Antibody molecules (Igs), built of heavy (H) and light (L) chains, are made by B-lymphocytes. L chains are composed of two μH chains of five Ig domains. The amino terminal domains of H and L chains are variable in amino acid sequence (VH and VL) particularly at the three hypervariable regions that form the antigen combining site. Assembled H-L chain pairs are stabilized by a disulfide bridge between the constant domain of the L chain and the first constant region domain of the H chain (CH1). In addition, noncovalent interactions, especially between the VH and VL domains, are involved in H-L chain associations (1). The genes encoding Ig H and L chains are assembled by stepwise somatic rearrangements of gene segments encoding different part of the V regions (2). During B lymphocyte development in bone marrow of adult mice and humans, DH segments first are rearranged to JH segments, followed by VH segments to DHJH segments on the H chain loci and, finally, VL-segment rearrangements to JL segments on the L chain loci. Between the segments, at the joints, nontemplated N regions can be inserted, which all encode parts of the third complementary-determining regions (CDR3) of H chains, and also of L chains in humans, but not in mice. A wide variety of CDR3 amino acid sequences can be generated, as long as the nucleotide sequences remain in-frame and no stop codons were generated. It follows that these rearrangement processes also generate out-of-frame, nonfunctional H and L chain gene loci.
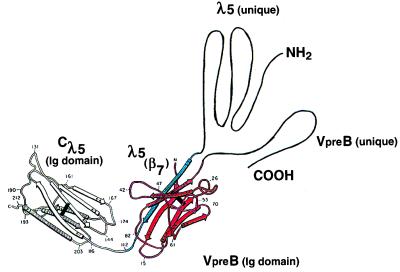
Different stages of pre-B cell development can be characterized by the rearrangement status of the Ig gene loci (3, 4). Even before VH segments are rearranged to DHJH segments, and well before VL segments are rearranged to JL segments, pre-B cells express a set of B lineage-specific genes called VpreB and λ5. They encode two proteins that can assemble with each other to form an L-chain-like structure, the surrogate L chain (5–8). Although the three-dimensional structures of the VpreB and λ5 proteins have not yet been determined, the nucleotide sequences of the genes encoding them have allowed predictions of their structures (Fig. 1). Thus, VpreB appears to have a V domain-like structure, but it lacks the last β-strand (β7) of a typical V domain. Instead it has a carboxyl terminal end that shows no sequence homologies to any other proteins. On the other side, λ5 appears to have a constant region—the domain with strong homologies to λL chains. Furthermore, it has, toward its amino terminal end, two functionally distinct regions. One, situated near Cλ5, has strong homology to J regions of the λL chains, i.e., to the β7-strand of Vλ domains. The other, i.e., the amino terminal region of the λ5 protein, is unique in that it shows only marginal sequence homologies to Ig domain structures (5). From these sequence homologies, it has been proposed that the β7 strand within the amino terminal portion of the λ5 protein could provide the structure that is missing in VpreB to form a complete, however noncovalently assembled, Ig domain and, thus, could mediate assembly of the two proteins (Fig. 1). As reported by Minegishi, Hendershot, and Conley in this issue of the Proceedings (9), deletion of the β7 strand in the amino terminal portion of the λ5 protein completely abrogates the formation of the surrogate L chain. By measuring proper or improper folding of the protein components of the surrogate L chains and their mutants, Minegishi et al. conclude that complementation of the incomplete VpreB domain by the extra β7-strand of λ5 is necessary and sufficient for folding and assembly of the surrogate L chain (10–12).
Figure 1.
Structure of associated VpreB and λ5 protein as proposed by Kudo et al. (6), the Ig-like portions of VpreB (β-strands 1–6 in red) and λ5 (Cλ5, β-strands 1–7, amino terminal pair β7-strand in blue) are assumed to fold as Ig domains. The figure is modified from that published in ref. 6.
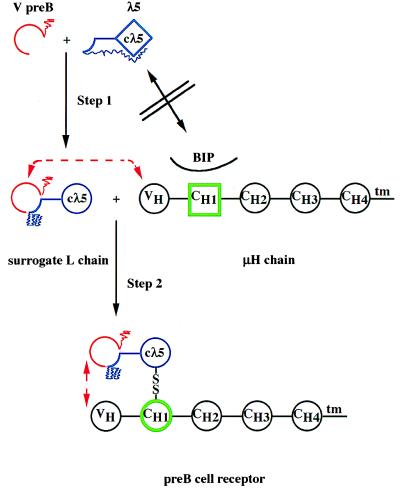
Minegishi et al. extend their studies to show, by deletional mutagenesis of the unique amino terminal portion of λ5 and the carboxyl terminal portion of VpreB, that these two parts of the proteins do not mediate assembly of the surrogate L chain, as the two deleted forms of the proteins still assemble. On the other hand, the unique amino terminal region of the λ5 protein appears to act as an intramolecular chaperone in preventing a proper folding of the Cλ5 domain, when it is made in the absence of its partner, VpreB (Fig. 2). Observations from our own laboratory support this finding, as a whole series of μH chains do not associate with λ5 protein expressed in the absence of VpreB protein in the same cell (T. Seidl and F.M., unpublished work). It is tempting to speculate that the putative β-strands of the unique portion of the λ5 protein may, in fact, disturb the proper folding of the Cλ5 domain by assembling into the Cλ5 fold.
Figure 2.
Stepwise formation of the pre-B cell receptor.
Isolated λ5 protein has an improperly folded structure in
which Cλ5 (◊, blue) has not yet attained an Ig-domain
structure (○) and is associated with the amino terminal
unique portion of λ5, which acts as an intramolecular
chaperone ( ). The
β7 strand of the amino terminal λ5 portion
is folded in an Ig-domain- like fashion
(
). The
β7 strand of the amino terminal λ5 portion
is folded in an Ig-domain- like fashion
( ). The VpreB
protein is folded as an Ig-like domain, missing the
β7-strand with a unique, non-Ig carboxyl terminal portion
(
). The VpreB
protein is folded as an Ig-like domain, missing the
β7-strand with a unique, non-Ig carboxyl terminal portion
( , red). The isolated
λ5-protein cannot displace BIP and associate with the
CH1-domain of the μH chain, because neither
λ5 (◊) nor CH1 (□, green) are
properly Ig-domain-folded. Association of VpreB with
λ5 induces an Ig-domain-structure in Cλ5
and displaces the intramolecular chaperone
(
, red). The isolated
λ5-protein cannot displace BIP and associate with the
CH1-domain of the μH chain, because neither
λ5 (◊) nor CH1 (□, green) are
properly Ig-domain-folded. Association of VpreB with
λ5 induces an Ig-domain-structure in Cλ5
and displaces the intramolecular chaperone
( ). VpreB
interacts with VH of the μH chain, Cλ5
displaces BIP and induces an Ig-domain structure in CH1
(○, green), forms a disulfide bridge and thus, the pre-B
cell receptor.
). VpreB
interacts with VH of the μH chain, Cλ5
displaces BIP and induces an Ig-domain structure in CH1
(○, green), forms a disulfide bridge and thus, the pre-B
cell receptor.
Once the surrogate L chain has been assembled properly it awaits the
production of a μH chain from a productively
VHDHJH-rearranged
H chain allele. Initially the μH chains bind the hsp70 chaperone BIP
via their CH1 domains (13, 14). This prevents the
CH1 domain from folding properly, and the μH
chain is retained in the endoplasmic reticulum until BIP is replaced by
a constant domain of a L chain (Hellman
 , unpublished work as quoted
by Minegishi et al. in ref. 9). It can be expected that the
properly folded Cλ5 domain in the assembled
surrogate L chain can do the same to the CH1
domain of the μH chain, whereas free λ5
protein, improperly folded in the absence of
VpreB, cannot do so (Fig. 2).
, unpublished work as quoted
by Minegishi et al. in ref. 9). It can be expected that the
properly folded Cλ5 domain in the assembled
surrogate L chain can do the same to the CH1
domain of the μH chain, whereas free λ5
protein, improperly folded in the absence of
VpreB, cannot do so (Fig. 2).
However, not all μH chains, which initially are formed in pre-B cells can assemble with surrogate L chains (15, 16). In fact, to our surprise, nearly half of all newly generated μH chains were found not to be able to pair. The inability of a μH chain to pair may have more than one structural reason. Some CDR3 regions may either be too bulky or repulsive for proper interactions with surrogate L chains. Certain VH segments carried in the H chain locus, such as the VH81x and VHQ52 segments, may have germ-line-encoded structural features within the V domains that do not allow proper pairing with surrogate L chains. At present, we have no good explanation why such nonfitting V-region segments would be inherited and used in the generation of μH chains.
Those μH chains that fit to surrogate L chains can form a pre-B cell receptor and display it on the cell surface. Once this has been achieved several events take place in that pre-B cell. The lymphocyte-specific components of rearrangement machinery, i.e., the genes encoding RAG-1, RAG-2, and TdT are turned off (17). That is part of the mechanism by which allelic exclusion is secured, ensuring that one of the two H chain alleles in a B lineage cell expresses a protein, i.e., one pre-B cell expresses only one type of pre-B cell receptor. Furthermore, the expression of the genes encoding the surrogate L chain genes is turned off, and remains turned off, at least, until a mature B cell has been made (5). That limits the number of surrogate L-chain molecules available in a pre-B cell for forming a pre-B cell receptor. If pairing or nonpairing of surrogate L chain with μH chains is not an all or nothing phenomenon, hence, if different avidities exist between individual μH chains and the surrogate L chain, then different concentrations of associated pre-B cell receptors may exist in and on individual pre-B cells.
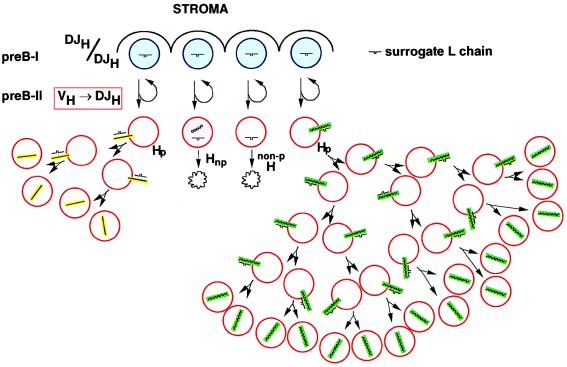
The formation of pre-B cell receptors on the surface of the cells also induces them to enter cell cycle and divide several times. As the clones of pre-B cells expand they dilute the surrogate L chain molecules initially present in the first cell of the clone. If the number of surrogate L chain molecules has been reduced so much that a critical number of pre-B cell receptors is no longer available in and on a given pre-B cell proliferation ceases. The higher the avidity of interaction between a μH chain and the surrogate L chain, the longer pre-B cells expressing them will expand by proliferation (Fig. 3).
Figure 3.
Formation of the pre-B cell receptor and
its effect on B cell development. DJH-rearranged pre-B I
cells (5) in contact with bone marrow stromal cells undergo asymmetric
divisions that yield one cell, which retains contact with stroma and,
thus, pre-B I differentiation status. The other cell loses contact to
stroma, is induced to differentiate to a pre-B II cell in rearranging a
VH to DHJH segment on the H chain
loci. Nonproductive (np) rearrangements leads to μH chain-negative
cells that die ( ). Productive
rearrangements can lead to μH chain that cannot pair with surrogate L
chain (15). Cells with nonpairing μH chains cannot form a pre-B cell
receptor and die. On the other hand, pairing μH chain leads to pre-B
cell receptor expression on the surface, which induces pre-B II cell
proliferation. The higher the avidity, the longer is the proliferative
phase (high avidity = green, low avidity = yellow). In the
simplest form of this model, the surrogate L chain assumes a ligand
function for the μH chain to induce proliferation, hence,
an external ligand binding to VpreB and/or VH
must not necessarily be present to induce clonal expansion of pre-B
cell receptor-expressing pre-B II cells.
). Productive
rearrangements can lead to μH chain that cannot pair with surrogate L
chain (15). Cells with nonpairing μH chains cannot form a pre-B cell
receptor and die. On the other hand, pairing μH chain leads to pre-B
cell receptor expression on the surface, which induces pre-B II cell
proliferation. The higher the avidity, the longer is the proliferative
phase (high avidity = green, low avidity = yellow). In the
simplest form of this model, the surrogate L chain assumes a ligand
function for the μH chain to induce proliferation, hence,
an external ligand binding to VpreB and/or VH
must not necessarily be present to induce clonal expansion of pre-B
cell receptor-expressing pre-B II cells.
Once the pre-B cell exits the cell cycle the rearrangement machinery is turned on again. However, now the κ and λL chain gene loci have become accessible for VL to JL rearrangements, while the H chain loci, which have not yet been VHDHJH-rearranged, must have previously been closed to avoid VH to DHJH rearrangements on the second allele, i.e., to maintain allelic exclusion. Once L chains are expressed from individual, productively VLJL-rearranged L chain loci in clones of μH chain-expressing pre-B cells, they are tested for their capacity to pair with a given μH chain in a given cell. The properly folded CL domain of κL or λL chains now displaces BIP on μH chain, induces proper folding of the CH1 domain of the μH chain and finally forms a disulfide-bonded H-L chain complex, part of the Ig molecule that then is displayed as B cell receptor for antigen on the surface. In λ5-defective mutant mice the fitness of the VH domains of the μH chain expressed in pre-B cells cannot be screened until L chains are expressed. VH repertoire changes in μH chains using VH81x and VHQ52 segments occur in λ5-mutant mice at the transition of a precursor to an immature surface Ig-expressing B cell, as they occur in wild-type mice at the transition from pre-B I to a pre-B II cell (Fig. 3). It remains to be seen whether surrogate L chain as stereotype for an L chain can muster all VH domains for fitness to all VL domains.
Acknowledgments
I thank Drs. K. Karjalainen and A.R. Rolink for critical reading of the commentary. The Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche Ltd., Basel, Switzerland.
Footnotes
A commentary on this article begins on page 3041.
References
- 1.Branden C I, Lindqvist Y, Schneider G. Acta Crystallogr B. 1991;47:824–835. doi: 10.1107/s0108768191007127. [DOI] [PubMed] [Google Scholar]
- 2.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 4.Melchers F, Rolink A. In: B-Lymphocyte Development and Biology. Paul W E, editor. Philadelphia: Lippincott; 1999. [Google Scholar]
- 5.Melchers F, Karasuyama H, Haasner D, Bauer S, Kudo A, Sakaguchi N, Jameson B, Rolink A. Immunol Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]
- 6.Kudo A, Bauer S R, Melchers F. Prog Immunol. 1989;7:339–347. [Google Scholar]
- 7.Pillai S, Baltimore D. Nature (London) 1987;329:172–174. doi: 10.1038/329172a0. [DOI] [PubMed] [Google Scholar]
- 8.Kerr W G, Cooper M D, Feng L, Burrows P D, Hendershot L M. Int Immunol. 1989;1:355–361. doi: 10.1093/intimm/1.4.355. [DOI] [PubMed] [Google Scholar]
- 9.Minegishi Y, Hendershot L M, Conley M E. Proc Natl Acad Sci USA. 1999;96:3041–3046. doi: 10.1073/pnas.96.6.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guelpa-Fonlupt V, Bossy D, Alzari P, Fumoux F, Fougereau M, Schiff C. Mol Immunol. 1994;31:1099–1108. doi: 10.1016/0161-5890(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 11.Bornemann K D, Brewer J W, Perez E, Doerre S, Sita R, Corley R B. J Immunol. 1997;158:2551–2557. [PubMed] [Google Scholar]
- 12.Minegishi Y, Coustan-Smith E, Wang Y H, Cooper M D, Campana D, Conley M E. J Exp Med. 1998;187:71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberini C M, Bet P, Milstein C, Sitia R. Nature (London) 1990;347:485–487. doi: 10.1038/347485a0. [DOI] [PubMed] [Google Scholar]
- 14.Sitia R, Neuberger M, Alberini C, Bet P, Fra A, Valetti C, Williams G, Milstein C. Cell. 1990;60:781–790. doi: 10.1016/0092-8674(90)90092-s. [DOI] [PubMed] [Google Scholar]
- 15.ten Boekel E, Melchers F, Rolink A G. Immunity. 1997;7:357–368. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- 16.Kline G H, Hartwell L, Beck-Engeser G B, Keyna U, Zaharevitz S, Klinman N R, Jack H M. J Immunol. 1998;161:1608–1618. [PubMed] [Google Scholar]
- 17.Grawunder U, Leu T M, Schatz D G, Werner A, Rolink A G, Melchers F, Winkler T H. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]