at the Igh locus [5] and shapes its conformation [18, 20]. The loading of cohesin at active promoters forms developmentally regulated cohesin binding sites [8] that can engage in cell type-specific long-range interactions to assist recombination [17]. Recent studies using high-resolution chromatin conformation capture (3C) support the idea that CTCF and cohesin control the rearrangement of Ig and TCR loci via long-range interactions [16–18]. In the examples described to date, they do this by regulating lineage- and developmental stage-specific transcription, and there is as yet no evidence that they act directly to promote synapsis between distal gene segments within Ig or TCR loci. The quest continues for factors that address the challenge how to bring lymphocyte receptor coding regions together over large genomic distances.
Acknowledgments
We apologize to those colleagues who’s work we could not discuss due to space restrictions.
Footnotes
The authors have no financial conflict of interest.
References
- 1.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 2.Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, Dillon N, Gray D, Perry RP, Merkenschlager M, Fisher AG. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 3.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 4.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011 Jan 11;21(1):12–24. doi: 10.1016/j.cub.2010.12.004. Epub 2010 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 11.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degner-Leisso SC, Feeney AJ. Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Semin Immunol. 2010 Dec;22(6):346–352. doi: 10.1016/j.smim.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro de Almeida C, Stadhouders R, de Bruijn MJ, Bergen IM, Thongjuea S, Lenhard B, van Ijcken W, Grosveld F, Galjart N, Soler E, Hendriks RW. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity. 2011;35:501–513. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira- Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Featherstone K, Wood AL, Bowen AJ, Corcoran AE. The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J Biol Chem. 2010;285:9327–9338. doi: 10.1074/jbc.M109.098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degner SC, Verma-Gaur J, Wong TP, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roldan E, Fuxa M, Chong W, et al. Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting Edge: Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondilis-Mangum HD, Shih HY, Mahowald G, Sleckman BP, Krangel MS. Regulation of TCRβ Allelic Exclusion by Gene Segment Proximity and Accessibility. J Immunol. 2011;187:6374–6381. doi: 10.4049/jimmunol.1102611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang Y, Zhou X, Hewitt SL, Skok JA, Garrard WT. A multifunctional element in the mouse Igκ locus that specifies repertoire and Ig loci subnuclear location. J Immunol. 2011;186:5356–5366. doi: 10.4049/jimmunol.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin Immunol. 2010;22:353–361. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]