Abstract
To elucidate the genes involved in the formation of white and black plumage in ducks, RNA from white and black feather bulbs of an F2 population were analyzed using RNA-Seq. A total of 2,642 expressed sequence tags showed significant differential expression between white and black feather bulbs. Among these tags, 186 matched 133 annotated genes that grouped into 94 pathways. A number of genes controlling melanogenesis showed differential expression between the two types of feather bulbs. This differential expression was confirmed by qPCR analysis and demonstrated that Tyr (Tyrosinase) and Tyrp1 (Tyrosinase-related protein-1) were expressed not in W-W (white feather bulb from white dorsal plumage) and W-WB (white feather bulb from white-black dorsal plumage) but in B-B (black feather bulb from black dorsal plumage) and B-WB (black feather bulb from white-black dorsal plumage) feather bulbs. Tyrp2 (Tyrosinase-related protein-2) gene did not show expression in the four types of feather bulbs but expressed in retina. C-kit (The tyrosine kinase receptor) expressed in all of the samples but the relative mRNA expression in B-B or B-WB was approximately 10 fold higher than that in W-W or W-WB. Additionally, only one of the two Mitf isoforms was associated with plumage color determination. Downregulation of c-Kit and Mitf in feather bulbs may be the cause of white plumage in the duck.
Introduction
Identification of genes controlling plumage color and their associated inheritance patterns are important topics in poultry science research. Plumage color control is essential for the uniform appearance of birds in the poultry industry. White plumage is the most favorable color for producers of meat-type commercial birds not only because ducks with unpigmented feathers are easy to clean (comparing with unpigmented feather body, pigmented feather bulbs or follicles left in skin showing black dots and make carcass look dirty ), but also genes involved in melanogenesis may have pleiotropic effects on other phenotypes [1]. It has been reported that multiple genes exist at different loci controlling plumage color in ducks [2]. These loci include white neck MR, extended black E, blue dilution G, dominant white belly S, head cheek decorated R, white skin and mouth Y, and recessive white c. Compared to studies of plumage color in chicken and quail, few gene or pathway identification studies have been conducted in ducks. A 6-bp deletion that inactivated Tyrosinase (Tyr) in a line of albino chickens was reported [3]. Chang et al. [4] found that the causal mutation for the recessive white allele in chickens is the insertion of a complete avian retroviral sequence in intron 4 of the tyrosinase gene. The Mitf (Microphthalmia-associated transcription factor) gene encodes a transcription factor of Tyr family genes with important roles in pigmentation. MITF seems to be primarily associated with loss of pigmentation and patterning, i.e., white spotting in both dogs and cattle [5]–[7] as opposed to hyperpigmentation, which in the Silky was recently shown that the higher expression of Mitf is a downstream effect of increased EDN3 expression [8]. Higher expression of Mitf, which is associated with hyperpigmentation, was observed in Silky Fowl [9]. A stop codon caused by a 2-bp deletion in exon 11 of Mitf was found to be responsible for the “silver” plumage color in Japanese quail [10]. Mitf expression can be regulated by Scf-Kit signaling and can itself activate the transcription of the Tyr genes [11], [12]. c-Kit is required during the feather growth cycle for melanocyte activation in humans [13]. Mutations in c-Kit can cause coat color change in mammals [14]–[16]. Allele-specific genetic interactions between Mitf and c-Kit were also reported to affect melanocyte development in humans [17]. The expression pattern of c-Kit was investigated during embryonic development in chicken and quail [18], [19]. Mutations in other genes were also found to be associated with plumage color in these systems. Gunnarsson et al. [20] reported an 8.3-kb deletion upstream of Sox10 that caused dark-brown plumage in chickens. Other genes, including Mc1r, Asip, and Pmel17, also contribute to plumage color [21]–[23]. Few recent studies have focused on the genetic mechanisms involved in duck plumage color formation. High-throughput genomic approaches are promising ways to identify genes and pathways involved in plumage color formation. Due to the unavailability of an assembled reference sequences, high-throughput expression tools have not been widely used in ducks, although one study used chicken microarrays for genome-wide expression analysis to identify genes related to sperm storage [24].
The white Liancheng is an egg-type duck and white Kaiya is a meat-type duck in South China. In our previous study, 80% of individuals in an F1 population from a Kaiya × Liancheng cross had a phenotype of grey plumage on their heads, wings, backs or tails, with a white belt running from neck to chest. The F2 population was segregated, individuals with white, black, and black-white plumage were found. We reported a new autosomal locus (designated T) that may control plumage color in ducks [25]. However, the identity and number of genes involved in plumage color control in these ducks is not clear.
This study is the first genome-wide expression analysis to use RNA-Seq to find differentially expressed genes related to black and white plumage color in ducks. A large number of genes was found to be differentially expressed between white and black feather bulbs. Our analysis found that important genes and pathways associated with pigment formation are differentially regulated between black and white feather bulbs. We further characterized the expression of a few key genes related to pigmentation.
Results
Overview of RNA-Seq Data
To maximize the coverage of duck feather bulb mRNA by RNA sequencing, libraries were constructed by pooling RNA isolated from 6 white feather bulbs (3 from white dorsal plumage and the other 3 from white-black dorsal plumage) as sample W-1 library, and 6 black feather bulbs (3 from black dorsal plumage and the other 3 from white-black dorsal plumage) as sample B-1 library. RNA-Seq yielded 5,000,000 raw reads from each library. Low-quality reads (i.e., tags containing only adaptors and ambiguously called bases (reads that has many Ns)) were removed, resulting in 4,887,399 and 4,867,376 clean tags, 217,133 and 235,874 of which were distinct (i.e., non-identical), from white and black feather bulbs, respectively. These distinct clean tags were mapped to 9,009 and 8,498 genes in Ensemble Gallus gallus databases, and 1,458 and 1,584 genes in an Anas platyrhynchos EST library for W-1 and B-1 libraries, respectively. In total, 10,467 and 10,082 distinct clean tags were mapped to genes, accounting for 4.43% and 4.64% of the total distinct clean tags in the white and black feather bulb RNA libraries, respectively. A summary of sequencing tags and matched genes is shown in Table 1.
Table 1. RNA-Seq data summary and annotation results.
| RNA-Seq sample | Black feather bulb (B-1) | White feather bulb (W-1) | ||||
| Total tags (raw data) | 5,000,000 | 5,000,000 | ||||
| Clean tags | 4,887,399 | 4,867,376 | ||||
| Total distinct clean tags | 217,133 | 235,874 | ||||
| Mapping to gene | Reference | Chicken | Duck | Chicken | Duck | |
| DCT | 8,498 | 1,584 | 9,009 | 1,458 | ||
| total | 10,082 | 10,467 | ||||
| % of TDCT | 4.64% | 4.43% | ||||
| Total unknown DCT | 207,051 | 225,407 | ||||
| % unknown of TDCT | 95.36% | 235,874 | ||||
Note: DCT- distinct clean tag; TDCT- Total distinct clean tag; Chicken- Gallus gallus; Duck- Anas platyrhynchos.
Tag reads analysis showed that more than 83% of the tags were present in 1 to 5 reads, while less than 2% of the tags were present in more than 100 reads. Tags with different numbers of reads between black and white feather bulb libraries matched 5,240 annotated genes. Details of these genes and their related sequence counts in the W-1 and B-1 libraries are listed in Table S1.
Differentially Expressed Genes in White and Black Feather Bulbs
In this study, a rigorous algorithm was used to identify differentially expressed genes in the two samples based on “The significance of digital gene expression profiles” [26]. A total of 2,642 tags were found to be differentially expressed (DETs). Among these tags, only 186 mapped to annotated genes, yielding 133 differentially expressed genes (log2Ratio≥1, P<0.01, FDR<0.001) (see Tables S2 and S3) [26], [27]. Compared to black feather bulbs, white feather bulbs showed 82 downregulated and 51 upregulated genes according to statistical criteria for raw reads and TMP (number of transcripts per million clean tags).
Gene Ontology Analysis of Differentially Expressed Genes
These 133 genes that are differentially expressed between white and black feather bulbs could be grouped into 94 pathways by gene ontology (GO) analysis. The pathways and differentially expressed genes between the two types of feather bulbs are shown in Table S4. Among the pathways, melanogenesis (c-Kit/Tyr/Tyrp1) and tyrosine metabolism (Tyr/Tyrp1) were directly related to bird plumage pigmentation. A summary of this pathway analysis is shown in Table 2. GO analysis also identified the MAPK (Mitogen-Activated Protein Kinase) signaling pathway, which can link the functions of Kit and Mitf. Differential expression of the Mitf isoforms is dependent on the activation of MEK1-ERK2 in the MAPK signaling pathway [28]. Interestingly, GO analysis also showed enrichment of pathways involving p53 signaling, apoptosis, Toll-like receptor signaling and immune function. Compared to unpigmented feather bulbs, pigmented feather bulbs have normal melanocytes, in which a series gene cooperated with each other to perform melanogenesis, melanin formation and transportation. Defects occurring in any part of this process may cause feather unpigmentation. The melanogenic pathway which involved in melanogenesis and Tyrosine metabolism was also enriched in the 94 pathways in GO analysis. In addition, we found that pigmented and unpigmented feather bulbs may have differences in many physiological and biochemical processes, including apoptosis, cell cycle, immune response, metabolism, and signaling transduction, etc. It is a complicated regulation network. It could be that genes involved in several processes including melanogenesis and immunity are co-expressed. However, earlier studies showed that some of these pathways may be related to pigmentation [29], which is similar to our results. For example, up-regulation of genes in the Toll like receptor signaling pathway can be associated with melanocytes cell growth and melanogenesis [30]. Our result may provide further evidence for a relationship between TLRs and melanogenesis.
Table 2. Digital differential expression analysis of c-Kit, Tyrp1, Tyr and Tyrp2 genes between black and white feather bulbs by RNA-Seq.
| Gene | Raw-B-1 | Raw-W-1 | TPM-B-1 | TPM-W-1 | Log2ratio(W-1/B-1) | P-Value | FDR | |||
| c-Kit | 160 | 16 | 32.74 | 3.29 | −3.31489 | 3.44E-31 | 5.00E-29 | |||
| Tyrp1 | 3250 | 0 | 664.89 | 0.01 | −16.021 | 0 | 0 | |||
| Tyr | 184 | 0 | 37.65 | 0.01 | −11.8784 | 5.96E-56 | 3.48E-54 | |||
| Tyrp2 | 0 | 0 | ||||||||
Note: Raw-B-1, Raw data of black feather bulb expression; Raw-W-1, Raw data of white feather bulb expression; TPM-B-1, Normalized expression level of genes in black plumage feather bulb library; TPM-W-1, Normalized expression level of genes in white plumage feather bulb library; Log2 ratio(W-1/B-1), log2(multiples of differentially expressed genes); P-value corresponds to differential gene expression test; FDR (False Discovery Rate) is used to determine the threshold P-value in multiple tests and analyses by manipulating the FDR value [27].
qPCR Confirmation of Differential Gene Expression in White and Black Feather Bulbs from Different Types of Ducks
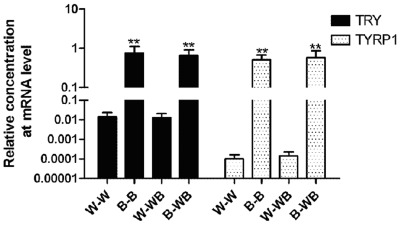
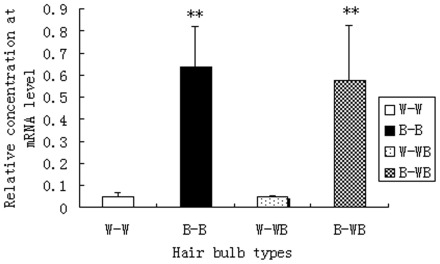
To confirm the differential gene expression from the RNA-Seq data, we used qPCR to measure the expression of Tyr, Tyrp1, and c-Kit, three genes in the melanogenesis pathway, in four combinations of feather bulbs and plumage types: W-W (white feather bulb from white plumage), W-WB (white feather bulb from white-black plumage), B-B (black feather bulb from black plumage), and B-WB (black feather bulb from white-black plumage). The results showed that the two critical genes in the melanogenesis pathway, Tyr and Tyrp1, had almost no expression in white feather bulbs from either white dorsal plumage or white-black dorsal plumage (Figure 1). We also found that Tyr and Tyrp1 were normally expressed in black feather bulbs from either black dorsal plumage or white-black dorsal plumage. The expression or absence of expression of Tyr and Tyrp1 genes indicated the pigmentation status of the feathers, regardless of whether the feather is from ducks with white, black or white-black plumages. C-Kit expression was significantly different (P<0.01, Figure 2) between W-W and B-B, or B-WB, as well as between W-WB and B-B, or B-WB samples. In contrast, c-Kit expression showed no significant difference between W-W and W-WB or between B-B and B-WB samples. C-Kit mRNA expression is approximately 10-fold higher in B-B and B-WB compared to W-W or W-WB samples.
Figure 1. The expression of Tyrp1 and Tyr genes in black and white feather bulb samples from different plumage types.
Figure 2. The expression of c-Kit gene in black and white feather bulb samples from different plumage types.
Expression Comparison of Mitf, Tyr, Tyrp1, c-Kit and Tyrp2 in Retinas and Feather Bulbs
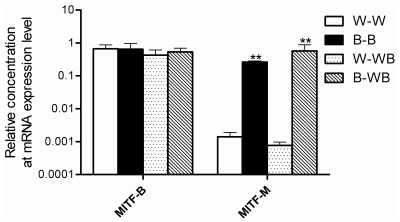
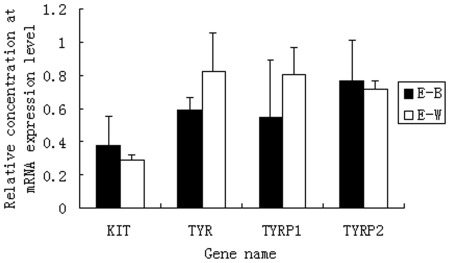
The expression of Tyr and Tyrp1 is regulated by Mitf. However, we did not find differential expression of Mitf in the RNA-Seq data. To investigate whether Mitf showed the same expression pattern as Tyr and Tyrp1, we performed cloning and qPCR analysis of this gene. The results showed that two isoforms of Mitf, M and B, exist. The B isoform was expressed in both black and white feather bulbs, while the M isoform was only expressed in black feather bulbs, regardless of whether they were collected from ducks with pure black or black-white plumage (Figure 3). We also used qPCR to test the relative expression of Tyr, Tyrp1, c-Kit and Tyrp2 in retinas, another organ in which melanogenesis occurs. The results showed that all 4 genes were expressed in retinas (Figure 4). Moreover, Tyrp2 is expressed only in retinas; no expression was detected for this gene in feather bulbs (Figure 5).
Figure 3. The expression of Mitf gene in black and white feather bulb samples from different plumage types.
Figure 4. The expression of Tyrp1, Tyr, c-Kit and Mitf genes in retina samples from black plumage and white plumage ducks.
Figure 5. Semi-RT-PCR measurement of Tyrp2 gene expression in retinas and feather bulbs from ducks with different plumage types.
Lanes 1–6: cDNA from 2 replicates of white, white-black and black plumage duck eyes; lanes 7–14: cDNA from 2 replicates of W-W, B-WB, W-WB and B-B feather bulbs; lanes 15–22: β-actin, samples are the same as lane 7–14, respectively. M: Marker I, six bands at 600, 500, 400, 300, 200, and 100 bp.
Discussion
Until now, there has been no study using genome-wide expression analysis in duck feather bulbs by RNA- sequencing. Our study offered new information related to gene expression profiles in black and white feather bulbs in the duck. The entire duck genomic sequence is not available; thus, our data analysis was based on the Ensemble Gallus gallus database and the Anas platyrhynchos EST database. Although duck and chicken coding sequences have high homology (up to 90% for many genes), using the Gallus gallus database to match duck sequences can be difficult, as most of the tags are from the 3′UTR of genes. Compared to more than 10 million tags, only 3,000 reference ESTs exist in the Anas platyrhynchos EST library, which is too few for annotation. In our study, only 133 genes were identified from 2,642 differentially expressed tags, while most tags did not match any annotated genes. The reasons may include: (1) The short tags were from 3′UTR of the mRNAs; (2) When using the chicken sequences as reference to annotate the genes, the tag will not be annotated to genes if there are 2-bp mismatches as the length of each tag is 17-bp. Thus, only a low proportion of differentially expressed tags could be matched to annotated genes. Also, this method is not sensitive enough to detect genes that are very weakly expressed. Fortunately, three genes in the melanogenesis pathway were identified, indicating that this pathway is crucial for duck plumage color determination.
Skin, coat, and feather color in mammals and birds are determined mainly by 2 melanins, eumelanin and pheomelanin [31]. In human, hair color (or lack thereof, i.e., white hair) is determined by whether the hair bulb has normal biosynthesis of melanin and its subsequent transfer from melanocyte to keratinocytes [32]. In colored feather bulbs, melanin can be synthesized at the first step as described by Korner [33]. For melanogenesis, Tyr, Tyrp1, and Tyrp2 were directly involved in the synthesis of melanins. In this study, Solexa sequencing showed low expression of Tyr and Tyrp1 in white feather bulbs from ducks with pure white or white-black plumage, while these genes showed normal expression in black feather bulbs from ducks with pure black or white-black plumage. These results demonstrated that a lack of Tyr and Tyrp1 expression led to a deficiency in the biosynthesis of melanin in white feather bulbs and is the direct cause of white duck plumage. Surprisingly, Tyrp2 was not expressed in white or black feather bulbs but was expressed in retinas, indicating that this gene may be responsible for retinal pigmentation. This result is similar to that in human [34]. We cloned this gene from duck eye retina mRNA, but the mechanism of its restricted gene expression is not clear.
Previous studies on coat or feather color mainly focused on the effects of nucleotide deletion, mutation, or insertion in single genes. Schmidt [35] found that a single point mutation in exon 1 of the mouse tyrosinase gene caused the dark-eyed albino phenotype. Tobita-Teramoto et al., [3] reported that a six-nucleotide deletion in the tyrosinase coding sequence caused chickens to be albino. Additionally, in chickens, a retroviral insertion in intron 4 of the tyrosinase gene, leading to a lack of exon 5, which encodes the carboxy-terminal membrane spanning domain, caused the recessive white phenotype [4], [36], [37]. In this study, our results showed that the duck Tyr gene was expressed normally in retinas from ducks with either black or white plumage. All the ducks in this study had normal, dark retinas. Thus, the Tyr gene has normal function ducks with black or white plumage, but the expression in white feather bulbs was suppressed. The genes that inhibit Tyr expression could be responsible for plumage color in this population.
It was reported that Mitf is a member of the bHLH-leucine zipper transcription factor family and played an important role in the development of retinal cells, mast cells, osteoclasts and melanocytes [38]. Alleles of Mitf have been associated with coat color in dogs [7] and mice [39], [40], as well as with plumage color in quail [41]. Minvielle [10] demonstrated that a 2-bp deletion in exon 11 of Mitf caused white plumage in Japanese quail. In this study, in contrast to Japanese quail, the Mitf-M isoform did not show expression in white plumage feather bulbs, although both Mitf-M and Mitf-B isoforms were normally expressed in retinas and black plumage feather bulbs. The difference in Mitf-M expression between white and black feather bulbs in this study suggests that Mitf-M is involved in determining feather pigmentation in the duck through either cis or trans acting regulatory elements as opposed to non-synonymous coding variants like in the quail. The expression pattern of the duck Pmel17 is the same as Mitf-M, Tyr, Tyrp1. Pmel17 plays a central role in the biogenesis of melanosomes. Studies in other animals showed that this gene is involved in the maturation of melanosomes [42]. Plumage variants in chicken are associated with insertion/deletion polymorphisms in the Pmel17 gene [23], [43], [44]. Also, silver coat color in horse is associated with a missense mutation in the Pmel17 gene [45]. Our study demonstrated Pmel17 may also play roles in duck pigmentation.
Although little is known about the role of c-Kit expression in the regulation of plumage color in birds, the roles of Scf and c-Kit signaling in melanoblast or melanocyte migration, proliferation and differentiation during embryogenesis and maintaining postnatal cutaneous melanogenesis were reported in mammals [13], [46], [47]. Additionally, the function of c-Kit in feather follicle melanogenesis [46], [48], [49] and the maintenance of human hair pigmentation [50] have been widely studied in various models. Nucleotide deletions from introns [14] and copy-number variation [51]–[53] of c-Kit were associated with pig coat color. C-Kit mutations in horses were associated with white coat color [16], [54]–[56], and exon skipping in the c-Kit gene in horses causes a Sabino spotting pattern [57]. Furthermore, c-Kit is a candidate for white spotting in cats [58], and the c-Kit signaling pathway is involved in post-developmental processes of mature cells [59]. Taken together, the c-Kit gene plays a critical role in animal coat color and is specifically associated with ‘white’. In this study, we found there was no significant difference in c-Kit expression in retinas from ducks with white or black plumage. W-W, B-B, W-WB, and B-WB feather bulbs all showed expression of the c-Kit gene, although the expression level in black feather bulbs was 10-fold higher than that in white feather bulbs. In white feather bulbs, this basal level of c-Kit expression may be able to maintain cell proliferation and differentiation but is not sufficient to promote pigmentation, although there is no in vitro confirmation work. In contrast, c-Kit expression in black feather bulbs is 10 times higher than that in white feather bulbs, allowing for cell proliferation and differentiation as well as maintenance of postnatal melanogenesis. It is possible that the lower level of c-Kit expression in the white feather bulb is a downstream consequence of few or no active melanocytes in the feather bulb, but not a genetic lesion at the c-Kit locus.
Conclusion
Plumage color variation in Kaiya-Liancheng F2 ducks was determined by whether melanin can be synthesized in the feather bulb. Our results provide solid evidence on some of the functional players in feather pigmentation in the duck, e.g. upregulation of c-Kit and Mitf in black feather bulbs may be responsible for black plumage formation.
Materials and Methods
Experimental Animals
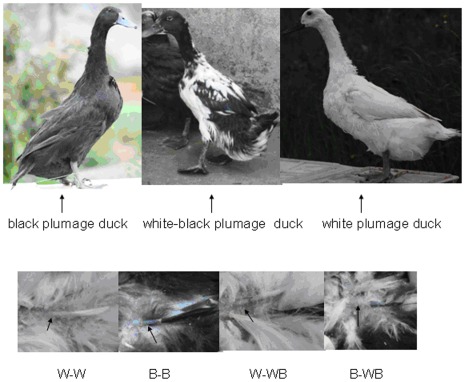
The genetic background of experimental ducks was described by Gong et al., [25]. Three white plumage ducks, three black plumage ducks and three white-black plumage ducks were randomly selected from a population of the Kaiya-Liancheng F2 generation. The three plumage patterns are shown in Figure 6. Three feather bulbs from the same individual were pooled as one sample. The white feather bulbs from white duck back were marked as W-W, whereas the black feather bulbs from black duck back were marked as B-B. White feather bulbs from white-black duck back were marked as W-WB and black feather bulbs from white-black duck back were marked as B-WB, respectively. The four feather bulb types are shown in Figure 6. All research involving animals were conducted according to the regulation (No. 5 proclaim of the Standing Committee of Hubei People’s Congress) approved by the Standing Committee of Hubei People’s Congress, and the ethics committee of Huazhong Agricultural University, P. R. China. The approved permit number for this study is “HBAC20091138”.
Figure 6. Plumage types and feather bulbs used in this study.
A: Three duck plumage types; B: Feather bulbs from four plumage types, W-W (white feather bulb from white plumage), B-B (black feather bulb from black plumage), W-WB (white feather bulb from white-black plumage), B-WB (black feather bulb from white-black plumage).
Total RNA Extraction from Feather Bulbs and RT-PCR
Feather bulbs were put into 2-mL tubes containing 1 mL TRIzol reagent (Invitrogen, San Diego, CA). One ceramic bead was added immediately to each tube. The tubes were then ground for 30 seconds by EASY GRIND. Total RNA was extracted according to the manufacturer’s protocol. The quality and quantity of RNA samples were checked by Spectrophotometer ND-1000 (Nano-Drop) and denaturing agarose gel electrophoresis. All RNA samples were treated with DNAse-I for later use.
RNA-Seq, Data Mining and Gene Ontology Analysis
Solexa sequencing of W-W and B-B pooled RNA was conducted in BGI, Shenzhen. Three databases were employed for sequence analysis, the Ensemble Gallus gallus databases, reference gene (ftp.ensembl.org/pub/release-59/fasta/gallus_gallus/cdna/Gallus gallus. WASHUC2.59.cdna. all. fa.gz), reference genomic DNA (ftp://ftp.ensembl.org/pub/release-59/fasta/gallus_gallus/dna), and the Anas platyrhynchos EST database (http://www.ncbi.nlm.nih.gov/nucest/?term=Anas%20platyrhynchos). In this study, a rigorous algorithm has been developed to identify differentially expressed genes between two samples by BGI based on “The significance of digital gene expression profiles” [26]. We used a P-value corresponding to a differential gene expression test at statistically significant levels [27]. “FDR (False Discovery Rate) ≤0.001 and the absolute value of log2Ratio≥1” were used to identify DEGs (Different Expression Genes) and DETs (Different Expression Tags). Pathways of differentially expressed genes were analyzed by DAVID v6.7. Gene function classification was conducted using the Gene Ontology FAT set term.
c-Kit, Tyr, Tyrp1 and Tyrp2 Gene Expression in Feather Bulb and Retina Samples by qPCR
To confirm the differential expression of genes revealed by RNA-Seq, the expression of genes in the melanogenesis pathway, including c-Kit, Tyr and Tyrp1, was measured by qPCR. In addition, the expression of Mitf and Tyrp2 was measured because they are directly involved in melanin biosynthesis. β-actin was used as a reference control. qPCR analysis was performed on Roche lightercycler® 480, using lightercycler® 480 SYBR Green I master detection reagents (Roche Diagnostics 11367523). All reactions were performed in triplicate within each PCR assay and under the same cycling conditions: denaturation at 95°C for 3 min, followed by 40 cycles of amplification (95°C for 20 s, 60°C for 20 s, and 72°C for 20 s) with a single acquisition of fluorescence at the end of the extension step. Melt curve analysis was performed over a range of 55∼95°C to verify single product generation at the end of the assay. qPCR data analysis was performed with the Light Cycler analysis software. Relative quantification analyses were performed in EXCEL using the comparative CT method. Comparisons between qPCR data sets were made with Student’s t-test. Differences were considered significant if P<0.05. Further, semi-RT-PCR measurements of Tyrp2 gene expression in retinas and feather bulbs from ducks with different plumage types were also conducted. All primers information is shown in Table 3.
Table 3. Primers used in Semi-RT-PCR and qPCR.
| Gene | Primers | Sequence (5′- 3′) | Size (bp) | AT | Function |
| β-actin | β-actin-Fβ-actin-R | AACTGGGATGACATGGAGAAGA ATGGCTGGGGTGTTGAAGGT | 189 | 60°C | Semi-RT PCR & qPCR |
| c-Kit | c-Kit-Fc-Kit-R | GCTGATGCTGCCAATGAGT TTTGCCACCTGGTAAGAGA | 151 | 60°C | qPCR |
| Tyr | E3-F1425E4NR | TTACATGGTCCCCTTTATTC CAATCACAGCTGCACCAACC | 182 | 60°C | qPCR |
| Tyrp1 | F1250R1439 | AATGAGATGTTTGTTACTG ACTGATCAGTGAGAAGAGG | 208 | 60°C | qPCR |
| Tyrp2 | F1496R1703 | CACCTATGCCATTGACCTGCC AGCAAGGAAACGAAGCAAGGG | 228 | 60°C | qPCR |
| Mitf-B | BF383MBR220 | CCCAGTTCATGCAGCAGAGAGT CCAGGCGGCATGACATGATCAC | 268 | 60°C | Semi-RT-PCR |
| Mitf-M | MF16MBR220 | TGCAGTCACTTCTCTCACAACC CCAGGCGGCATGACATGATCAC | 226 | 60°C | qPCR |
Note: AT = Annealing temperature.
Supporting Information
The information of genes that expressed in duck feather bulbs that have different reads.
(XLS)
Differentially expressed tags between white and black plumage feather bulb libraries.
(XLS)
Differentially expressed genes between white and black plumage feather bulb libraries.
(XLS)
Gene Ontology analysis of the differentially expressed genes.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Natural Science Foundation of China (31172201), Fundamental Research Funds for the Central Universities (2011QC006), and the Creative Team Project of Ministry of Education of China (IRT-0831). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ducrest AL, Keller L Roulin A. Pleiotropy in the melanocortin system, coloration and behavioural syndromes. Trends In Ecology & Evolution. 2008;23:502–510. doi: 10.1016/j.tree.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Lancaster FM. Bibliographia genetica XIX. p; 1963. The inheritance of plumage colour in the common duck. pp. 317–404. [Google Scholar]
- 3.Tobita-Teramoto T, Jang GY, Kino K, Salter DW, Brumbaugh J, et al. Autosomal albino chicken mutation (ca/ca) deletes hexanucleotide (-deltaGACTGG817) at a copper-binding site of the tyrosinase gene. Poult Sci. 2000;79:46–50. doi: 10.1093/ps/79.1.46. [DOI] [PubMed] [Google Scholar]
- 4.Chang CM, Coville JL, Coquerelle G, Gourichon D, Oulmouden A, et al. Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genomics. 2006;7:19. doi: 10.1186/1471-2164-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontanesi L, Scotti E Russo V. Animal Genetics no-no; 2011. Haplotype variability in the bovine MITF gene and association with piebaldism in Holstein and Simmental cattle breeds. [DOI] [PubMed] [Google Scholar]
- 6.Hayes BJ, Pryce J, Chamberlain AJ, Bowman PJ, Goddard ME. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 2010;6:e1001139. doi: 10.1371/journal.pgen.1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlsson EK, Baranowska I, Wade CM, Salmon HN, Zody MC, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nature Genetics. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- 8.Dorshorst B, Molin AM, Rubin CJ, Johansson AM Stromstedt L, et al. A complex genomic rearrangement involving the endothelin 3 locus causes dermal hyperpigmentation in the chicken. PLoS Genet. 2011;7:e1002412. doi: 10.1371/journal.pgen.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Zhu X, Yang L, Li J Lian Z, et al. Expression and network analysis of genes related to melanocyte development in the Silky Fowl and White Leghorn embryos. Molecular Biology Reports. 2011;38:1433–1441. doi: 10.1007/s11033-010-0248-2. [DOI] [PubMed] [Google Scholar]
- 10.Minvielle F, Bed’Hom B, Coville JL, Ito S, Inoue-Murayama M, et al. The “silver” Japanese quail and the MITF gene: causal mutation, associated traits and homology with the “blue” chicken plumage. BMC Genet. 2010;11:15. doi: 10.1186/1471-2156-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsujimura T, Morii E, Nozaki M, Hashimoto K, Moriyama Y, et al. Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood. 1996;88:1225–1233. [PubMed] [Google Scholar]
- 12.Hou L, Panthier JJ Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127:5379–5389. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- 13.Botchkareva NV, Khlgatian M, Longley BJ, Botchkarev VA, Gilchrest BA. SCF/c-kit signaling is required for cyclic regeneration of the hair pigmentation unit. Faseb Journal. 2001;15:645–658. doi: 10.1096/fj.00-0368com. [DOI] [PubMed] [Google Scholar]
- 14.Fontanesi L, D’Alessandro E, Scotti E, Liotta L, Crovetti A, et al. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Animal Genetics. 2010;41:478–492. doi: 10.1111/j.1365-2052.2010.02054.x. [DOI] [PubMed] [Google Scholar]
- 15.Fontanesi L, Tazzoli M, Russo V Beever J. Genetic heterogeneity at the bovine KIT gene in cattle breeds carrying different putative alleles at the spotting locus. Animal Genetics. 2010;41:295–303. doi: 10.1111/j.1365-2052.2009.02007.x. [DOI] [PubMed] [Google Scholar]
- 16.Haase B, Brooks SA, Schlumbaum A, Azor PJ, Bailey E, et al. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet. 2007;3:e195. doi: 10.1371/journal.pgen.0030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen B, Chen Y, Li H, Wang J, Shen J, et al. Allele-specific genetic interactions between Mitf and Kit affect melanocyte development. Pigment Cell Melanoma Res. 2010;23:441–447. doi: 10.1111/j.1755-148X.2010.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecoin L, Lahav R, Martin FH, Teillet MA, Le Douarin NM. Steel and c-kit in the development of avian melanocytes: a study of normally pigmented birds and of the hyperpigmented mutant silky fowl. Dev Dyn. 1995;203:106–118. doi: 10.1002/aja.1002030111. [DOI] [PubMed] [Google Scholar]
- 19.Niwa T, Mochii M, Nakamura A Shiojiri N. Plumage pigmentation and expression of its regulatory genes during quail development–histochemical analysis using Bh (black at hatch) mutants. Mech Dev. 2002;118:139–146. doi: 10.1016/s0925-4773(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 20.Gunnarsson U, Kerje S, Bed’Hom B, Sahlqvist AS, Ekwall O, et al. The Dark brown plumage color in chickens is caused by an 8.3-kb deletion upstream of SOX10. Pigment Cell Melanoma Res. 2011;24:268–274. doi: 10.1111/j.1755-148X.2011.00825.x. [DOI] [PubMed] [Google Scholar]
- 21.Hiragaki T, Inoue-Murayama M, Miwa M, Fujiwara A, Mizutani M, et al. Recessive black is allelic to the yellow plumage locus in Japanese quail and associated with a frameshift deletion in the ASIP gene. Genetics. 2008;178:771–775. doi: 10.1534/genetics.107.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerje S, Lind J, Schutz K, Jensen P, Andersson L. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Animal Genetics. 2003;34:241–248. doi: 10.1046/j.1365-2052.2003.00991.x. [DOI] [PubMed] [Google Scholar]
- 23.Kerje S, Sharma P, Gunnarsson U, Kim H, Bagchi S, et al. The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics. 2004;168:1507–1518. doi: 10.1534/genetics.104.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang HL, Cheng YS, Huang CW, Huang MC, Hsu WH. A novel genetic marker of the ovomucoid gene associated with hatchability in Tsaiya ducks (Anas platyrhynchos). Animal Genetics. 2011;42:421–427. doi: 10.1111/j.1365-2052.2010.02161.x. [DOI] [PubMed] [Google Scholar]
- 25.Gong Y, Yang Q, Li S, Feng Y, Gao C, et al. Grey plumage colouration in the duck is genetically determined by the alleles on two different, interacting loci. Animal Genetics. 2010;41:105–108. doi: 10.1111/j.1365-2052.2009.01967.x. [DOI] [PubMed] [Google Scholar]
- 26.Audic S Claverie JM. The significance of digital gene expression profiles. Genome Research. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural Brain Research. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 28.Primot A, Mogha A, Corre S, Roberts K, Debbache J, et al. ERK-regulated differential expression of the Mitf 6a/b splicing isoforms in melanoma. Pigment Cell Melanoma Res. 2010;23:93–102. doi: 10.1111/j.1755-148X.2009.00652.x. [DOI] [PubMed] [Google Scholar]
- 29.Solano F, Briganti S, Picardo M, Ghanem G. Hypopigmenting agents: an updated review on biological, chemical and clinical aspects. Pigment Cell Res. 2006;19:550–571. doi: 10.1111/j.1600-0749.2006.00334.x. [DOI] [PubMed] [Google Scholar]
- 30.Kang HY, Park TJ, Jin SH. Imiquimod, a Toll-like receptor 7 agonist, inhibits melanogenesis and proliferation of human melanocytes. Journal Of Investigative Dermatology. 2009;129:243–246. doi: 10.1038/jid.2008.184. [DOI] [PubMed] [Google Scholar]
- 31.Delfgaauw J, Duschl J, Wellbrock C, Froschauer C, Schartl M, et al. MITF-M plays an essential role in transcriptional activation and signal transduction in Xiphophorus melanoma. Gene. 2003;320:117–126. doi: 10.1016/s0378-1119(03)00817-5. [DOI] [PubMed] [Google Scholar]
- 32.Tobin DJ. Human hair pigmentation–biological aspects. Int J Cosmet Sci. 2008;30:233–257. doi: 10.1111/j.1468-2494.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 33.Korner A, Pawelek J. Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science. 1982;217:1163–1165. doi: 10.1126/science.6810464. [DOI] [PubMed] [Google Scholar]
- 34.Commo S, Gaillard O, Thibaut S, Bernard BA. Absence of TRP-2 in melanogenic melanocytes of human hair. Pigment Cell Res. 2004;17:488–497. doi: 10.1111/j.1600-0749.2004.00170.x. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt A, Beermann F. Molecular basis of dark-eyed albinism in the mouse. Proc Natl Acad Sci U S A. 1994;91:4756–4760. doi: 10.1073/pnas.91.11.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CM, Furet JP, Coville JL, Coquerelle G, Gourichon D, et al. Quantitative effects of an intronic retroviral insertion on the transcription of the tyrosinase gene in recessive white chickens. Animal Genetics. 2007;38:162–167. doi: 10.1111/j.1365-2052.2007.01581.x. [DOI] [PubMed] [Google Scholar]
- 37.Sato S, Otake T, Suzuki C, Saburi J, Kobayashi E. Mapping of the recessive white locus and analysis of the tyrosinase gene in chickens. Poult Sci. 2007;86:2126–2133. doi: 10.1093/ps/86.10.2126. [DOI] [PubMed] [Google Scholar]
- 38.Hallsson JH, Haflidadottir BS, Schepsky A, Arnheiter H, Steingrimsson E. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res. 2007;20:185–200. doi: 10.1111/j.1600-0749.2007.00373.x. [DOI] [PubMed] [Google Scholar]
- 39.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annual Review Of Genetics. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 40.Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nature Genetics. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 41.Mochii M, Mazaki Y, Mizuno N, Hayashi H, Eguchi G. Role of Mitf in differentiation and transdifferentiation of chicken pigmented epithelial cell. Developmental Biology. 1998;193:47–62. doi: 10.1006/dbio.1997.8800. [DOI] [PubMed] [Google Scholar]
- 42.Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Molecular Biology Of The Cell. 2001;12:3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natt D, Kerje S, Andersson L, Jensen P. Plumage color and feather pecking–behavioral differences associated with PMEL17 genotypes in chicken (Gallus gallus). Behavior Genetics. 2007;37:399–407. doi: 10.1007/s10519-006-9125-0. [DOI] [PubMed] [Google Scholar]
- 44.Karlsson AC, Kerje S, Andersson L, Jensen P. Genotype at the PMEL17 locus affects social and explorative behaviour in chickens. Br Poult Sci. 2010;51:170–177. doi: 10.1080/00071661003745802. [DOI] [PubMed] [Google Scholar]
- 45.Brunberg E, Andersson L, Cothran G, Sandberg K, Mikko S, et al. A missense mutation in PMEL17 is associated with the Silver coat color in the horse. BMC Genet. 2006;7:46. doi: 10.1186/1471-2156-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishikawa S, Kusakabe M, Yoshinaga K, Ogawa M, Hayashi S, et al. In utero manipulation of coat color formation by a monoclonal anti-c-kit antibody: two distinct waves of c-kit-dependency during melanocyte development. Embo Journal. 1991;10:2111–2118. doi: 10.1002/j.1460-2075.1991.tb07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida H, Hayashi S, Shultz LD, Yamamura K, Nishikawa S, et al. Neural and skin cell-specific expression pattern conferred by steel factor regulatory sequence in transgenic mice. Dev Dyn. 1996;207:222–232. doi: 10.1002/(SICI)1097-0177(199610)207:2<222::AID-AJA10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Kos L, Aronzon A, Takayama H, Maina F, Ponzetto C, et al. Hepatocyte growth factor/scatter factor-MET signaling in neural crest-derived melanocyte development. Pigment Cell Res. 1999;12:13–21. doi: 10.1111/j.1600-0749.1999.tb00503.x. [DOI] [PubMed] [Google Scholar]
- 49.Kurita K, Nishito M, Shimogaki H, Takada K, Yamazaki H, et al. Suppression of progressive loss of coat color in microphthalmia-vitiligo mutant mice. Journal of Investigative Dermatology. 2005;125:538–544. doi: 10.1111/j.0022-202X.2005.23861.x. [DOI] [PubMed] [Google Scholar]
- 50.Hachiya A, Sriwiriyanont P, Kobayashi T, Nagasawa A, Yoshida H, et al. Stem cell factor-KIT signalling plays a pivotal role in regulating pigmentation in mammalian hair. Journal of Pathology. 2009;218:30–39. doi: 10.1002/path.2503. [DOI] [PubMed] [Google Scholar]
- 51.Giuffra E, Tornsten A, Marklund S, Bongcam-Rudloff E, Chardon P, et al. A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mammalian Genome. 2002;13:569–577. doi: 10.1007/s00335-002-2184-5. [DOI] [PubMed] [Google Scholar]
- 52.Johansson MM, Chaudhary R, Hellmen E, Hoyheim B, Chowdhary B, et al. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mammalian Genome. 1996;7:822–830. doi: 10.1007/s003359900244. [DOI] [PubMed] [Google Scholar]
- 53.Seo BY, Park EW, Ahn SJ, Lee SH, Kim JH, et al. An accurate method for quantifying and analyzing copy number variation in porcine KIT by an oligonucleotide ligation assay. BMC Genet. 2007;8:81. doi: 10.1186/1471-2156-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haase B, Brooks SA, Tozaki T, Burger D, Poncet PA, et al. Seven novel KIT mutations in horses with white coat colour phenotypes. Animal Genetics. 2009;40:623–629. doi: 10.1111/j.1365-2052.2009.01893.x. [DOI] [PubMed] [Google Scholar]
- 55.Marklund S, Moller M, Sandberg K, Andersson L. Close association between sequence polymorphism in the KIT gene and the roan coat color in horses. Mammalian Genome. 1999;10:283–288. doi: 10.1007/s003359900987. [DOI] [PubMed] [Google Scholar]
- 56.Terry RR, Bailey E, Bernoco D, Cothran EG. Linked markers exclude KIT as the gene responsible for appaloosa coat colour spotting patterns in horses. Animal Genetics. 2001;32:98–101. doi: 10.1046/j.1365-2052.2001.00737.x. [DOI] [PubMed] [Google Scholar]
- 57.Brooks SA, Bailey E. Exon skipping in the KIT gene causes a Sabino spotting pattern in horses. Mammalian Genome. 2005;16:893–902. doi: 10.1007/s00335-005-2472-y. [DOI] [PubMed] [Google Scholar]
- 58.Cooper MP, Fretwell N, Bailey SJ, Lyons LA. White spotting in the domestic cat (Felis catus) maps near KIT on feline chromosome B1. Animal Genetics. 2006;37:163–165. doi: 10.1111/j.1365-2052.2005.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Motro B, van der Kooy D, Rossant J, Reith A, Bernstein A. Contiguous patterns of c-kit and steel expression: analysis of mutations at the W and Sl loci. Development. 1991;113:1207–1221. doi: 10.1242/dev.113.4.1207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The information of genes that expressed in duck feather bulbs that have different reads.
(XLS)
Differentially expressed tags between white and black plumage feather bulb libraries.
(XLS)
Differentially expressed genes between white and black plumage feather bulb libraries.
(XLS)
Gene Ontology analysis of the differentially expressed genes.
(DOC)