Abstract
Recent studies demonstrated that cancer stem cells (CSCs) have higher tumorigenesis properties than those of differentiated cancer cells and that transcriptional factor-SOX2 plays a vital role in maintaining the unique properties of CSCs; however, the function and underlying mechanism of SOX2 in carcinogenesis of lung cancer are still elusive. This study applied immunohistochemistry to analyze the expression of SOX2 in human lung tissues of normal individuals as well as patients with adenocarcinoma, squamous cell carcinoma, and large cell and small cell carcinoma and demonstrated specific overexpression of SOX2 in all types of lung cancer tissues. This finding supports the notion that SOX2 contributes to the tumorigenesis of lung cancer cells and can be used as a diagnostic probe. In addition, obviously higher expression of oncogenes c-MYC, WNT1, WNT2, and NOTCH1 was detected in side population (SP) cells than in non-side population (NSP) cells of human lung adenocarcinoma cell line-A549, revealing a possible mechanism for the tenacious tumorigenic potential of CSCs. To further elucidate the function of SOX2 in tumorigenesis of cancer cells, A549 cells were established with expression of luciferase and doxycycline-inducible shRNA targeting SOX2. We found silencing of SOX2 gene reduces the tumorigenic property of A549 cells with attenuated expression of c-MYC, WNT1, WNT2, and NOTCH1 in xenografted NOD/SCID mice. By using the RNA-Seq method, an additional 246 target cancer genes of SOX2 were revealed. These results present evidence that SOX2 may regulate the expression of oncogenes in CSCs to promote the development of human lung cancer.
Introduction
Cancer stem cells (CSCs) represent a very small population of cancer cells from which tumors originates. They possess the same unique character as embryonic stem (ES) cells, such as clonogenicity, pluripotency and self-renewal and thus have the ability to initiate a tumor, sustain its growth and be responsible for cancer recurrence [1]. Recent studies have shown that CSCs like cell subpopulations could be isolated from various cultured tumor cell lines or tissues by using the Hoechst33342 dye efflux method to separate side population (SP) cells [2] or by sorting cells expressing specific stem cell surface markers, such as CD133(+), CD44(+), CD34(+) and CD38(+) [3]–[5] et al.
Lung cancer represents the most common cause of cancer-related lethality in both men and women throughout the world with very low five-year survival rates, even after clinical therapy [6], [7]. This malignancy is usually divided into different histological types according to the phenotypes of cells from which the tumor arises, including squamous cell carcinoma (SCC), adenocarcinoma and neuroendocrine carcinoma, such as small cell lung cancer (SCLC) as well as large cell lung cancer [8]. Adenocarcinoma, SCC and large cell lung cancer are also collectively named non-small cell lung cancer (NSCLC), representing the most common types of lung cancer with lower growth rate and spread speed than those of SCLC. Among NSCLC, peripheral adenocarcinoma is the leading subtype which accounts for approximately 80% of cases in lung cancer patients [9]. Several studies showed that CD133 (+), CD44 (+) and CD87 (+) can be used as surface markers to identify CSCs in lung cancer [10]–[12]. Recent studies reported isolated SP from both a mouse tumor model [13] and a variety of lung cancer cell lines by using the Hoechst dye efflux method [14]–[16]. It was found that isolated SP cells show higher expression levels of stem cell genes, such as SOX2 and OCT4 and tumorigenesis properties than NSP cells [2].
The important function of transcription factor SOX2 in maintaining the unique properties of ES cells and CSCs has been extensively investigated. It was also established that induced pluripotent stem (iPS) or pluripotent cancer (iPC) cells could be generated by co-transfection of Sox2 cDNA with other transcription factors such as Oct4, Klf4 and c-Myc into fibroblast or cancer cells [17]–[20]. In fact, SOX2 was highly expressed in isolated CSCs like cells at both mRNA and protein levels. Extensive studies revealed that SOX2 regulates the complex transcriptional network to maintain the unique characteristics of stem cells [21] and the anti-apoptosis property of CSCs [15], [22]. Consequently, targeting of SOX2 is a promising strategy for tumor therapy. Although numerous investigations of clinically-derived tumor tissues reported the specific overexpression of SOX2 in certain types of tumor tissues, such as prostate and breast cancers [22], [23] and indicated its importance for tumorigenesis, the underlying mechanism for the tumorigenic property of SOX2 gene is still largely unknown.
Oncogenes play important roles in the development of carcinoma. Among them, NOTCH, WNT and c-MYC are well-established oncogenes in the initiation and progression of lung cancer cells. It was reported that WNT family proteins-WNT1, WNT2 and NOTCH proteins -NOTCH1, NOTCH3 as well as their downstream protein HES-1 are overexpressed in NSCLC cell lines or tissues [24]–[31]. Overexpression of these oncogenes or activation of their signal pathways induced lung carcinoma [32], [33]. As such, targeting of these genes by using siRNA/shRNA, mutation, specific inhibitors or monoclonal antibodies could inhibit tumor growth and induce apoptosis in lung cancer cell lines in experimental mouse models [4], [34]–[36]. Aside from their important role in tumorigenesis of lung cancer, these oncogenes also formed a functional interaction network. It was also reported that NOTCH1 induces the expression of c-MYC, in addition, both proteins regulate the expression of same target genes participating in cell growth regulation [37]. c-MYC was also revealed to be the downstream target of WNT/β-Catenin signaling and further studies showed the promoter of c-MYC to align with WNT/β-Catenin responsive enhancers [38], revealing a possible regulatory mechanism of WNT signaling on c-MYC.
In view of the important contributions of SOX2 in maintaining the stemness property of CSCs, we hypothesize that SOX2 might govern the transcriptional network of oncogenes to affect the tumorigenesis of lung carcinoma. Our results thus far demonstrate that SOX2 regulates the transcriptional network of oncogenes, including WNT1, WNT2, NOTCH1 and c-MYC and promotes the tumorigenesis of human lung cancer cells. This study also supports the notions that targeting of SOX2 is an effective strategy for lung cancer therapy.
Materials and Methods
Ethics Statement
All animal experiments were performed strictly under the guidelines on laboratory animals of Nankai University and were approved by the Institute Research Ethics Committee at the Nankai University (Permit number: 10011). Mice were anesthetized with a mixture of oxygen/isoflurane before each experiment and all efforts were made to minimize their suffering. For human samples, the commercialized high-density tissue microarrays were purchased and the use of the human tissue in this study was approved by the Human Research Committee of Nankai University.
Vector Construction
shRNA sequence for silencing human SOX2 gene was searched and blasted using RNAi designer from the invitrogen website (https://rnaidesigner.invitrogen.com/rnaiexpress/index.jsp). One shRNA targeting human SOX2 was designed and chemically synthesized as shRNA-SOX2 (AAAAGGGACATGAT CAGCATGTATTGGATCCAATACATGCTGATCATGTCCC), and a scrambled sequence (AAAAGCTACACTATCGAGCAATTTTGGATCCAAAATTGCTCGATAGTGTAGC) was used as control for knockdown analysis. The palindromic DNA oligos were annealed to each other to form a double-strand oligo and ligated to the linearized pLV-H1TetO-GFP-Bsd (Cat# SORT-C03, Biosettia Inc., San Diego, CA) and pLV-H1-EF1α-puro (Cat# SORT-B19, Biosettia Inc., San Diego, CA) vector to generate circled pLV-H1TetO-shRNA-SOX2-GFP-Bsd and pLV-H1-EF1α-shRNA-SOX2-puro plasmid separately. The puromycin resistant cDNA from pLV-H1-EF1α-shRNA-SOX2-puro plasmid was cut by Nhe Ι and Sal Ι enzyme to replace the GFP-Bsd fragment in pLV-H1TetO-shRNA-SOX2-GFP-Bsd to generate the pLV-H1TetO-shRNA-SOX2-puro plasmid.
Cell Culture
Wild types (Wt) of A549 and H460 cells were obtained from ATCC. A549-Wt cells were infected with lentivirus encoding the firefly luciferase (FL) gene (Cat. # GlowCell-14b-1, Biosettia, SanDiego, CA) and selected by 10 µg/ml Blasticidin (Bsd) to generate A549 cells with stable overexpression of FL (A549-FL). A549-Wt, H460-Wt and A549-FL cells were infected with lentivirus carrying pLV-H1TetO-shRNA-SOX2-puro plasmid (Biosettia, SanDiego, CA), followed by clonal selection using puromycin (10 µg/ml for A549 and 2 µg/ml for H460) to generate a polyclone of A549, H460 or A549-FL cells with stable expression of Dox inducible shRNA-SOX2 (A549-H1tetO-shRNA-SOX2, H460-H1tetO-shRNA-SOX2 or A549-FL/H1tetO-shRNA-SOX2). A549 and H460 cells were maintained in F12K and RPMI 1640 media supplemented with 10% fetal bovine serum in the presence of 100 u/ml penicillin and 0.1 mg/ml streptomycin separately. For control purposes, A549-Wt, H460-Wt and A549-FL were infected with lentivirus carrying scrambled shRNA and subjected to identical clone selection procedures to generate the stable control cell lines A549-H1tetO-shRNA-Con, H460-H1tetO-shRNA-Con and A549-FL/H1tetO-shRNA-Con.
Immunohistochemistry and Tissue Microarrays
The expression of SOX2 in high-density tissue microarrays (Cat. # BC04119b, LC727, LC2085b, LC2161, Alenabio, Xi’an, China PR) was detected by the standard biotin-avidin-complex method with monoclonal mouse antibody against SOX2 (Cat. # ab75485, Abcam Inc, Cambridge, UK) at a 1∶200 dilution. The images were recorded by Olympus BX51 Epi-fluorescent microscopy under a 10× or 40× objective (Olympus Co. Tokyo, Japan).
Flow Cytometry Analysis and Sorting
A549-Wt cells were harvested and resuspended in pre-warmed DMEM+ buffer (DMEM with 2% FBS and 10 mM HEPES buffer) at a density of 1.0×106 cells/ml. Hoechst 33342 dye was added at a final concentration of 10 µg/ml in the presence or absence of 10 µM Fumitremorgin C (FTC). Cell samples were placed in a 37°C water bath for 60 minutes (min) and mixed every 10 min. Cells were collected and resuspended in cold HBSS+ buffer (Hanks’ Balanced Salt Solution with 2% FBS and 10 mM HEPES buffer). At the end of the staining period, cells were resuspended in cold HBSS+ buffer containing 2 µg/ml propidium iodide (PI) for dead cell discrimination. The Hoechst dye was excited with a UV laser at 355 nm, and its fluorescence measured with a 460/50 BP filter (Hoechst Blue) and a 670/30 BP filter (Hoechst Red).
RT-PCR and Real-time RT-PCR
Total mRNAs from A549 cells were isolated by TRIzol reagent (Cat. #15596-018, Invitrogen Inc, Carlsbad, CA) and reverse-transcribed into cDNAs with MMLV reverse transcriptase (Promega, Madison, MI). Following this, semi-quantitive RT-PCR was performed to detect the mRNA expression levels of NOTCH1, WNT1, WNT2, c-MYC, SOX2, OCT4, NANOG, ABCB1 and ABCG2. For an equal loading control, mRNA of human β-actin was tested at the same time. Primers used for both experiments are summarized in Table 1 . Real time RT-PCR was performed on Opticon (Bio-Rad, Hercules, CA) in 25 µl reaction volumes by using TransStart Green qPCR SuperMix Kit (TransGen Biotech, Beijing, China, PR). The 2−ΔΔCt method was used to determine the relative mRNA folding changes. Statistical results were averaged from three independent experiments performed in triplicate.
Table 1. The primers used for RT-PCR and real-time RT-PCR.
| WNT1 | Forward primer 5′- CCCGGTTATTCGCCCACCCG -3′ |
| Backward primer 5′- CAAGGGGTCTCCCGCGGAGA -3′ | |
| WNT2 | Forward primer 5′- ACAGCAGGCCGTGTGTGCAA -3′ |
| Backward primer 5′- AGGCAGTCCTGACAGCGCAC -3′ | |
| NOTCH1 | Forward primer 5′- CGTCCGTGCCCCTCAACCAC -3′ |
| Backward primer 5′- CAGGACGGTGCTGGTGCCAG -3′ | |
| c-MYC | Forward primer 5′- CGCCCTCCTACGTTGCGGTC -3′ |
| Backward primer 5′- CGTCGTCCGGGTCGCAGATG -3′ | |
| SOX2 | Forward primer 5′- AAAACAGCCCGGACCGCGTC -3′ |
| Backward primer 5′- CTCGTCGATGAACGGCCGCT -3′ | |
| NANOG | Forward primer 5′- ACCTCAGCCTCCAGCAGATGCA -3′ |
| Backward primer 5′- GGTGCTGAGGCCTTCTGCGT -3′ | |
| OCT4 | Forward primer 5′- AAGCGATCAAGCAGCC -3′ |
| Backward primer 5′- GGAAAGGGACCGAGGAGTA -3′ | |
| ABCB1 | Forward primer 5′- TTGAAGGGGACCGCAATGGAGGA -3′ |
| Backward primer 5′- GTCCAGCCCCATGGATGATGGC -3′ | |
| ABCG2 | Forward primer 5′- CACCAATGGCTTCCCCGCGAC -3′ |
| Backward primer 5′- GGGTCCCAGGATGGCGTTGAGA -3′ | |
| β-actin | Forward primer 5′- GGCATCCACGAAACTACCTT -3′ |
| Backward primer 5′- CTCGTCATACTCCTGCTTGC -3′ |
Western Blotting
Cell lysates from A549 and H460 cell lines were prepared with RIPA buffer in the presence of protease inhibitor cocktails as described previously [22]. Protein (20 µg) was loaded onto 5–12% Tris-Acrylamide gels and blotted with antibodies that included: polyclonal anti-SOX2, WNT2 (Cat.# sc-20088, sc-50361, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), WNT1 (Cat.# ab85060, Abcam Inc, Cambridge, UK), monoclonal anti-NOTCH1 (Cat.# 3608, Cell Signal Technology Inc, Danvers, MA), c-MYC, β-actin (Cat.# sc-40, sc-47778, Santa Cruz Biotechnology, Inc. Santa Cruz, CA), and horseradish peroxidase-conjugated secondary antibodies. Blotting results were detected by an ECL chemiluminescence kit (Millipore, Billerica, MA).
Tumor Xenograft
Male NOD/SCID mice at 6–8 weeks age were separated randomly into two groups (n = 5 for each group). 1×106 A549-FL/H1tetO-shRNA-Con or A549-FL/H1tetO-shRNA-SOX2 cells were xenografted into each mouse through tail vein injection. All mice were fed with 0.2 mg/ml Dox plus 0.05% sucrose in the drinking water from the first day after inoculation.
Bioluminescent Image
Mice were anesthetized with a mixture of oxygen/isoflurane and received luciferin (Cat. # 119222, Caliper Life Sciences, Hopkinton, MA) i.p at 150 µg/g body weight. Bioluminescent signals were measured 10 min later with an IVIS 100 imaging system and quantified with Living Image Software (Xenogen, Alameda, CA).
RNA-Seq
The A549-H1tetO-shRNA-SOX2 and its control cells were collected after being incubated with Dox for 4 days. 4 µg total RNA from each sample was extracted by TRIzol reagent. Oligo(dT) magnetic beads adsorption method was used to purify mRNA, their transcriptome data were profiled and compared following standard protocols (digital gene expression, DGE, 3 million reads, Beijing Genomics Institute at Shenzhen, China). The cancer genes were selected from 7 databases summarized in the website: http://microb230.med.upenn.edu/protocols/cancergenes.html and those genes with false discovery rate (FDR) <0.001 were selected out as the target genes of SOX2. RNA-Seq data have been deposited to NCBI Gene Expression Omnibus (GEO) database (GSE36597; reviewer access link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=druvhickewcwcvy&acc=GSE36597).
Statistical Analysis
Values were expressed as Means ± S.E.M. Significance was determined by χ2 test in Table 2 and hypergeometric test in Table 3 , others were determined by Student’s t-test. A value of p<0.05 was used as the criterion for statistical significance. * indicates significant difference with p<0.05, ** indicates significant difference with p<0.01.
Table 2. The correlation of SOX2 with clinical status of patient with lung cancer.
| SOX2 (+) cell counts | ≤10% | 11–30% | 31–49% | ≥50% | p Value |
| Tumorigenesis | <0.001 | ||||
| Normal/Paracarcinoma | 31 | 6 | 1 | 0 | |
| Tumor | 9 | 42 | 82 | 284 | |
| Adenocarcinoma | 4 | 25 | 44 | 127 | |
| SCC | 0 | 15 | 31 | 104 | |
| Large cell carcinoma | 3 | 1 | 5 | 22 | |
| SCLC | 2 | 1 | 2 | 31 | |
| Gender | 0.322 | ||||
| Male | 32 | 35 | 66 | 201 | |
| Female | 8 | 13 | 17 | 83 | |
| Age at diagnosis | 0.024 | ||||
| <65 | 9 | 33 | 50 | 207 | |
| ≥65 | 0 | 9 | 32 | 77 | |
| Histological phenotype | 0.011 | ||||
| NSCLC | 7 | 41 | 80 | 253 | |
| SCLC | 2 | 1 | 2 | 31 | |
| TNM stage | |||||
| NSCLC | 0.705 | ||||
| Ι-ΙΙ | 7 | 34 | 60 | 182 | |
| ΙΙΙ-ΙV | 0 | 5 | 10 | 33 | |
| SCLC | 0.327 | ||||
| Ι-ΙΙ | 1 | 0 | 1 | 20 | |
| ΙΙΙ-ΙV | 0 | 1 | 1 | 7 | |
| Histological grade | |||||
| Adenocarcinoma | 0.002 | ||||
| Grade 1 | 2 | 11 | 11 | 15 | |
| Grade 2 | 1 | 6 | 16 | 54 | |
| Grade 3 | 1 | 4 | 13 | 50 | |
| Localization of SOX2 | <0.001 | ||||
| Cytoplasma | 40 | 19 | 26 | 53 | |
| Nuclear + Cytoplasma | 0 | 29 | 57 | 231 |
Table 3. The most enriched 20 signaling pathways of SOX2 target genes in KEGG database.
| # | Pathway | Count of DEG with pathway annotation | Pvalue |
| 1 | Ribosome | 27 | 8.18E-09 |
| 2 | Aminoacyl-tRNA biosynthesis | 14 | 8.31E-06 |
| 3 | Protein processing in endoplasmic reticulum | 24 | 0.00067961 |
| 4 | Renal cell carcinoma | 15 | 0.00114849 |
| 5 | Metabolic pathways | 129 | 0.00132165 |
| 6 | Bladder cancer | 11 | 0.0027594 |
| 7 | Insulin signaling pathway | 23 | 0.0031735 |
| 8 | Vibrio cholerae infection | 12 | 0.00394879 |
| 9 | Gap junction | 16 | 0.00487479 |
| 10 | Pathways in cancer | 45 | 0.00773737 |
| 11 | Lysosome | 20 | 0.00824269 |
| 12 | Glycosaminoglycan biosynthesis - keratan sulfate | 5 | 0.00859688 |
| 13 | Protein export | 6 | 0.00925981 |
| 14 | Vasopressin-regulated water reabsorption | 9 | 0.01188259 |
| 15 | Arginine and proline metabolism | 10 | 0.01337271 |
| 16 | DNA replication | 8 | 0.01393426 |
| 17 | Shigellosis | 12 | 0.01882112 |
| 18 | Phosphatidylinositol signaling system | 13 | 0.01897797 |
| 19 | Glycerolipid metabolism | 8 | 0.0191644 |
| 20 | Alanine, aspartate and glutamate metabolism | 7 | 0.01921653 |
DEG: differentially expressed genes.
Results
SOX2 is Specifically Expressed in Human Lung Cancer Tissues
To determine whether SOX2 contributes to tumorigenesis of lung cancer, an immunohistochemical method was used to detect expression of SOX2 in human lung tissues of normal/paracarcinoma (n = 38), adenocarcinoma (n = 200), SCC (n = 150), SCLC (n = 36) and large cell lung cancer (n = 31). The correlation of SOX2 with clinical status of patients with lung cancer was summarized in Table 2 . Predominantly higher expression of SOX2 was found in both NSCLC and SCLC than in normal/paracarcinoma tissues (p<0.001), indicating the important role of SOX2 in the tumorigenesis of lung cancer. Statistically results significant showed the number of SOX2 positive cells to be positively correlated with age at diagnosis (p = 0.024). Moreover, SCLC tissues revealed a higher expression level of SOX2 than NSCLC tissues (p = 0.011). There was a positive correlation between SOX2 and the pathological degree of human adenocarcinoma (p = 0.002), indicating that SOX2 may inhibit the differentiation of adenocarcinoma cells. From results of immunohistochemical staining ( Fig. 1 ), we found either cytosol or both nuclei and cytosol localization of SOX2 in cancer cells, which was consistent with our previous findings in human prostate cancer cells [22]. In addition, most of the tumor tissues with high expression level of SOX2 showed both nuclear and cytosol localization of SOX2; in contrast, those tissue with lower expression level of SOX2 always revealed only cytosol localization. The correlation between SOX2 expression level and its localization was significant (p<0.001); however, there was no statistically significant relationship between SOX2 positive cancer cell counts and TNM stages or gender.
Figure 1. Immunohistochemical staining of SOX2 (brown) in human lung tissue of normal individuals, and patients with squamous lung carcinoma, adenocarcinoma lung cancer, small cell lung cancer and large cell lung cancer in paraffin-embedded human lung cancer tissue microarray.
The representative staining of SOX2 in these tissues was subjected separately to microscopy images under 10× (A) and 40× objective (B).
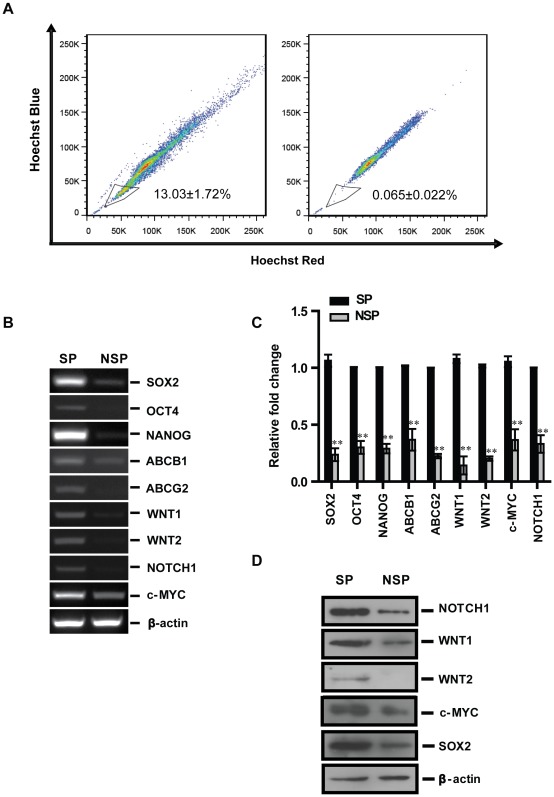
Oncogenes WNT1, WNT2, NOTCH1 and c-MYC are Overexpressed at Both mRNA and Protein Levels in SP Cells of the A549 Cell Line
Following identification of specific overexpression of SOX2 in human lung tumor tissues, the expression of SOX2 and well-established oncogenes were compared between SP and none side population (NSP) in A549 cells. SP cells are CSCs-like cells that show higher tumorigenesis and chemo-resistant properties than those of NSP cells [39], [40]. Since they express high expression levels of ATP-binding cassette (ABC) transporter family members, which help them to extrude the Hoechst 33342 dye and some drugs more effectively, SP cells could be isolated by the Hoechst 33342 efflux method using FACS. As shown in Fig. 2A , approximately, 13% of cells could be isolated as SP by the Hoechst dye efflux method. Stemness of isolated SP cells was proven by their specific overexpression of SOX2, OCT4, NANOG, ABCB1 and ABCG2 genes. Significantly higher expression levels of WNT1, WNT2, NOTCH1 and c-MYC were detected in SP cells than those observed in NSP cells at both mRNA ( Fig. 2B , Fig. 2C ) and protein levels ( Fig. 2D ), suggesting the molecular mechanism underlying the high tumorigenic property of CSCs.
Figure 2. Oncogenes WNT1, WNT2, c-MYC and NOTCH1 are specifically overexpressed in SP of human lung adenocarcinoma cell line A549.
A. SP cells in A549 were detected and separated by FACS using the Hoechst33342 dye efflux method (left) and verified by their failure to efflux this dye after incubation with FTC, a specific inhibitor of multidrug transporter-ABCG2 (right). This figure represents 1 of 3 experiments. The mRNA expression levels of oncogenes and stem cell genes were compared between the SP and NSP cells by using semi-quantitive RT-PCR, real-time RT-PCR in B and C separately, n = 3. D. Western blotting results of WNT1, WNT2, NOTCH1, c-MYC and SOX2 proteins in SP and NSP of A549 cells.
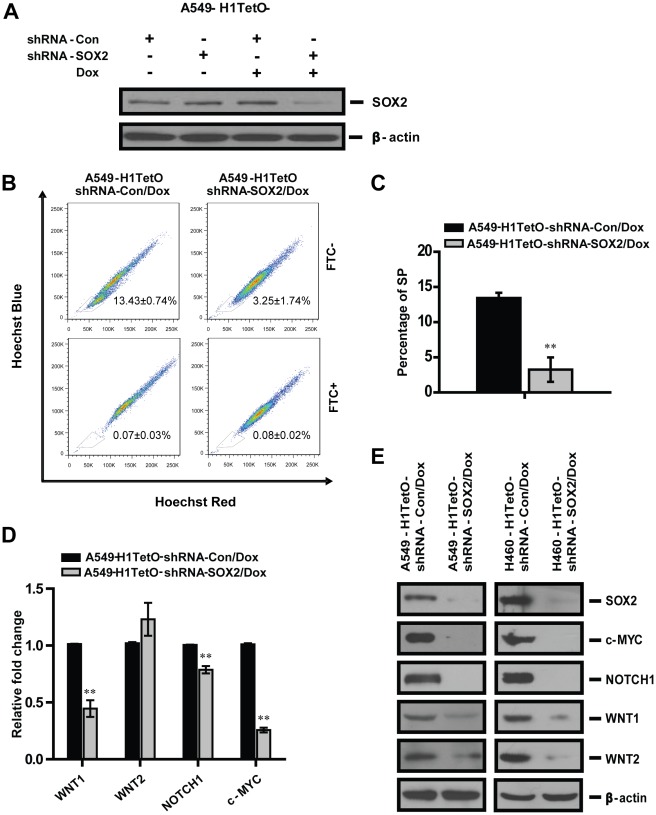
SOX2 Regulates the Expression of Oncogenes in A549 and H460 Cells
Since SOX2 is an important transcriptional factor that maintains the function of CSCs, we hypothesized that SOX2 might regulate the transcription of certain oncogenes in tumor cells. To test this contention, A549 cells were established with stable expression of doxycycline (Dox) inducible shRNA targeting SOX2 (A549-H1tetO-shRNA-SOX2). The inducible knockdown of SOX2 was confirmed by Western blotting after 4 days’ incubation of A549 cells with Dox, and 70% silencing efficiency of shRNA was demonstrated in the expression of SOX2 gene ( Fig. 3A ). At the same time, significant reduced percentage of SP in A549 cells with SOX2 gene silencing was revealed ( Fig. 3B and 3C ), demonstrating the important function of SOX2 in maintaining the CSC properties of A549 cells. In addition, this finding also supported the contribution of SOX2 to the tumorigenesis capacity of lung cancer cells since it was well established that the SP has higher tumorigenesis property than NSP in various types of lung cancer cells [16]. By using real time RT-PCR and Western blotting, significant reduced expressions of WNT1, NOTCH1 and c-MYC at both mRNA and protein levels were shown in A549 cells with SOX2 down-regulation ( Fig. 3D and 3E ). An interesting phenomenon observed was that although mRNA expression of WNT2 was slightly elevated upon silencing of SOX2, its protein expression level decreased, indicating a translational inhibition of WNT2 in A549 cells with SOX2 silencing. The same regulatory effects of SOX2 on the expression of WNT1, WNT2, NOTCH1 and c-MYC were also observed in the human large cell lung carcinoma cell line H460 with SOX2 silencing ( Fig. 3E ).This finding indicates a universal regulatory mechanism of SOX2 on the oncogene network of both human lung cell lines. These findings support the notion that SOX2 may regulate the transcription of key oncogenes in lung cancer to promote tumorigenesis.
Figure 3. SOX2 regulates the expression of oncogenes in both A549 and H460 cells.
A. A549-H1tetO-shRNA-SOX2 and control cells were incubated with Dox for 4 days and silencing of SOX2 gene was confirmed by Western blotting. B. FACS analysis of the SP in A549 cells with or without SOX2 silencing. The SP cells in A549 were confirmed by their ability to efflux Hoechst 33342 dye in the absence of FTC (up panel, FTC-), but fail to efflux the dye after incubation with FTC (low panel, FTC+). C. The percentage of SP in A549 cells from each experiment group was averaged from three independent experiments and plotted. D. Real-time RT-PCR was used to compare mRNA expression levels of oncogenes in A549 cell line with or without down-regulation of SOX2, n = 3. E. Protein expression levels of WNT1, WNT2, NOTCH1 and c-MYC genes were detected in A549 and H460 cells with SOX2 silencing by Western blotting.
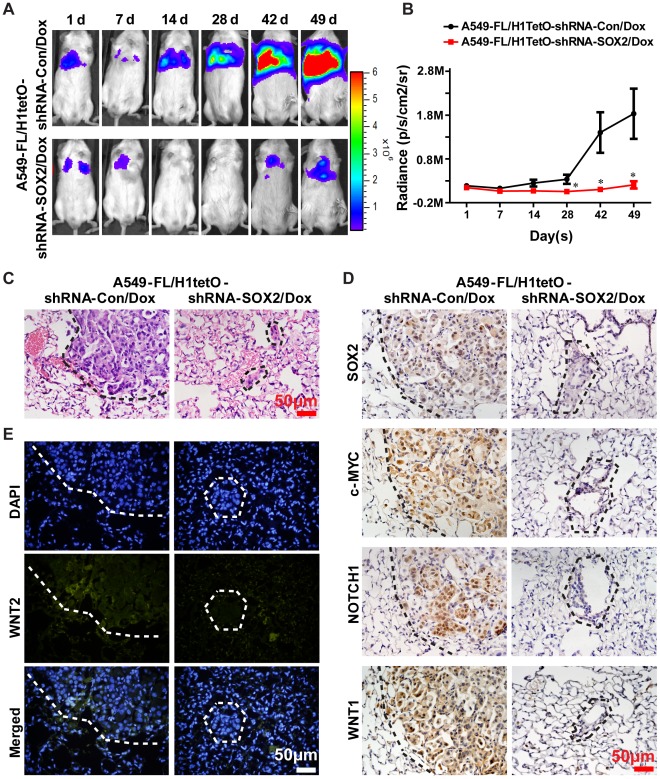
Silencing of SOX2 Attenuates Tumorigenesis of Human Lung Cancer Cells in Xenografted NOD/SCID Mice
Following the identification of SOX2’s function on tumorigenesis and its regulatory effect on expression of oncogenes in human lung cancer cells, we next explored its function in vivo. For this purpose, A549-FL/H1tetO-shRNA-SOX2 and control cells were inoculated into the NOD/SCID mice by tail vein injection. The presence of A549-FL cells was monitored by noninvasive bioluminescence imaging. The mice xenografted with A549-FL/H1tetO-shRNA-SOX2 and A549-FL/H1tetO-shRNA-Con cells were fed with Dox through drinking water from the first day after inoculation to induce expression of shRNA. As observed in Fig. 4A and 4B , same amount of bioluminescence signals could be detected in mice 1 day after injection, indicating that the same number of lung tumor cells became trapped in lung capillaries in both experimental groups. Subsequent decrease in bioluminescence signals was detected in the 7 following days due to a failing in survival of xenografted lung cancer cells [41]. Progressively increasing signals could be observed in mice xenografted with A549-FL/H1tetO-shRNA-Con cells after 14 days of treatment, indicating that these cells had succeeded in homing and proliferating. Twenty-one days after tumor cell challenge, 5 of 5 A549-FL/H1tetO-shRNA-Con xenografted mice developed tumor, but only 2 of 5 A549-FL/H1tetO-shRNA-SOX2 xenografted mice developed tumor 42 days after initial tumor cell challenge. Xenografted lung tumors in NOD/SCID mice were confirmed by hematoxylin and eosin (HE) stainings as shown in Fig. 4C . The protein expression levels of SOX2 and oncogenes in murine lung tissues were assayed by immunohistochemistry or immunofluorescence methods. We found a reduced tumorigenesis potential of A549 cells in vivo as well as a significantly attenuated expression of WNT1, WNT2, NOTCH1 and c-MYC after silencing of the SOX2 gene ( Fig. 4D and Fig. 4E ). Taken together, these results supported the tumorigenic property of SOX2 and its regulatory effect on oncogenes in vivo.
Figure 4. Silencing of SOX2 inhibits tumorigenesis and regulates expression of oncogenes in vivo.
A. A549 cells were injected via the tail vein into NOD/SCID mice and the bioluminescence images of xenografted tumor were taken at the times indicated. B. Bioluminescence intensity was measured and plotted, n = 5. C. HE staining was used to detect xenografted tumors in lung tissues of xenografted NOD/SCID mice. D. Immunohistochemistry staining of SOX2, c-MYC, NOTCH1 and WNT1 (all shown in brown color) from lung tissues of xenografted NOD/SCID mice in situ. E. Immunofluorescence of WNT2 (green) in xenografted murine lung tissues. All microscopy images were recorded under a 40× objective. The figures in C, D and E represent 1 of 5 experiments. The tumor regions in C, D and E were all circulated by dash lines.
SOX2 Regulates the Transcriptional Network of Oncogenes
Having identified the function of SOX2 in the tumorigenesis property of lung cancer cells and its target oncogenes, we then tested all the other targeted genes by RNA-Seq and found 246 cancer genes. Among them, 74 genes (30%) were up-regulated ( Fig. 5A ) and 172 genes (70%) were down-regulated ( Fig. 5B ) in A549 cells with SOX2 silencing. We noticed the transcription levels of NOTCH3 and WNT 7B are significantly reduced, revealing the comprehensive effects of SOX2 on the transcription of WNT and NOTCH gene families. In addition, the mRNA expression levels of well-established oncogenes such as KLF4, EGFR, BCL10, JUN, YAP1 and JAK1 et al. were significantly reduced when compared with control cells, but the transcriptions of EGF, RHOC et al. were greatly increased, demonstrating the complicated molecular mechanism in the tumorigenesis process of SOX2. In addition, our RNA-Seq result revealed 840 differentially expressed genes (DEG) with pathway annotation after SOX2 silencing in A549 cells. Through enrichment analysis of SOX2 target genes in the cell signaling pathway database of Kyoto Encyclopedia of Genes and Genomes (KEGG), we found the pathway in cancer is one of the most significantly enriched ones ( Table 3 and Fig. S1). This result further supports the important function of SOX2 in development of tumor.
Figure 5. Cluster image of the other target cancer genes of SOX2.
Standardized TPM (transcripts per million clean tags) values were applied to compare the other target cancer genes expression levels between A549 cells with SOX2 silencing (shRNA-SOX2) and their control (shRNA-Con). The transcription of each gene is represented by a square with a color that codes for the values of Lg TPM. Specifically, bright green represents low expression, bright red represents strong expression. The target genes whose transcriptions are up-regulated with the silencing of SOX2 were presented in A and those down-regulated genes were presented in B.
Discussion
In this study, we used immunohistochemistry to systemically analyze the expression of SOX2 in various types of lung cancers and found that SOX2 is predominantly overexpressed in adenocarcinoma, SCC, large cell carcinomas and SCLC tissues, indicating that SOX2 can be used as a universal marker for the diagnosis of human lung cancer. Here we also present evidence indicating that SOX2 plays an important role in carcinogenesis of lung cancer.
Since the concept of CSC has been established, many studies supported this concept by demonstrating that CSCs or CSCs like cells are highly tumorigenic [16]; however, few studies have been performed to investigate the molecular mechanism of this phenomenon. Here we demonstrated a higher expression of SOX2 and oncogenes- NOTCH1, WNT1, WNT2 as well as c-MYC in SP cells than those in NSP cells, thus supporting a possible contribution of these oncogenes to the stemness property of CSCs.
Recent studies have revealed the complicated interaction between SOX2 and WNT signaling pathway. For example, it was reported that SOX2 antagonizes the WNT signaling to inhibit the differentiation of adult stem cells (ASCs) [42] and osteoblast lineage [43] and enhance the tumorigenesis and self renewal property of osteosarcomas [44] by promoting the transcription of negative regulator of WNT signaling, such as DKK1, APC and GSK3β. SOX2 was also reported to synergistically interacted with β-catenin, the downstream molecule of the WNT signal pathway, to regulate the transcription of a target gene to promote cell proliferation and tumorigenesis of human breast cancer [23]. However, as an important stem cell gene, the underlying molecular mechanism of SOX2’s function in tumorigenesis is comprehensive and still needs to be intensively investigated. In this regard, our results demonstrated that silencing of SOX2 significantly reduces the protein expression level of oncogenes-WNT1, WNT2, c-MYC and NOTCH1 in human lung cancer and further revealed other target cancer genes of SOX2. It is thus possible that SOX2 can cooperate with these important oncogenes to promote tumor occurrence. Recent study showed down-regulation of SOX2 gene inhibits proliferation and induces apoptosis in tumor cells [15], [45], [46], we observed the same phenomenon in human lung cancer cell lines (data not shown), those may also be important factors that finally lead to the suppressed tumor growth in A549 cells; however, this study focused on the transcriptional changes of oncogenes and did not address other mechanisms.
Bioluminescence imaging is a novel diagnostic system that provides an easy and noninvasive visualization of the size and location of tumor cells in xenografted animals. Two types of models are usually applied to explore tumorigenicity of lung cancer cells: one is the intravenous metastasis model and the other is the subcutaneous flank tumor model [47], [48]. The first model was used in our study to observe SOX2’s function in the tumorigenesis of lung cancer in vivo, which reflected the whole process of homing and proliferation of tumor cells in lung. It was interesting to find that after 42 days of tumor cell inoculation, lung tumors could be detected in 2 of the 5 A549-FL/H1tetO-shRNA-SOX2 xenografted mice. From the immunohistochemistry stain of tumor tissues, the possibility that the tumor formed from “escaper” cells that not responsive to shRNA-SOX2 could be excluded, since the stain intensity of SOX2 was obviously decreased in the shRNA-SOX2 group. It is thus possible that the transcription of other oncogenes may not be affected or could even be unregulated by SOX2 and this may be the key reason leading to tumor occurrence in NOD/SCID mice. This possibility was verified by RNA-Seq screening of the targeted cancer genes of SOX2 by showing the up-regulated transcription of oncogenes after SOX2 gene silencing. Discovery of these genes’ function in the occurrence of tumor after SOX2 down-regulation and use of combined therapies targeting them and the SOX2 gene may improve the outcome of lung tumor therapy.
In summary, this study broadens our understanding of the molecular mechanism of tumorigenesis of both CSCs and SOX2. Our study supports the notion that silencing of the SOX2 gene is an effective strategy for human lung cancer therapy.
Supporting Information
Pathway in cancer, one of the most significantly enriched pathways of SOX2 targets from KEGG cell signaling pathway database. The frame showed the SOX2 target that was down-regulated (green) or up-regulated (red) upon SOX2 silencing.
(DOC)
Acknowledgments
We thank Dr. Azucena Gomez-Cabrero (Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA, USA) for helpful discussion and comments and Junhong Wang (FACS Facility, Institute of Hematology, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin, China) for helpful instructions in cell sorting.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by National Science Foundation of China grant 31000616 (to NL), China Junior Faculty Funds grant 20090031120044 (to NL) China Fundamental Research Funds for the Central Universities grant 65011241 (to NL), National Science Foundation of China grant 30830096 (to RX), Major State Basic Research Development Program of China (China 973 Program) 2007CB914804 (to RX), Support Project of Tianjin Scientific & Technological Commission for 973 Program 08QTPTJC28700 (to RX), and Key Project of Tianjin Scientific & Technological Commission for China-Sweden Cooperation Research Program 09ZCZDSF04000 (to RX). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Garcia-Barrado MJ, Sancho C, Iglesias-Osma MC, Moratinos J. Effects of verapamil and elgodipine on isoprenaline-induced metabolic responses in rabbits. Eur J Pharmacol. 2001;415:105–115. doi: 10.1016/s0014-2999(01)00808-1. [DOI] [PubMed] [Google Scholar]
- 2.Kruger JA, Kaplan CD, Luo Y, Zhou H, Markowitz D, et al. Characterization of stem cell-like cancer cells in immune-competent mice. Blood. 2006;108:3906–3912. doi: 10.1182/blood-2006-05-024687. [DOI] [PubMed] [Google Scholar]
- 3.Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- 4.You L, He B, Xu Z, Uematsu K, Mazieres J, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23:6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
- 5.Du L, Wang H, He L, Zhang J, Ni B, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 6.Kato H, Ichinose Y, Ohta M, Hata E, Tsubota N, et al. A randomized trial of adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. N Engl J Med. 2004;350:1713–1721. doi: 10.1056/NEJMoa032792. [DOI] [PubMed] [Google Scholar]
- 7.Winton T, Livingston R, Johnson D, Rigas J, Johnston M, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 8.Pine SR, Marshall B, Varticovski L. Lung cancer stem cells. Dis Markers. 2008;24:257–266. doi: 10.1155/2008/396281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jemal A, Siegel R, Ward E, Murray T, Xu J, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 10.Le QT, Chen E, Salim A, Cao H, Kong CS, et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin Cancer Res. 2006;12:1507–1514. doi: 10.1158/1078-0432.CCR-05-2049. [DOI] [PubMed] [Google Scholar]
- 11.Gutova M, Najbauer J, Gevorgyan A, Metz MZ, Weng Y, et al. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS One. 2007;2:e243. doi: 10.1371/journal.pone.0000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5:e14062. doi: 10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagi S, Kishimoto H, Kawahara K, Sasaki T, Sasaki M, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Invest. 2007;117:2929–2940. doi: 10.1172/JCI31854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 15.Xiang R, Liao D, Cheng T, Zhou H, Shi Q, et al. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br J Cancer. 2011;104:1931. doi: 10.1038/bjc.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatsugawa M, Takahashi A, Hirohashi Y, Torigoe T, Inoda S, et al. SOX2 is overexpressed in stem-like cells of human lung adenocarcinoma and augments the tumorigenicity. Lab Invest. 2011;91:1796–1804. doi: 10.1038/labinvest.2011.140. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 19.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A. 2010;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 22.Jia X, Li X, Xu Y, Zhang S, Mou W, et al. SOX2 promotes tumorigenesis and increases the anti-apoptotic property of human prostate cancer cell. J Mol Cell Biol. 2011;3:230–238. doi: 10.1093/jmcb/mjr002. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Shi L, Zhang L, Li R, Liang J, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–17978. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Thiagalingam A, Chopra H, Borges MW, Feder JN, et al. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci U S A. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins BJ, Kleeberger W, Ball DW. Notch in lung development and lung cancer. Semin Cancer Biol. 2004;14:357–364. doi: 10.1016/j.semcancer.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 27.Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, et al. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 2010;116:5676–5685. doi: 10.1002/cncr.25551. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Sun PL, Li JZ, Jheon S, Lee CT, et al. Aberrant Wnt1/beta-Catenin Expression is an Independent Poor Prognostic Marker of Non-small Cell Lung Cancer After Surgery. J Thorac Oncol. 2011;6:716–724. doi: 10.1097/JTO.0b013e31820c5189. [DOI] [PubMed] [Google Scholar]
- 30.Iwakawa R, Kohno T, Kato M, Shiraishi K, Tsuta K, et al. MYC amplification as a prognostic marker of early-stage lung adenocarcinoma identified by whole genome copy number analysis. Clin Cancer Res. 2011;17:1481–1489. doi: 10.1158/1078-0432.CCR-10-2484. [DOI] [PubMed] [Google Scholar]
- 31.Allen TD, Zhu CQ, Jones KD, Yanagawa N, Tsao MS, et al. Interaction between MYC and MCL1 in the genesis and outcome of non-small-cell lung cancer. Cancer Res. 2011;71:2212–2221. doi: 10.1158/0008-5472.CAN-10-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacheco-Pinedo EC, Durham AC, Stewart KM, Goss AM, Lu MM, et al. Wnt/beta-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J Clin Invest. 2011;121:1935–1945. doi: 10.1172/JCI44871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasai K, Sukezane T, Yanagita E, Nakagawa H, Hotta A, et al. Oncogene-mediated human lung epithelial cell transformation produces adenocarcinoma phenotypes in vivo. Cancer Res. 2011;71:2541–2549. doi: 10.1158/0008-5472.CAN-10-2221. [DOI] [PubMed] [Google Scholar]
- 34.Lin L, Mernaugh R, Yi F, Blum D, Carbone DP, et al. Targeting specific regions of the Notch3 ligand-binding domain induces apoptosis and inhibits tumor growth in lung cancer. Cancer Res. 2010;70:632–638. doi: 10.1158/0008-5472.CAN-09-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu D, Kadota K, Ueno M, Nakashima N, Yokomise H, et al. Eur J Cancer; 2011. Adenoviral vector expressing short hairpin RNA targeting Wnt2B has an effective antitumour activity against Wnt2B2-overexpressing tumours. doi: 10.1016/j.ejca.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Sato N, Yamabuki T, Takano A, Koinuma J, Aragaki M, et al. Wnt inhibitor Dickkopf-1 as a target for passive cancer immunotherapy. Cancer Res. 2010;70:5326–5336. doi: 10.1158/0008-5472.CAN-09-3879. [DOI] [PubMed] [Google Scholar]
- 37.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yochum GS. Multiple Wnt/ss-catenin responsive enhancers align with the MYC promoter through long-range chromatin loops. PLoS One. 2011;6:e18966. doi: 10.1371/journal.pone.0018966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moserle L, Ghisi M, Amadori A, Indraccolo S. Side population and cancer stem cells: therapeutic implications. Cancer Lett. 2010;288:1–9. doi: 10.1016/j.canlet.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Guo LP, Chen LZ, Zeng YX, Lu SH. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;67:3716–3724. doi: 10.1158/0008-5472.CAN-06-4343. [DOI] [PubMed] [Google Scholar]
- 41.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SB, Seo KW, So AY, Seo MS, Yu KR, et al. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2012;19:534–545. doi: 10.1038/cdd.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo E, Basu-Roy U, Zavadil J, Basilico C, Mansukhani A. Distinct functions of Sox2 control self-renewal and differentiation in the osteoblast lineage. Mol Cell Biol. 2011;31:4593–4608. doi: 10.1128/MCB.05798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu-Roy U, Seo E, Ramanathapuram L, Rapp TB, Perry JA, et al. Oncogene; 2011. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. doi: 10.1038/onc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hussenet T, Dali S, Exinger J, Monga B, Jost B, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto S, Tanaka F, Sato K, Kimura S, Maekawa T, et al. Monitoring with a non-invasive bioluminescent in vivo imaging system of pleural metastasis of lung carcinoma. Lung Cancer. 2009;66:75–79. doi: 10.1016/j.lungcan.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Jenkins DE, Oei Y, Hornig YS, Yu SF, Dusich J, et al. Bioluminescent imaging (BLI) to improve and refine traditional murine models of tumor growth and metastasis. Clin Exp Metastasis. 2003;20:733–744. doi: 10.1023/b:clin.0000006815.49932.98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pathway in cancer, one of the most significantly enriched pathways of SOX2 targets from KEGG cell signaling pathway database. The frame showed the SOX2 target that was down-regulated (green) or up-regulated (red) upon SOX2 silencing.
(DOC)