Tks5, a master regulator of invadopodia in cancer cells, is also crucial for osteoclast cell–cell fusion.
Abstract
Osteoclasts fuse to form multinucleated cells during osteoclastogenesis. This process is mediated by dynamic rearrangement of the plasma membrane and cytoskeleton, and it requires numerous factors, many of which have been identified. The underlying mechanism remains obscure, however. In this paper, we show that Tks5, a master regulator of invadopodia in cancer cells, is crucial for osteoclast fusion downstream of phosphoinositide 3-kinase and Src. Expression of Tks5 was induced during osteoclastogenesis, and prevention of this induction impaired both the formation of circumferential podosomes and osteoclast fusion without affecting cell differentiation. Tyrosine phosphorylation of Tks5 was attenuated in Src−/− osteoclasts, likely accounting for defects in podosome organization and multinucleation in these cells. Circumferential invadopodia formation in B16F0 melanoma cells was also accompanied by Tks5 phosphorylation. Co-culture of B16F0 cells with osteoclasts in an inflammatory milieu promoted the formation of melanoma–osteoclast hybrid cells. Our results thus reveal an unexpected link between circumferential podosome/invadopodium formation and cell–cell fusion in and beyond osteoclasts.
Introduction
Cell fusion, which results in the formation of multinucleated cells, is a fundamental biological event that is required for various developmental and homeostatic processes, including fertilization, myogenesis, giant cell formation, and osteoclastogenesis. Multinucleation may enhance the function of the fused cells, but the precise mechanisms that govern this process remain incompletely understood. Osteoclasts are derived from monocytes, which are recruited to the bone surface to resorb the bone matrix. Hyperactivity of osteoclasts is characteristic of bone destructive disorders, such as osteoporosis, rheumatoid arthritis, and bone metastasis, whereas hypoactivity of these cells is associated with osteopetrosis. Osteoclast precursors on the bone surface are stimulated by macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) secreted by osteoblasts, with this stimulation being essential for activation of signaling cascades that underlie osteoclastogenesis. NFATc1 (nuclear factor of activated T cells cytoplasmic 1) is a master transcription factor that is induced and activated downstream of the receptor activator of NF-kB on osteoclast precursors (Takayanagi et al., 2002). NFATc1 promotes the transcription of various osteoclast-specific genes, including those for tartrate-resistant acid phosphatase (TRAP), cathepsin K, and β3 integrin (Matsuo et al., 2004; Negishi-Koga and Takayanagi, 2009), which allow the cells to degrade the bone matrix as well as to establish a fusion-competent state.
Activated osteoclast precursors develop columnar actin puncta, known as podosomes, at their ventral surface (Jurdic et al., 2006; Saltel et al., 2008), and similar structures, known as invadopodia, also appear in invasive cancer cells. In the early phase of osteoclast differentiation, these actin puncta organize into dynamic rings, and as the cells undergo fusion, these circumferential podosomes eventually mature into stabilized structures known as sealing belts (Jurdic et al., 2006). Efficient bone resorption is then achieved through the secretion of protons and acid-dependent proteases at the ruffled border formed inside these belts (Zhao and Patrick Ross, 2007).
The tyrosine kinase Src regulates podosome organization, as revealed by the observation that osteoclasts from Src−/− mice manifest impaired formation of sealing belts and reduced bone resorption activity, resulting in the development of severe osteopetrosis (Soriano et al., 1991; Lakkakorpi et al., 2001). Furthermore, Src has been shown to regulate actin polymerization in podosomes (Destaing et al., 2008) as well as osteoclast spreading and fusion (Winograd-Katz et al., 2011). Osteoclast fusion involves a highly intricate orchestration of both lipids and proteins, given that lipid bilayer membranes do not spontaneously fuse, whereas they are often deformed to generate membrane nanotubes (Hase et al., 2009) or pinched off to generate vesicles that are transmitted to neighboring cells (Camussi et al., 2010). Despite the identification of diverse molecules as essential participants in cell–cell fusion (Mbalaviele et al., 1995; Saginario et al., 1998; Yagi et al., 2005; Lee et al., 2006; Yang et al., 2008; Gonzalo et al., 2010), the precise functions of and interactions among those molecules, in particular their relation to Src signaling, as well as the mechanism of force generation required to overcome the energetic barrier for fusion remain uncharacterized.
In addition to multinucleated osteoclasts, osteoclast–cancer cell hybrid cells have been detected in bone metastasis lesions in individuals with myeloma (Andersen et al., 2007, 2010). Moreover, leukocyte–cancer cell fusion has been implicated in malignant cancer progression, including metastasis (Pawelek and Chakraborty, 2008b). Although the biological relevance of osteoclast– or leukocyte–cancer cell hybrids remains unknown, it is possible that unique patterns of gene expression in such hybrid cells contribute to cancer progression. Unraveling the molecular basis for cell–cell fusion may thus provide insight into both physiological and pathological processes.
We now show that the expression of Tks5 is induced during osteoclastogenesis. Tks5 is an adaptor protein with an N-terminal phox homology (PX) domain and was originally identified as a Src substrate (Lock et al., 1998). It plays a pivotal role in invadopodia formation in Src-transformed cells as well as in cancer cells, thereby contributing to matrix degradation and cancer cell invasion, as well as in neural crest cell migration during embryonic development (Abram et al., 2003; Seals et al., 2005; Oikawa et al., 2008; Diaz et al., 2009; Gianni et al., 2009; Stylli et al., 2009; Murphy et al., 2011). We found that Tks5 is essential not only for circumferential podosome formation in osteoclasts but for cell–cell fusion of them, acting downstream of phosphoinositide (PI) 3-kinase and Src. Observation of live cells revealed that osteoclast fusion is mediated by plasma membrane extensions associated with circumferential podosomes, with Tks5 likely regulating osteoclast fusion by controlling podosome formation. The finding that circumferential podosomes in osteoclasts mediate cell–cell fusion is analogous to the recent observation that myoblast fusion is mediated by invasive podosome-like structures (Sens et al., 2010). Moreover, we show that the combination of RANKL, TGF-β, and TNF-α stimulates B16F0 murine melanoma cells to generate circumferential invadopodia in association with Tks5 accumulation and thereby promotes the formation of melanoma–osteoclast hybrids. Our results thus identify Src-Tks5 signaling as a new target in the development of therapeutics for the treatment of bone-destructive diseases, including osteolytic bone metastasis.
Results
Localization of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 at sites of osteoclast fusion
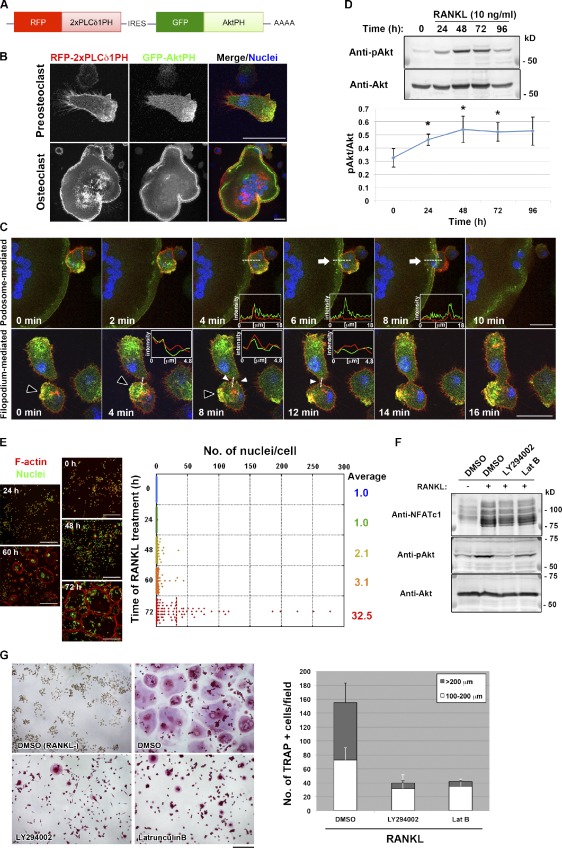
To investigate the mechanisms that underlie the dynamic interaction between plasma membranes during cell–cell fusion, we examined the localization of PIs, which is indicative of cell polarity, in RANKL-induced osteoclastogenic cultures with the use of the pleckstrin homology (PH) domains of PLCδ1 and Akt. The PH domain of PLCδ1 (PLCδ1PH), which binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), was tagged with RFP, whereas the PH domain of Akt (AktPH), which selectively binds both PtdIns(3,4)P2 and PtdIns(3,4,5)P3, was tagged with GFP (Fig. 1 A). Both PLCδ1PH and AktPH constructs were coexpressed in RANKL-stimulated RAW264.7 mouse macrophages under the control of a doxycycline (Dox)-responsive promoter. PLCδ1PH was distributed uniformly along the plasma membrane, whereas AktPH was localized at the leading edge of migrating mononuclear cells or at the sealing belt of multinucleated cells (Fig. 1 B). Live imaging of osteoclastogenesis revealed that cell–cell fusion was often mediated by PtdIns(3,4)P2- or PtdIns(3,4,5)P3-enriched membrane extensions. Those membrane extensions were categorized either as podosome related, which emerged from the sealing belt of large multinucleated cells (Fig. 1 C and Video 1), or filopodium like, which often formed after a transient expansion of podosome-like protrusions (Fig. 1 C and Video 2). Of a total of 50 fusion events observed, 34 (68%) occurred via podosome-related protrusions, and seven (14%) occurred via filopodium-like protrusions, both of which were associated with AktPH accumulation. In contrast, no obvious accumulation of AktPH was observed in the remaining 18%, with the fusion mediated by long projections (Video 3).
Figure 1.
Polarized membrane extensions mediate osteoclast fusion. (A) Schematic representation of the mRNA encoding the reporter constructs used for detection of PIs. IRES, internal ribosome entry site. (B) Live imaging of a mononuclear cell (top) and a multinuclear osteoclast (bottom) in RANKL-stimulated cultures of RAW264.7 cells expressing the reporter constructs in A. Cells were exposed to DAPI at 1 µg/ml (blue) for 1 h before observation to visualize nuclei. AktPH showed a polarized distribution, whereas PLCδ1PH was localized uniformly along the plasma membrane. (C) Live imaging of cells as in B during osteoclastogenesis. Arrows indicate a podosome-related protrusion in a multinucleated osteoclast. Closed and open arrowheads indicate a transient membrane expansion and filopodium-like protrusions, respectively. Pixel intensities on the traversing dashed lines were measured using LAS AF software and shown in the boxed areas (arbitrary units). See also Videos 1, 2, 3, and 4. (D, top) Immunoblot analysis of lysates of RAW264.7 macrophages stimulated with RANKL for the indicated times with antibodies to Ser473-phosphorylated (p) or total forms of Akt. (bottom) The pAkt/Akt signal intensity ratio was determined. Means ± SD from three independent experiments are indicated. *, P < 0.05 versus 0 h (Student’s t test). (E, left) RAW264.7 macrophages stimulated with RANKL for the indicated times were stained with rhodamine-phalloidin (red) to visualize F-actin and with DAPI (green) to visualize nuclei. (right) The distribution of the number of nuclei per cell was determined, with >100 cells counted at each time point. (F) RAW264.7 macrophages were stimulated with RANKL for 72 h, with the addition of 10 µM LY294002 or 0.1 µg/ml Latrunculin B (Lat B) for the last 16 h. DMSO was used as a vehicle control. Cell lysates were then subjected to immunoblot analysis with antibodies to NFATc1 or to Ser473-phosphorylated or total forms of Akt. (G, left) RAW264.7 macrophages treated with RANKL and inhibitors as in F were stained for TRAP activity. (right) The number of TRAP-positive cells with the indicated diameters was also determined. Data are means ± SD from three independent experiments. Bars: (B and C) 25 µm; (E) 250 µm; (G) 200 µm.
We could detect both PtdIns(3,4)P2 and PtdIns(3,4,5)P3 at the sites of fusion by using specific probes, the PH domains of Tapp1 (Tapp1PH) and Grp1 (Grp1PH), respectively (Video 4; Furutani et al., 2006). Given that PtdIns(3,4)P2 and PtdIns(3,4,5)P3 production is required for invadopodium formation in Src-transformed cells (Oikawa et al., 2008), we examined whether these PIs also participate in cell–cell fusion mediated by protrusions originating from podosomes or sealing belts in osteoclasts. First, we found that the phosphorylation of Akt at Ser473 was increased from 48 to 72 h after exposure of RAW264.7 macrophages to RANKL (Fig. 1 D), suggesting the presence of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 on the plasma membrane downstream of PI 3-kinase. Second, cell–cell fusion occurred most frequently between 60 and 72 h after exposure to RANKL, hereafter referred to as the frequent fusion period (Fig. 1 E). Third, inhibition of PI 3-kinase activity or actin polymerization with LY294002 or Latrunculin B, respectively, during the frequent fusion period (Fig. 1 F) resulted in a reduced fusion rate (Fig. 1 G), although neither the expression of NFATc1 (Fig. 1 F) nor the activity of TRAP was affected (Fig. 1 G). These data thus suggested that extension of the polarized membrane is required for cell–cell fusion during osteoclastogenesis. The membrane extensions are accompanied by podosome-related or filopodium-like lateral membrane protrusions, suggesting that the sites of cell–cell fusion are enriched in molecules engaged in the regulation of both actin polymerization and membrane deformation. We hereafter refer to these protrusions as fusion-competent protrusions.
Tks5 as a potential mediator of PtdIns(3,4)P2- or PtdIns(3,4,5)P3-enriched fusion-competent protrusion formation
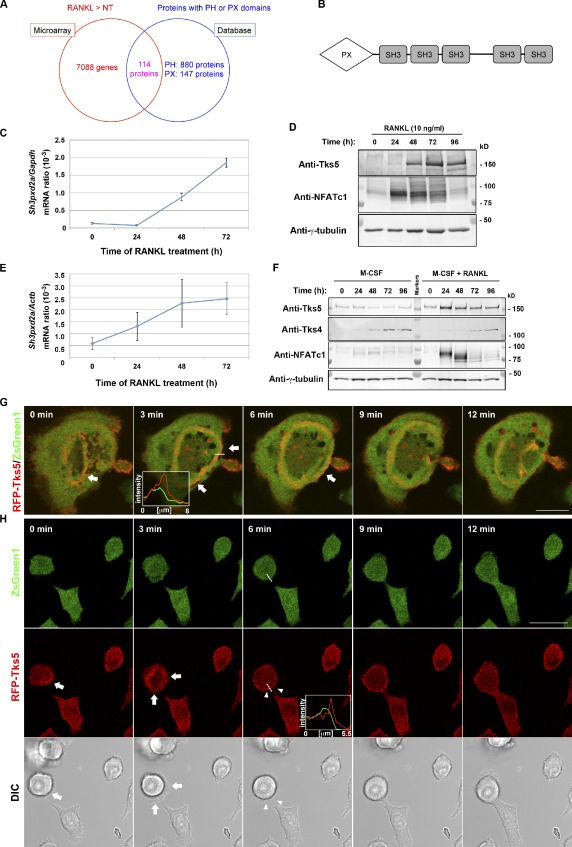
To identify molecules responsible for the formation of fusion-competent protrusions downstream of PtdIns(3,4)P2 or PtdIns(3,4,5)P3, we first compared gene expression profiles between nontreated and RANKL-treated RAW264.7 macrophages by microarray analysis (Fig. 2 A). Among the genes whose expression was up-regulated (>1.3-fold increase) by RANKL (Table S1), we searched in silico for gene products that contain a PH domain or PX domain that might confer affinity for PtdIns(3,4)P2 or PtdIns(3,4,5)P3 (Fig. 2 A and Table S2). After confirming RANKL-dependent up-regulation of its expression by quantitative RT-PCR and immunoblot analysis, we selected Tks5 (encoded by Sh3pxd2a; Fig. 2 B) for further investigation because of its emerging importance in the formation of specialized membrane structures, such as invadopodia, in Src-transformed cells (Abram et al., 2003; Seals et al., 2005; Oikawa et al., 2008) and in cancer cells (Seals et al., 2005; Stylli et al., 2009). The amounts of Tks5 mRNA and protein were increased in response to RANKL treatment both in RAW264.7 cells (Fig. 2, C and D) and in primary bone marrow macrophages (Fig. 2, E and F). In contrast, the expression of Tks4, a close relative of Tks5 that was also identified as a regulator of invadopodia (Buschman et al., 2009), was not affected by RANKL (Fig. 2 F). Live imaging of osteoclasts revealed that Tks5 was predominantly concentrated at circumferential podosomes, especially at the outer edge of the expanding ring (Fig. 2 G). Tks5 was also localized at the plasma membrane of mononuclear osteoclasts undergoing fusion, which followed a transient expansion of podosome-like protrusions (Fig. 2 H and Video 5). The formation of thin filopodium-like protrusions also appeared synchronized with Tks5 accumulation at the plasma membrane (Fig. 2 H and Video 5); penetration of one such filopodium into an adjacent cell resulted in fusion. In contrast, a deletion mutant of Tks5, which lacks the PX domain (Tks5-ΔPX), did not localize to the sites of cell–cell fusion (Video 6). These data thus suggested that Tks5 is a regulator of the formation of fusion-competent protrusions during osteoclastogenesis.
Figure 2.
Tks5 as a potential mediator of fusion-competent protrusion formation. (A) Venn diagram summarizing the results of screening for molecules required for osteoclast fusion downstream of PI 3-kinase. Microarray analysis of RAW264.7 cells stimulated with 10 ng/ml RANKL for 72 h or not treated (NT) revealed 7,088 genes whose expression was up-regulated by RANKL, and database analysis yielded 1,027 proteins that contain a PH or PX domain; 114 proteins were present in both datasets. See also Tables S1 and S2. (B) Domain organization of Tks5. SH3, Src homology 3 domain. (C and D) RAW264.7 macrophages cultured in the presence of 10 ng/ml RANKL for the indicated times were subjected either to quantitative RT-PCR analysis of the amount of Sh3pxd2a mRNA (normalized by that of Gapdh mRNA; C) or to immunoblot analysis with antibodies to Tks5, to NFATc1, or to γ-tubulin (loading control; D). Quantitative data are means ± SD from three independent experiments. (E and F) Mouse bone marrow–derived macrophages cultured in the presence of 10 ng/ml M-CSF with or without 10 ng/ml RANKL for the indicated times were subjected either to quantitative RT-PCR analysis of the amount of Sh3pxd2a mRNA (normalized by that of Actb mRNA; E) or to immunoblot analysis with the indicated antibodies (F). Quantitative data are means ± SD from three independent experiments. (G and H) Live imaging of RAW264.7 macrophages expressing RFP-tagged mouse Tks5. Images were obtained after stimulation with 10 ng/ml RANKL for 48 h. ZsGreen1 was separately expressed as an internal control, and differential interference contrast (DIC) images were also obtained. Arrows indicate the outer edge of an expanding podosome (G) or transient expansion of a podosome-related protrusion (H). Arrowheads in H represent a fusing plasma membrane manifesting RFP-Tks5 accumulation at the plasma membrane. Pixel intensities on the traversing dashed lines were measured using LAS AF or FluoView software and shown in boxed areas (arbitrary units). Bars, 25 µm. See also Videos 5 and 6.
Tks5 is required for cell–cell fusion during osteoclastogenesis
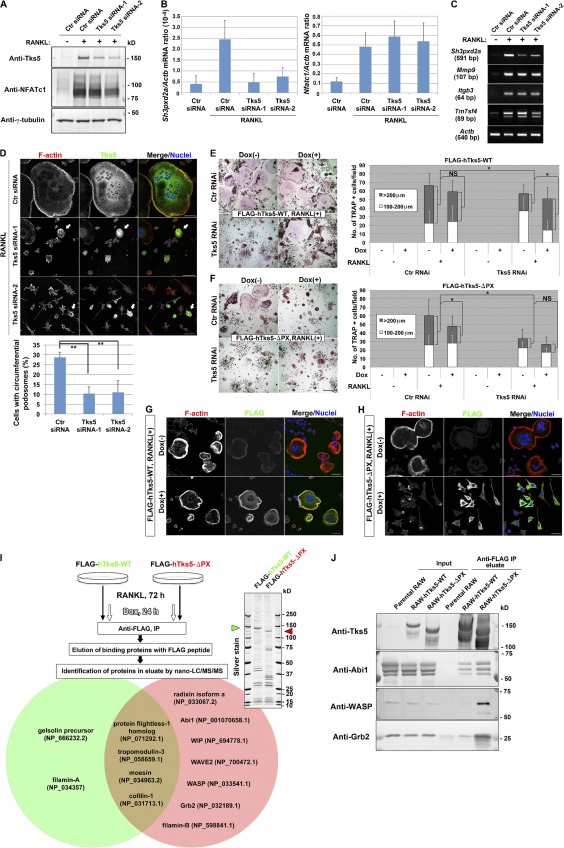
We next examined whether Tks5 is required for cell–cell fusion during osteoclastogenesis with the use of RNAi. Two siRNAs, each specific for Tks5, effectively suppressed Tks5 expression at the mRNA and protein levels without interfering with that of NFATc1 (Fig. 3, A and B) or target genes of NFATc1 (Fig. 3 C) in RAW264.7 macrophages. The RNAi-induced depletion of Tks5 in RANKL-treated RAW264.7 macrophages resulted in marked impairment of podosome formation, whereas cells that escaped from Tks5 knockdown exhibited circumferential podosomes (Fig. 3 D). Knockdown efficiency of Tks5 protein was almost complete in ∼70% of the cells, which correlated with the rate of suppression of circumferential podosome formation (Fig. 3 D, bottom). Furthermore, Tks5 knockdown abolished multinucleation (Fig. 3 D) without affecting two-dimensional migration and polarity formation (Fig. S1 A and Video 7). These results suggested that Tks5 is required for the generation of fusion-competent protrusions.
Figure 3.
Tks5 plays an essential role in osteoclast fusion by generating circumferential podosomes. (A–D) RAW264.7 macrophages were transfected on a consecutive 2 d with control (Ctr) or one of two Tks5 siRNAs and were then stimulated with 10 ng/ml RANKL for 72 h. The cells were then subjected to immunoblot analysis (A), to quantitative RT-PCR analysis (B), or to semiquantitative RT-PCR analysis (C) as indicated. Data in B are means ± SD from three independent experiments. (D) The cells were also stained with rhodamine-phalloidin to visualize F-actin, with antibodies to Tks5, and with DAPI to visualize nuclei. Arrows indicate cells still expressing Tks5 that generate circumferential podosomes. (bottom) The percentage of cells with circumferential podosomes among >100 cells scored was also determined. Data are means ± SD from three independent experiments. **, P < 0.005 (Student’s t test). Bars, 50 µm. See also Fig. S1 A and Video 7. (E and F) RAW264.7 macrophages that express FLAG-tagged hTks5-WT or hTks5-ΔPX under the control of a Dox-responsive promoter were transfected with control or Tks5 siRNAs as described for A–D. They were then cultured in the absence or presence of 10 ng/ml RANKL or 1 µg/ml Dox for 72 h. TRAP-positive multinucleated osteoclasts of the indicated diameters were then identified and counted. Bars, 200 µm. Quantitative data are means ± SD from three independent experiments. *, P < 0.05 (Student’s t test). See also Fig. S1 (B–D). (G and H) RAW264.7 macrophages harboring Dox-sensitive FLAG–hTks5-WT (G) or FLAG–hTks5-ΔPX (H) constructs were cultured in the presence of RANKL with or without Dox for 72 h and were then stained with rhodamine-phalloidin (red), antibodies to FLAG, and DAPI (blue). Bars, 25 µm. (I) Binding proteins of hTks5-WT or hTks5-ΔPX were identified as described in the Materials and methods. A portion of the binding proteins was subjected to SDS-PAGE and silver staining. Green and red arrowheads indicate hTks5-WT and hTks5-ΔPX, respectively. Among the binding proteins identified, cytoskeletal proteins that associated with hTks5-WT or with hTks5-ΔPX are listed in the green and red circles, respectively. Accession numbers were obtained from the NCBI Protein database. (J) The binding proteins eluted as in I as well as the original cell lysates (Input) were also subjected to immunoblot analysis with the indicated antibodies. See also Fig. S1 E and Fig. S2 A. IP, immunoprecipitation; LC-MS/MS, liquid chromatography with tandem mass spectrometry; RAW, RAW264.7.
We also established stable RAW264.7 cell lines expressing wild-type (WT) human Tks5 (hTks5; hTks5-WT) or a deletion mutant thereof lacking the PX domain (hTks5-ΔPX) under the control of a Dox-responsive promoter (Fig. S1 B). We then introduced Tks5 siRNA into these cell lines to suppress the expression of the endogenous mouse protein in the absence of Dox (Fig. S1, C and D), resulting in inhibition of RANKL-induced cell–cell fusion (Fig. 3, E and F). The induction of hTks5 expression after depletion of the mouse protein revealed that hTks5-WT, but not hTks5-ΔPX, again rendered the cells fusion competent (Fig. 3, E and F). Furthermore, hTks5-ΔPX acted as a dominant-negative mutant in cells transfected with a control siRNA and suppressed cell–cell fusion (Fig. 3 F), whereas the expression of hTks5-WT in control siRNA-transfected cells had no effect (Fig. 3 E).
Staining for F-actin revealed that well-organized and ring-shaped podosomes were apparent in cells expressing hTks5-WT (Fig. 3 G). In contrast, expression of hTks5-ΔPX abolished formation of circumferential podosomes, with dot-shaped actin aggregations being frequently observed instead (Fig. 3 H). Together, these results indicated that the localization of Tks5 to membrane domains enriched in PtdIns(3,4)P2 or PtdIns(3,4,5)P3 depends on the PX domain during osteoclastogenesis.
A comprehensive mass spectrometric analysis of proteins bound to FLAG-tagged hTks5-WT or hTks5-ΔPX in osteoclasts revealed that a greater number of actin-regulatory proteins was associated with the ΔPX mutant than with the WT protein (Fig. 3 I), with some of these differential interactions being validated by immunoblot analysis (Fig. 3 J). These proteins included Abi1, WASP, WIP, and Grb2, which are indispensable for adhesion-mediated cell spreading (Ring et al., 2011), podosome formation (Linder et al., 1999; Chou et al., 2006; Oikawa et al., 2008), or polarized cell motility (Tsuboi, 2006). It is therefore possible that hTks5-ΔPX impairs podosome formation by mediating the mislocalization of these actin-regulatory molecules because of promiscuous binding, whereas the interaction between hTks5-WT and such molecules is tightly regulated in time and space.
In support of this scenario, Tks5 was shown to form a multisubunit complex, a presumably inactive form, in resting NIH 3T3 cells, and such complex formation was attenuated in Src-transformed NIH 3T3 (NIH-src) cells (Fig. S1 E). It is thus likely that Tks5 exists in a “masked” form in the absence of stimulatory signals, such as PtdIns(3,4)P2 or PtdIns(3,4,5)P3, as well as Src-mediated phosphorylation (see following paragraph; Lock et al., 1998; Stylli et al., 2009). Such stimulation may result in exposure of the five SH3 domains of Tks5 and facilitate its interaction with actin-regulatory molecules in a spatially and temporally restricted manner. Our results thus indicated that Tks5 is responsible for the formation of fusion-competent protrusions through regulation of circumferential podosomes and that it might act downstream of active Src to drive cell–cell fusion.
Phosphorylation by Src is essential for Tks5 function during osteoclastogenesis
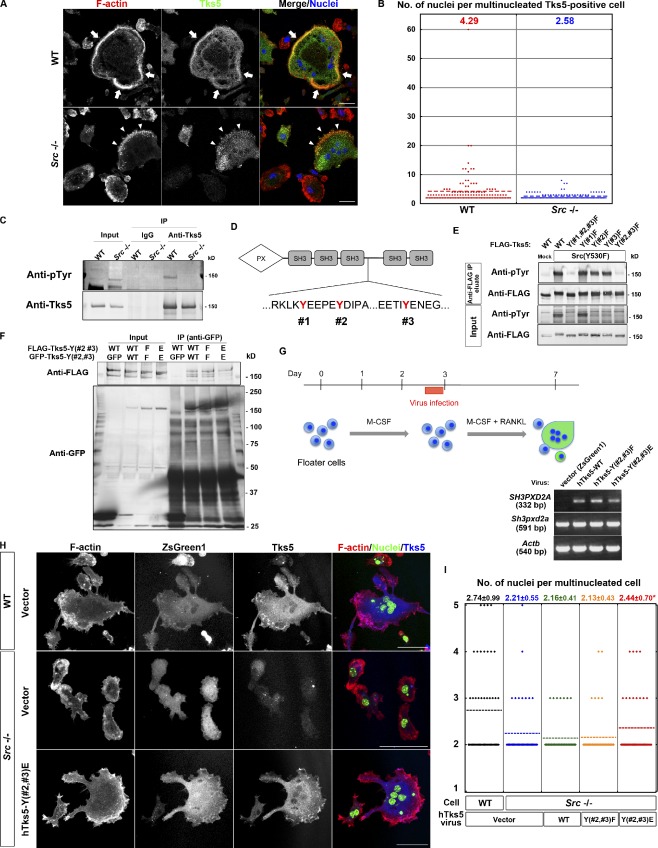
Src is essential for the dynamic organization of circumferential podosomes and osteoclast function both in vitro and in vivo (Soriano et al., 1991; Destaing et al., 2008; Winograd-Katz et al., 2011). Given that Tks5 was originally identified as a substrate for Src (Lock et al., 1998), we evaluated the relevance of the Src-Tks5 axis in osteoclast fusion. Consistent with previous observations, osteoclasts differentiated from spleen macrophages lacking Src showed impaired formation of sealing belts. Sporadic formation of actin puncta was observed in multinucleated Tks5-positive Src−/− cells, whereas the sealing belt was organized at the periphery of multinucleated Tks5-positive WT cells (Fig. 4 A). Furthermore, the mean number of nuclei in multinucleated Tks5-positive Src−/− cells was smaller than that in multinucleated Tks5-positive WT cells (Fig. 4 B), suggesting that Src functions upstream of Tks5 both in circumferential podosome formation and in osteoclast fusion. Indeed, Tks5 was tyrosine-phosphorylated in WT osteoclasts, whereas such phosphorylation was markedly attenuated in Src−/− osteoclasts (Fig. 4 C). To clarify the relevance of this phosphorylation, we generated mutant forms of hTks5 with phenylalanine substitutions at tyrosine residues (#1, #2, and #3) that constitute putative Src phosphorylation sites (Fig. 4 D). When expressed with an active form of Src, Src(Y530F), the triple mutant Y(#1,#2,#3)F did not undergo tyrosine phosphorylation, whereas two of the single mutants, Y(#2)F and Y(#3)F, showed a reduced level of phosphorylation compared with the WT protein (Fig. 4 E). Given that the double mutant Y(#2,#3)F was also not phosphorylated, we conclude that Tks5 is phosphorylated at Y#2 and Y#3 by Src (Fig. 4 E). Tks5 was recently shown to interact with the adaptor protein Nck in a manner dependent on the tyrosine phosphorylation of Tks5, and this interaction was found to be essential for matrix degradation by cancer cells (Stylli et al., 2009). As we could also detect Tks5 interaction with an adaptor, Nck, in osteoclasts (Fig. S2 A), it is possible that tyrosine phosphorylation of Tks5 triggers its activation by rearranging the binding proteins. To examine this possibility, we generated a glutamate substitution mutant of hTks5, Y(#2,#3)E, to mimic the effect of phosphorylation by Src. When expressed in NIH 3T3 cells, this mutant did not form a multisubunit complex, whereas both the WT protein and the Y(#2,#3)F mutant did (Fig. 4 F). These results suggested that tyrosine phosphorylation of Tks5 at Y#2 and Y#3 by Src functions in concert with the PX–PI interaction to induce a conformational change in Tks5 and thereby to trigger its activation. We next asked whether the Y(#2,#3)E mutant of hTks5 was able to correct the defects of Src−/− osteoclasts. WT, Y(#2,#3)F, and Y(#2,#3)E forms of hTks5 were introduced separately together with ZsGreen1 into primary spleen macrophages by retroviral infection, and the cells were then induced to differentiate into osteoclasts by exposure to RANKL (Fig. 4 G). Expression of the Y(#2,#3)E mutant partially reversed the morphological defects of Src−/− osteoclasts (Fig. 4, H and I), whereas neither the WT protein nor the Y(#2,#3)F mutant had a marked effect (Fig. 4, H and I), indicating that Tks5 is a key downstream effector of Src. Live imaging analysis revealed that both Y(#2,#3)F and Y(#2,#3)E mutants localized at podosomes and the sites of cell–cell fusion, indicating that phosphorylation by Src does not affect the localization of Tks5 (Fig. S2 B and Videos 8 and 9).
Figure 4.
Phosphorylation by Src is required for Tks5 to induce circumferential podosomes and osteoclast fusion. (A) Representative images of multinucleated osteoclasts differentiated from spleen macrophages of WT or Src−/− mice. The cells were stained with rhodamine-phalloidin to visualize F-actin, with antibodies to Tks5, and with DAPI to visualize nuclei. Mononuclear cells tended to express little Tks5, whereas Tks5 was abundant in multinuclear cells. Arrows indicate a sealing belt, and arrowheads indicate actin puncta formed at the cell periphery. Bars, 25 µm. (B) Quantification of the number of nuclei per cell in multinucleated Tks5-positive osteoclasts differentiated from spleen macrophages of WT or Src−/− mice. More than 100 cells were counted, and the mean values are indicated. (C) Lysates of osteoclasts differentiated from spleen macrophages of WT or Src−/− mice were subjected to immunoprecipitation with antibodies to Tks5 or control IgG. The resulting precipitates, as well as the original cell lysates (Input), were then subjected to immunoblot analysis with antibodies to phosphotyrosine (pTyr) or to Tks5. (D) The amino acid sequences of hTks5 containing putative Src phosphorylation sites (#1, #2, and #3). (E) NIH 3T3 cells were cotransfected with a vector for FLAG-tagged hTks5-WT or mutants thereof with the indicated tyrosine residues replaced with phenylalanine together with a vector for an active form of Src, Src(Y530F), or the corresponding empty vector (Mock). Cell lysates were subjected to immunoprecipitation with agarose-conjugated antibodies to FLAG, and the precipitated proteins were eluted with the FLAG peptide. The eluted proteins, as well as the original cell lysates (Input), were subjected to immunoblot analysis with the indicated antibodies. (F) NIH 3T3 cells coexpressing GFP-tagged and FLAG-tagged forms of hTks5-WT or the indicated phenylalanine or glutamate substitution mutants were subjected to immunoprecipitation with antibodies to GFP. The resulting precipitates, as well as the original cell lysates (Input), were then subjected to immunoblot analysis with the indicated antibodies. See also Fig. S2 A. (G, top) Protocol for virus infection during osteoclastogenesis from primary spleen macrophages. (bottom) The infected osteoclasts were subjected to semiquantitative RT-PCR analysis to confirm the expression of hTks5 constructs. (H) Representative images of virus-infected (ZsGreen1 positive) osteoclasts differentiated from either WT or Src−/− spleen macrophages. The cells were stained with rhodamine-phalloidin (red), antibodies to Tks5, and DAPI (green). Bars, 50 µm. (I) Quantification of the number of nuclei per cell in multinucleated osteoclasts with ZsGreen1 expression. Means ± SD from three independent experiments are indicated. *, P < 0.05 versus Src−/− cells with the vector (Student’s t test). See also Fig. S2 B and Videos 8 and 9. IP, immunoprecipitation.
Induction of Tks5 potentiates the invasion activity of the cells
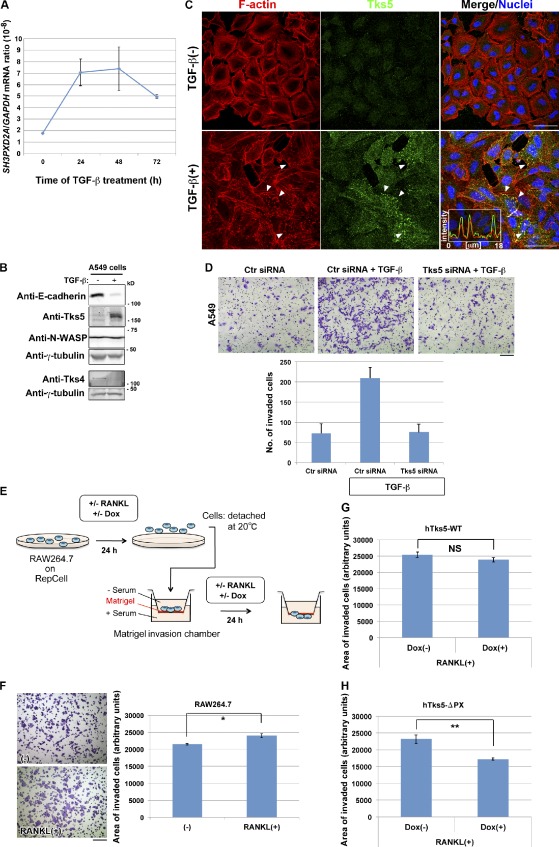
The amounts of Tks5 mRNA and protein were increased by exposure of A549 human pulmonary carcinoma cells to TGF-β, a potent inducer of the epithelial–mesenchymal transition (Fig. 5, A and B). This up-regulation of Tks5 expression was associated with the formation of invadopodia-like structures that accumulated Tks5 (Fig. 5 C and Video 10) and was required for the TGF-β–induced increase in invasive activity (Fig. 5 D and Fig. S3 A), as it has been shown to be for the invasive activity of other types of cancer cells (Seals et al., 2005) or Src-transformed fibroblasts (Oikawa et al., 2008). Similarly, exposure of RAW264.7 macrophages to RANKL for 24 h before transfer to a Matrigel chamber (Fig. 5 E) increased the invasive activity of these cells (Fig. 5 F). Expression of the ΔPX mutant of hTks5, but not that of hTks5-WT, suppressed the RANKL-induced increase in invasive activity (Fig. 5, G and H), mirroring the effects of these proteins on osteoclast fusion. These results prompted us to test the hypothesis that cell–cell fusion can be driven by an increased invasive activity of cells associated with Tks5 up-regulation or activation and the formation of fusion-competent protrusions related to podosomes or invadopodia.
Figure 5.
Induction of Tks5 is required for the promotion of invasive activity. (A and B) A549 pulmonary carcinoma cells cultured in the presence of 5 ng/ml TGF-β for the indicated times (A) or 72 h (B) were subjected to quantitative RT-PCR analysis of SH3PXD2A mRNA (A) or to immunoblot analysis with the indicated antibodies (B). Quantitative data are means ± SD from three independent experiments. (C) Representative images of A549 cells cultured in the absence or presence of TGF-β for 48 h. The cells were stained with rhodamine-phalloidin to visualize F-actin, with antibodies to Tks5, and with DAPI to visualize nuclei. Arrowheads indicate invadopodia-like structures showing Tks5 accumulation. Pixel intensities on the traversing dashed line were measured using LAS AF software and shown in a boxed area (arbitrary units). Bars, 50 µm. See also Video 10 for live imaging. (D) A549 cells transfected with control (Ctr) or Tks5 siRNAs were replated in Matrigel chambers and assayed for invasive activity in the absence or presence of TGF-β. Cells that had invaded through the Matrigel at 18 h after plating were stained with crystal violet (top), and those in three different sampling areas were counted. Bar, 200 µm. Quantitative data are means ± SD from three independent experiments. See also Fig. S3 A for immunoblot. (E) Experimental protocol for Matrigel invasion assays with RAW264.7 macrophages. (F–H) Control RAW264.7 macrophages (F) or those harboring Dox-sensitive constructs for hTks5-WT (G) or hTks5-ΔPX (H) were subjected to invasion assays as in E. Cells that invaded through the Matrigel were stained with crystal violet (F), and the area occupied by the invaded cells in three different sampling regions in a single chamber was quantified. Bar, 200 µm. The minus sign indicates no stimulus added to the culture. Data are means ± SEM for six independent experiments. *, P < 0.02; **, P < 0.002 (Student’s t test).
Circumferential podosomes/invadopodia potentiate fusion activity and invasion activity
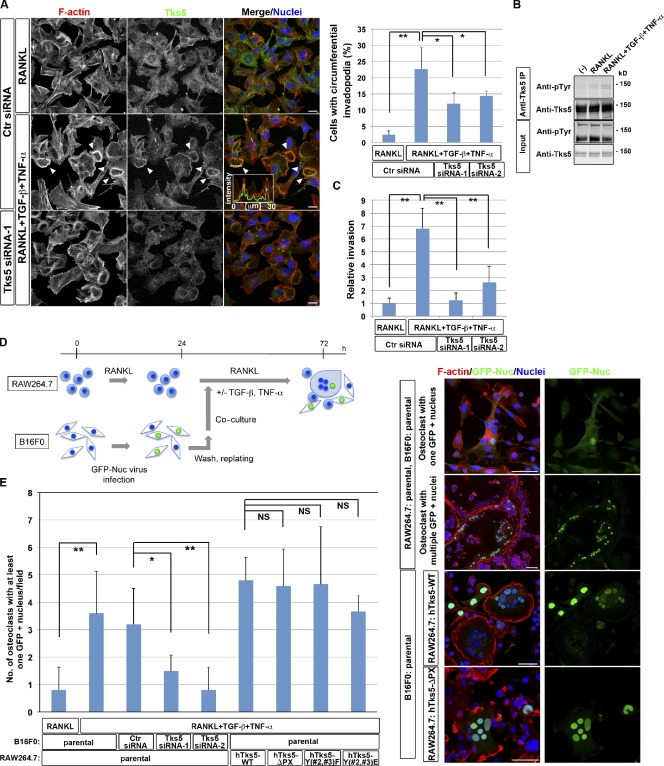
Among the cancer cells tested, we found that simultaneous stimulation of B16F0 murine melanoma cells with RANKL, TGF-β, and TNF-α induced the formation of circumferential invadopodia depending on Tks5 (Fig. 6 A and Fig. S3 B) as well as the tyrosine phosphorylation of Tks5 (Fig. 6 B). Furthermore, such stimulation increased the invasive activity of B16F0 cells, which also depended on Tks5 expression (Fig. 6 C). When RAW264.7 macrophages were co-cultured with B16F0 melanoma cells expressing GFP tagged with a nuclear localization sequence (GFP-Nuc), GFP-positive nuclei were detected in multinuclear osteoclasts (Fig. 6 D). The number of osteoclasts harboring B16F0 nuclei in normal osteoclastogenic cultures was increased when they were co-cultured in the presence of TGF-β and TNF-α, which was canceled when the expression of Tks5 was suppressed in B16F0 cells (Fig. 6 E). On the other hand, the expression of hTks5-ΔPX in osteoclasts had virtually no effect on heterocellular fusion (Fig. 6, D and E). These data suggest that B16F0 cells have the potential to serve as fusion donors, which require Tks5 expression, whereas osteoclasts consist of both donors and acceptors. This finding is consistent with the results of previous studies showing that chronic inflammation promotes heterocellular fusion in vivo (Johansson et al., 2008; Nygren et al., 2008). Our observations thus indicate that Tks5-mediated circumferential podosomes/invadopodia are able to function as a fusion machinery, and they shed light on the mechanism of osteoclast–cancer cell fusion.
Figure 6.
Circumferential podosomes/invadopodia mediate invasive and fusion activities. (A) B16F0 murine melanoma cells transfected with control (Ctr) or Tks5 siRNAs were cultured in the presence of 10 ng/ml RANKL alone or together with 5 ng/ml TGF-β and 5 ng/ml TNF-α for 48 h. (left) The cells were then stained with rhodamine-phalloidin to visualize F-actin, with antibodies to Tks5, and with DAPI to visualize nuclei. Arrowheads indicate circumferential invadopodia. Pixel intensities on the traversing dashed line were measured using LAS AF software and shown in a boxed area (arbitrary units). Bars, 25 µm. (right) The percentage of cells with circumferential invadopodia among >100 cells scored was also determined. Data are means ± SD from three independent experiments. *, P < 0.05; **, P < 0.001 (Student’s t test). See also Fig. S3 B for immunoblot. (B) B16F0 cells cultured in the presence of 10 ng/ml RANKL alone or together with 5 ng/ml TGF-β and 5 ng/ml TNF-α for 48 h were subjected to immunoprecipitation with antibodies to Tks5, and the resulting precipitates as well as the original cell lysates (Input) were subjected to immunoblot analysis with the indicated antibodies. The minus sign indicates no stimulus added to the culture. (C) B16F0 cells transfected with control (Ctr) or Tks5 siRNAs were cultured in the presence of 10 ng/ml RANKL alone or together with 5 ng/ml TGF-β and 5 ng/ml TNF-α for 48 h. The cells were then replated in Matrigel chambers and assayed for invasive activity in the same conditions. Cells that had invaded through the Matrigel at 24 h after plating were stained with crystal violet, and those in three different sampling areas were counted. Data are means ± SD from three independent experiments. **, P < 0.001 (Student’s t test). (D, left) Experimental protocol for co-culture of B16F0 melanoma cells and osteoclasts derived either from parental RAW264.7 macrophages or from those expressing hTks5-WT, hTks5-ΔPX, hTks5-Y(#2,#3)F, or hTks5-Y(#2,#3)E. See also the Materials and methods section for details. (right) The cultures were then stained with rhodamine-phalloidin (red), antibodies to GFP, and DAPI (blue). Bars, 50 µm. (E) Quantification of the number of osteoclasts harboring at least one GFP-positive nucleus after co-culture with B16F0 cells on an 11 × 22–mm coverslip as in D. Data are means ± SD from more than three independent experiments. *, P < 0.05; **, P < 0.01 (Student’s t test). IP, immunoprecipitation.
Discussion
Microarray analysis of genes whose expression is up-regulated during osteoclastogenesis followed by an in silico database search identified several molecules, including Tks5, as candidates for mediators of fusion downstream of PI 3-kinase. Transcription of the Tks5 gene might be directly induced by NFATc1 given that both the human and mouse genes (SH3PXD2A and Sh3pxd2a, respectively) contain a conserved NFATc1 recognition sequence located 1,238 and 1,163 bp upstream of the initiation codon, respectively (unpublished data). Knockdown of Tks5 in osteoclasts resulted in impaired formation of circumferential podosomes and cell–cell fusion, whereas polarized membrane extensions seemed to be unaffected. These observations thus suggested that circumferential podosomes may be required for osteoclast fusion.
Tks5 is expressed in invasive human cancer cells, including breast cancer and melanoma (Seals et al., 2005). Furthermore, such expression tends to correlate with malignancy, as observed during the epithelial–mesenchymal transition of cancer cells (Fig. 5) and in human tumor lesions (Seals et al., 2005), suggesting that Tks5 promotes cell invasion. Thus, osteoclast fusion might be a manifestation of the invasive properties of the cells. Indeed, the ΔPX mutant of Tks5 suppressed both osteoclast fusion and invasion into Matrigel, with deletion of the PX domain altering the interactions of Tks5 with other proteins.
The dynamic nature of circumferential podosomes distinguishes them from more stable structures, such as the sealing belts of mature osteoclasts, and this difference is likely reflected in their molecular composition and regulation. Although previous studies have suggested that podosome formation per se is not required for osteoclast fusion (Chellaiah et al., 2000; Tehrani et al., 2006), these studies did not distinguish circumferential podosomes and sealing belts. For example, RNAi-mediated depletion of cortactin in osteoclasts resulted in the loss of sealing belts without an effect on multinucleation (Tehrani et al., 2006). Cortactin is a substrate for Src and functions as a regulator of the cortical actin cytoskeleton in various cellular processes (Ammer and Weed, 2008). The expression of cortactin is induced in the final phase of osteoclastogenesis (Tehrani et al., 2006), suggesting that it does not affect the formation of fusion-competent protrusions but rather that it contributes to the stabilization of sealing belts by binding to and cross-linking actin filaments of fused osteoclasts. Osteoclasts from gelsolin knockout mice also appear to fuse normally without the formation of sealing belts (Chellaiah et al., 2000), suggesting that gelsolin is not required for the formation of fusion-competent protrusions. Indeed, our mass spectrometric analysis revealed that gelsolin interacts with hTks5-WT (inactive) but not with hTks5-ΔPX. Thus, gelsolin presumably functions in maturation of sealing belts in fused osteoclasts. Consistently, knockdown of Src in RAW264.7 cell–derived osteoclasts results not only in disruption of podosomes but in a reduced rate of cell–cell fusion (Winograd-Katz et al., 2011), as we observed with osteoclasts derived from Src−/− mice.
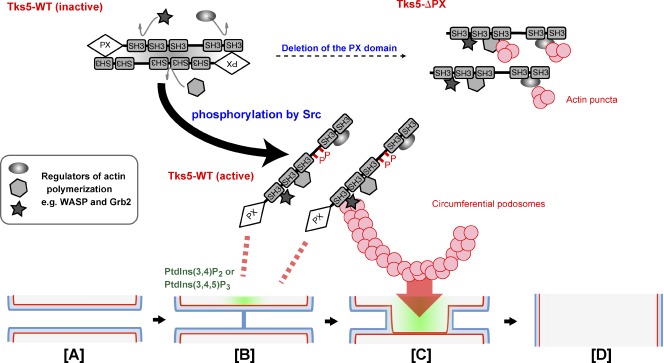
We propose a model for cell–cell fusion based on the Tks5-dependent formation of circumferential podosomes/invadopodia, depicted along with the classical model for membrane fusion via rupture of a hemifusion diaphragm (Fig. 7, [A]–[D]). Tks5 localizes to and is activated by PtdIns(3,4)P2 or PtdIns(3,4,5)P3 (Fig. 7 [B]). Interaction with these PIs as well as phosphorylation by Src induces Tks5 to adopt an “open” conformation that facilitates interaction with other proteins and allows Tks5 to serve as a molecular hub for regulators of actin polymerization, such as WASP and Grb2. The assembly of such a complex then promotes the formation of circumferential podosomes/invadopodia (Fig. 7 [C]).
Figure 7.
Model for Tks5 activation and cell–cell fusion mediated by circumferential podosomes/invadopodia. Tks5 is activated by binding to PtdIns(3,4)P2 or PtdIns(3,4,5)P3 through its PX domain as well as by phosphorylation by Src, resulting in the adoption of an “open” conformation amenable to interaction with other proteins. Activated Tks5 then promotes the formation of circumferential podosomes/invadopodia. Membranes thus are brought into close proximity [A], and the two outer leaflets (blue) fuse to form a hemifusion stalk [B]. The hemifusion stalk then expands to form a hemifusion diaphragm, with the inner leaflets (red) being in contact [C]. Rupture of the hemifusion diaphragm results in formation of the fusion pore [D]. Production of PtdIns(3,4)P2 or PtdIns(3,4,5)P3 (green) may trigger the latter steps by inducing the Tks5-dependent formation of circumferential podosomes/invadopodia.
How does a circumferential podosome/invadopodium mediate cell–cell fusion? Membrane fusion during various biological processes requires tethering of the apposing membranes followed by their destabilization and fusion without leakage of vesicular or cytosolic contents (Wickner and Schekman, 2008). Tks5-dependent circumferential podosomes in osteoclasts or invadopodia in cancer cells may allow stable apposition of the physically unstable membrane microdomains, such as those of high curvature, and thereby facilitate formation of the hemifusion stalk and hemifusion diaphragm (Fig. 7, [B] and [C]). Macrophage- or osteoclast-specific membrane proteins, such as macrophage fusion receptor (Saginario et al., 1998), DC-STAMP (Yagi et al., 2005), and OC-STAMP (Yang et al., 2008), may minimize the energy of hydration repulsion between the apposing membrane leaflets by mediating local tethering of the membranes. The expansion of circumferential podosomes by activated Tks5 might be linked to cell–cell fusion, in particular, to opening of the fusion pore (Fig. 7 [D]; Engel and Walter, 2008). In this regard, B16F0 melanoma cells appeared similar to osteoclasts in their ability to form circumferential invadopodia. Of the cancer cell lines tested, B16F0 was the only one that generated circumferential invadopodia in addition to dot-shaped invadopodia such as those observed in A549 cells. B16F0 cells also underwent fusion with osteoclasts.
Our finding that melanoma cells with circumferential invadopodia fuse with osteoclasts may shed light on the mechanism of cancer cell–osteoclast fusion (Pawelek and Chakraborty, 2008a) that is thought to occur at sites of bone metastasis. More than 30% of osteoclast nuclei from myeloma patients were found to contain translocated chromosomes derived from myeloma cells (Andersen et al., 2007). Given that bone is thought to be rich in active TGF-β (Tang et al., 2009), cancer cells on the bone surface may be stimulated to generate invadopodia, resulting in fusion with osteoclasts. Fusion of cancer cells with osteoclasts may contribute to evasion of immune surveillance, to the provision of a microenvironment for cancer development through the secretion of chemokines, or to up-regulation of bone resorption activity, resulting in osteolytic bone metastasis. Future studies are warranted to characterize the precise mechanism of podosome/invadopodium-mediated fusion as well as the properties of osteoclast–cancer cell hybrids.
Materials and methods
Antibodies
The following antibodies were used for immunofluorescence or immunoblot analysis: rabbit anti-Tks5 (sc-30122) and mouse monoclonal anti-NFATc1 (Santa Cruz Biotechnology, Inc.); mouse monoclonal anti-Grb2 and anti–E-cadherin (BD); rabbit anti-Tks4, anti-Nck, and mouse monoclonal antiphosphotyrosine (4G10; Millipore); rabbit anti-Akt, antiphospho-Akt (S473), and anti-WASP as well as mouse monoclonal anti-HA (Cell Signaling Technology); mouse monoclonal anti–γ-tubulin and anti-FLAG as well as rabbit anti-Abi1 (Sigma-Aldrich); rabbit anti-GFP (MBL International); rabbit anti–neural WASP; and various Alexa Fluor–conjugated secondary antibodies (Invitrogen).
Plasmid construction, mutagenesis, and retroviral gene transduction
All Tks5 constructs were generated by PCR with mouse or human cDNA as the template, and their identity was confirmed by sequencing. Of several transcriptional variants, the longest one (available from GenBank/EMBL/DDBJ under accession no. CAI13962.1) was the one we cloned from MDA-MB231 cells as an hTks5. Site-directed mutagenesis was performed with hTks5 cDNA as a template. Therefore, Y(#2,#3)F and Y(#2,#3)E mutants represent Y(558,620)F and Y(558,620)E, respectively. The cDNAs were cloned into pEGFP-C-1 (Takara Bio Inc.), pCMV-2B (Agilent Technologies), pCMV-HA, or pRetroX-IRES-ZsGreen1 (Takara Bio Inc.) with or without an N-terminal RFP sequence. For inducible expression, cDNA encoding full-length or truncated Tks5 was cloned with an N-terminal FLAG tag with or without an additional RFP sequence into pRetroX-Tight-Pur (Takara Bio Inc.). The cDNAs for the PH domains of PLCδ1 and Akt (Furutani et al., 2006) were also cloned into pRetroX-Tight-Pur. The cDNAs for the PH domains of Tapp1 and Grp1 (Furutani et al., 2006) were cloned into pRetroX-IRES-ZsGreen1 with an N-terminal RFP sequence and pEGFP-C-1, respectively. Retroviruses with the VSV-G envelope were produced by transfection of GP2-293 cells (Takara Bio Inc.) with pRetroX constructs and pVSV-G (Takara Bio Inc.) using Lipofectamine LTX (Invitrogen). mCherry tagged with LifeAct (Riedl et al., 2008) was a gift from S. Kurisu and T. Takenawa (Kobe University, Kobe, Japan) and was used to visualize F-actin in live cells.
Cell culture
RAW264.7 macrophages were cultured under 5% CO2 at 37°C in α-MEM supplemented with 10% FBS and penicillin–streptomycin. Polyclonal cell lines capable of inducible expression of hTks5 or the PH domains of PLCδ1 and Akt were generated by infection of RAW264.7 macrophages with the corresponding retrovirus together with the rtTA retrovirus (Takara Bio Inc.) followed by selection with 5 µg/ml puromycin and 500 µg/ml geneticin (G418). Bone marrow or spleen cells were isolated from C57BL/6J mice or from mice lacking c-Src (Src−/− mice) and were cultured for 6 h to overnight in α-MEM supplemented with 10% FBS for collection of nonadherent cells. For stromal cell–free osteoclast formation, nonadherent cells were plated at a density of 106 per well (bone marrow) or 3 × 106 per well (spleen) in 6-well plates and were cultured for 3 d in α-MEM supplemented with 10% FBS and recombinant human M-CSF (R&D Systems) at 10 ng/ml. The resulting M-CSF–dependent macrophages were used as osteoclast precursors. Osteoclast differentiation was induced by culture for 3–4 d in the presence of recombinant human M-CSF and recombinant mouse RANKL (R&D Systems), each at 10 ng/ml. B16F0 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, and A549 cells were grown in RPMI 1640 medium supplemented with 10% FBS. For inhibition of PI 3-kinase activity or actin polymerization, cells were treated with 10 µM LY294002 (EMD) or 0.1 µg/ml Latrunculin B (Sigma-Aldrich), respectively, for 16 h.
Microarray analysis
Microarray processing was performed by the Core Instrumentation Facility of the Keio University School of Medicine. Total RNA was isolated from RAW264.7 macrophages upon the indicated treatments using TRIZOL reagent (Invitrogen) following the manufacturer’s protocol and further purified with the RNeasy Mini kit (QIAGEN). RNA was quantified using a spectrophotometer (NanoDrop 1000; Thermo Fisher Scientific), and quality was monitored with a bioanalyzer (2100 Bioanalyzer; Agilent Technologies). RNA had an A260/280 ratio ≥2; RNA integrity number was ≥6.5. Cy3-labeled cRNA probes were prepared using a labeling kit (Quick Amp; Agilent Technologies) according to the manufacturer’s instructions. 1,650 ng Cy3-labeled cRNA was hybridized to a Whole Mouse Genome Microarray kit (4 × 44,000; design ID 014868; Agilent Technologies). Raw intensity data for each experiment were analyzed with GeneSpring GX software (Agilent Technologies).
RNAi
For siRNA transfection, RAW264.7 macrophages, B16F0 cells, or A549 cells were plated at 15–20% confluence in 6-well plates, cultured overnight, and then incubated for 6 h in the presence of 160 pmol siRNA and 4 µl RNAiMAX (Invitrogen) in Opti-MEM (Invitrogen). They were then cultured in complete medium for 18 h before a second round of siRNA transfection. The RAW264.7 macrophages, B16F0 cells or A549 cells were then cultured with 10 ng/ml RANKL, 5 ng/ml recombinant human TGF-β (PeproTech), and/or 5 ng/ml recombinant human TNF-α (R&D Systems) and analyzed. Stealth siRNAs (Invitrogen) targeted to mouse (5′-CAGUGUGACGAAGUCUUCCGGUUCU-3′ or 5′-CCAUCACCAAGAAAGAGAUCAGCUU-3′) or human (5′-AUCCUCUGAAGACUCACCCACCUUG-3′) Tks5 mRNAs were used for RNAi experiments. The control siRNA sequences were designed by scrambling the targeted siRNA sequences.
RT-PCR analysis
Total RNA was extracted from cells with the use of the TRIZOL reagent (Invitrogen), and portions (0.5–1 µg) of the RNA were subjected to RT with polymerase (SuperScript II; Invitrogen). RT-PCR primers (TaqMan) for Sh3pxd2a, Nfatc1, Gapdh, Actb, SH3PXD2A, and GAPDH were obtained from Applied Biosystems for quantitative PCR. The abundance of target mRNAs was normalized by that of mRNAs derived from the housekeeping genes Gapdh, Actb, or GAPDH. Quantitative PCR was performed with the use of a real-time PCR system (7500 Fast; Applied Biosystems). The primers (forward and reverse, respectively) for semiquantitative RT-PCR analysis included those for SH3PXD2A, 5′-CTCCGAGGCTCCCAGTGA-3′ and 5′-GCCTGCCCTTGGCGGGCA-3′; Sh3pxd2a, 5′-CAGTCCTGCATTTCTGACGG-3′ and 5′-GTTTTTCACGGCAAGGGACA-3′; and Actb, 5′-CTCTTTGATGTCACGCACGAT-3′ and 5′-GTGGGCCGCTCTAGGCACCAA-3′. TaqMan RT-PCR primers for Mmp9, Itgb3, and Tm7sf4 (DC-STAMP gene) were also obtained from Applied Biosystems for semiquantitative RT-PCR analysis.
Matrigel invasion assay
RAW264.7 macrophages (1.0 × 105 cells) cultured on RepCell (CellSeed, Inc.) dishes with or without 10 ng/ml RANKL or 1 µg/ml Dox for 24 h were replated in a Matrigel invasion chamber (BioCoat; BD) and cultured for 24 h under the same conditions. B16F0 cells (2.0 × 105 cells) cultured on normal dishes with 10 ng/ml RANKL or with 10 ng/ml RANKL, 5 ng/ml TGF-β, and 5 ng/ml TNF-α for 48 h were isolated by exposure to trypsin, replated in a BioCoat Matrigel invasion chamber, and cultured for 24 h under the same conditions. A549 cells (5.0 × 104 cells) cultured on normal dishes with or without 5 ng/ml TGF-β for 24 h were isolated by exposure to trypsin, replated in a BioCoat Matrigel invasion chamber, and cultured for 24 h under the same conditions. All cells were then fixed with 3.7% formaldehyde in PBS for 30 min and washed with PBS, and the invaded cells were stained with crystal violet. After washing the cells with PBS at least five times, cells in three different regions on the lower surface of the filter were either counted or had their area quantified with the use of ImageJ software (National Institutes of Health).
Co-culture
RAW264.7 macrophages harboring Dox-inducible hTks5 constructs or parental RAW264.7 macrophages were plated at a density of 3 × 105 per well in 6-well plates and cultured in the presence of 10 ng/ml RANKL with or without 1 µg/ml Dox for 24 h, respectively, before co-culture. The B16F0 cells were infected with a virus for GFP tagged with a nuclear localization signal (GFP-Nuc; BacMam 2.0; Invitrogen) for 15–18 h followed by extensive washing with PBS. They were then trypsinized and plated at a density of 2 × 105 per well in 6-well plates with RAW264.7 macrophages in the presence of 10 ng/ml RANKL with or without 1 µg/ml Dox, 5 ng/ml TGF-β, and 5 ng/ml TNF-α for 48 h.
Immunofluorescence analysis and TRAP staining
Cells cultured on coverslips were fixed with 3.7% formaldehyde in PBS or 2% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and then incubated with primary antibodies for ≥60 min. They were then washed with PBS and incubated with Alexa Fluor (488, 568, 594, 633, or 647)–conjugated secondary antibodies for 30 min. Cells were also stained with rhodamine-phalloidin (Invitrogen) to detect F-actin. The cells finally were washed with PBS and mounted onto slides using mounting medium (PermaFluor; Thermo Fisher Scientific). For TRAP staining, differentiated osteoclasts were fixed with 4% paraformaldehyde in PBS and with 1:1 (vol/vol) ethanol/acetone before detection of TRAP activity in the presence of 20 mM tartrate with the use of a kit (Leukocyte Acid Phosphatase (TRAP) Kit; Sigma-Aldrich).
Image acquisition and processing
All photographic images of cells were acquired using microscopes and corresponding confocal microscopy systems with software, including DMI 6000 with TCS SP5 and LAS AF software (Leica), Axiovert 200M with LSM 510 META and LSM software (Carl Zeiss), and FluoView FV10i with FluoView software (Olympus). A 20× objective with an NA of 0.7 (Leica), a 40× oil immersion objective with an NA of 1.25 (Leica), a 63× oil immersion objective with an NA of 1.4 (Leica), a 60× water immersion objective with an NA of 1.35 (Olympus), or a 63× oil immersion objective with an NA of 1.4 (Carl Zeiss) was used. Videos were taken under 5% CO2 at 37°C. Acquired images were processed with Photoshop (Adobe). Gels or blots were scanned with a densitometer (GS-800; Bio-Rad Laboratories) and analyzed with ImageJ software. Images were assembled with Photoshop. For each plate, fluorescence photographs were cropped, and each fluorochrome was adjusted identically for brightness and contrast to represent the observed images.
Immunoprecipitation, silver staining, and mass spectrometry
RAW264.7 macrophages with Dox-sensitive FLAG–hTks5-WT or FLAG–hTks5-ΔPX construct were incubated with 10 ng/ml RANKL for 72 h and in the additional presence of 1 µg/ml Dox for 24 h. They were then washed with ice-cold PBS and lysed with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitors and phosphatase inhibitors (Sigma-Aldrich). The cell lysates were subjected to immunoprecipitation with agarose bead–conjugated M2 antibodies to FLAG (Sigma-Aldrich), and bead-bound proteins were eluted with the FLAG peptide (Sigma-Aldrich) at 100 µg/ml and were analyzed by SDS-PAGE and silver staining with a kit (Dodeca; Bio-Rad Laboratories). The eluted proteins were also digested with trypsin, desalted with the use of a C18 device (ZipTip; Millipore), concentrated, and injected into a direct nanoflow liquid chromatography system (DiNa; KYA Technologies) coupled to a quadruple time-of-flight tandem mass spectrometer (QSTAR Elite; AB SCIEX) as described previously (Oyama et al., 2009). In brief, after applying the peptide mixture to a C18 column (800-µm inner diameter × 3 mm long), reversed-phase separation of the captured peptides was performed on a column (150-µm inner diameter × 75 mm long) filled with C18 (3-µm particles, 120-Å pores; HiQ sil; KYA Technologies) using DiNa. The peptides were eluted with a linear 5–65% gradient of acetonitrile containing 0.1% formic acid over 120 min at a flow rate of 200 nl/min and sprayed into QSTAR Elite. The mass spectrometric signals were compared with the RefSeq (National Center for Biotechnology Information) mouse protein database (36,417 sequences, as of July 5 2010) with the use of the Mascot algorithm (version 2.2.04; Matrix Science) and with the following parameters: variable modifications of oxidation (Met), acetylation (protein N terminus), pyroglutamination (Gln), and phosphorylation (Ser, Thr, and Tyr); two maximum missed cleavages; peptide mass tolerance of 200 ppm; and tandem mass spectrometry tolerance of 0.5 D. Protein identification was based on the criterion of at least one tandem mass spectrometry data with a Mascot score greater than the threshold (P < 0.05).
Statistics
Quantitative data are presented as means ± SD or means ± SEM and were compared with Student’s t test. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the effect of Tks5 knockdown on migration speed, establishment of inducible hTks5 cell lines, and multisubunit complex formation of Tks5. Fig. S2 shows the interaction of Tks5 with Nck and establishment of inducible RFP-tagged hTks5 mutant cell lines. Fig. S3 shows immunoblot analysis of Tks5 knockdown in A549 cells or B16F0 cells. Video 1 shows the localization of PIs during podosome-mediated cell–cell fusion. Video 2 shows the localization of PIs during filopodium-mediated cell–cell fusion. Video 3 shows the localization of PIs during cell–cell fusion with long projections. Video 4 shows the localization of PtdIns(3,4)P2 or PtdIns(3,4,5)P3 during cell–cell fusion. Video 5 shows the localization of Tks5-WT during cell–cell fusion. Video 6 shows the localization of Tks5-ΔPX during cell–cell fusion. Video 7 shows the localization of PIs during cell–cell fusion of osteoclasts with reduced Tks5 expression. Video 8 shows the localization of Tks5-Y(#2,#3)F during cell–cell fusion. Video 9 shows the localization of Tks5-Y(#2,#3)E during cell–cell fusion. Video 10 shows the localization of Tks5 in invadopodia-like structures of the A549 cell. Table S1 shows a list of genes whose expression was up-regulated in RAW264.7 macrophages treated with RANKL for 72 h. Table S2 shows a list of proteins with PX or PH domains among those encoded by the genes listed in Table S1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111116/DC1.
Acknowledgments
We thank Drs. S. Kurisu and T. Takenawa for the LifeAct construct and A. Oide, A. Nakamura, and M. Fujiwara (Core Instrumentation Facility, Keio University School of Medicine) for technical assistance.
This work was supported by the Promotion of Environmental Improvement for Independence of Young Researchers (Kanrinmaru Project) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a Grant-in-Aid for Young Scientists (A; grant 23689020) from the Ministry of Education, Culture, Sports, Science, and Technology, Takeda Science Foundation, and a Keio University Grant-in-Aid for Encouragement of Young Medical Scientists.
Footnotes
Abbreviations used in this paper:
- Dox
- doxycycline
- hTks5
- human Tks5
- M-CSF
- macrophage colony-stimulating factor
- PH
- pleckstrin homology
- PI
- phosphoinositide
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PX
- phox homology
- RANKL
- receptor activator of NF-κB ligand
- TRAP
- tartrate-resistant acid phosphatase
- WT
- wild type
References
- Abram C.L., Seals D.F., Pass I., Salinsky D., Maurer L., Roth T.M., Courtneidge S.A. 2003. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J. Biol. Chem. 278:16844–16851 10.1074/jbc.M300267200 [DOI] [PubMed] [Google Scholar]
- Ammer A.G., Weed S.A. 2008. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton. 65:687–707 10.1002/cm.20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen T.L., Boissy P., Sondergaard T.E., Kupisiewicz K., Plesner T., Rasmussen T., Haaber J., Kølvraa S., Delaissé J.M. 2007. Osteoclast nuclei of myeloma patients show chromosome translocations specific for the myeloma cell clone: a new type of cancer-host partnership? J. Pathol. 211:10–17 10.1002/path.2078 [DOI] [PubMed] [Google Scholar]
- Andersen T.L., Søe K., Sondergaard T.E., Plesner T., Delaisse J.M. 2010. Myeloma cell-induced disruption of bone remodelling compartments leads to osteolytic lesions and generation of osteoclast-myeloma hybrid cells. Br. J. Haematol. 148:551–561 10.1111/j.1365-2141.2009.07980.x [DOI] [PubMed] [Google Scholar]
- Buschman M.D., Bromann P.A., Cejudo-Martin P., Wen F., Pass I., Courtneidge S.A. 2009. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol. Biol. Cell. 20:1302–1311 10.1091/mbc.E08-09-0949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camussi G., Deregibus M.C., Bruno S., Cantaluppi V., Biancone L. 2010. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 78:838–848 10.1038/ki.2010.278 [DOI] [PubMed] [Google Scholar]
- Chellaiah M., Kizer N., Silva M., Alvarez U., Kwiatkowski D., Hruska K.A. 2000. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J. Cell Biol. 148:665–678 10.1083/jcb.148.4.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.C., Antón I.M., Holt M.R., Curcio C., Lanzardo S., Worth A., Burns S., Thrasher A.J., Jones G.E., Calle Y. 2006. WIP regulates the stability and localization of WASP to podosomes in migrating dendritic cells. Curr. Biol. 16:2337–2344 10.1016/j.cub.2006.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Sanjay A., Itzstein C., Horne W.C., Toomre D., De Camilli P., Baron R. 2008. The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol. Biol. Cell. 19:394–404 10.1091/mbc.E07-03-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz B., Shani G., Pass I., Anderson D., Quintavalle M., Courtneidge S.A. 2009. Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci. Signal. 2:ra53 10.1126/scisignal.2000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A., Walter P. 2008. Membrane lysis during biological membrane fusion: collateral damage by misregulated fusion machines. J. Cell Biol. 183:181–186 10.1083/jcb.200805182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M., Tsujita K., Itoh T., Ijuin T., Takenawa T. 2006. Application of phosphoinositide-binding domains for the detection and quantification of specific phosphoinositides. Anal. Biochem. 355:8–18 10.1016/j.ab.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Gianni D., Diaz B., Taulet N., Fowler B., Courtneidge S.A., Bokoch G.M. 2009. Novel p47(phox)-related organizers regulate localized NADPH oxidase 1 (Nox1) activity. Sci. Signal. 2:ra54 10.1126/scisignal.2000370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo P., Guadamillas M.C., Hernández-Riquer M.V., Pollán A., Grande-García A., Bartolomé R.A., Vasanji A., Ambrogio C., Chiarle R., Teixidó J., et al. 2010. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev. Cell. 18:77–89 10.1016/j.devcel.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K., Kimura S., Takatsu H., Ohmae M., Kawano S., Kitamura H., Ito M., Watarai H., Hazelett C.C., Yeaman C., Ohno H. 2009. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol. 11:1427–1432 10.1038/ncb1990 [DOI] [PubMed] [Google Scholar]
- Johansson C.B., Youssef S., Koleckar K., Holbrook C., Doyonnas R., Corbel S.Y., Steinman L., Rossi F.M., Blau H.M. 2008. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat. Cell Biol. 10:575–583 10.1038/ncb1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurdic P., Saltel F., Chabadel A., Destaing O. 2006. Podosome and sealing zone: specificity of the osteoclast model. Eur. J. Cell Biol. 85:195–202 10.1016/j.ejcb.2005.09.008 [DOI] [PubMed] [Google Scholar]
- Lakkakorpi P.T., Nakamura I., Young M., Lipfert L., Rodan G.A., Duong L.T. 2001. Abnormal localisation and hyperclustering of αVβ3 integrins and associated proteins in Src-deficient or tyrphostin A9-treated osteoclasts. J. Cell Sci. 114:149–160 [DOI] [PubMed] [Google Scholar]
- Lee S.H., Rho J., Jeong D., Sul J.Y., Kim T., Kim N., Kang J.S., Miyamoto T., Suda T., Lee S.K., et al. 2006. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 12:1403–1409 10.1038/nm1514 [DOI] [PubMed] [Google Scholar]
- Linder S., Nelson D., Weiss M., Aepfelbacher M. 1999. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. USA. 96:9648–9653 10.1073/pnas.96.17.9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock P., Abram C.L., Gibson T., Courtneidge S.A. 1998. A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 17:4346–4357 10.1093/emboj/17.15.4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K., Galson D.L., Zhao C., Peng L., Laplace C., Wang K.Z., Bachler M.A., Amano H., Aburatani H., Ishikawa H., Wagner E.F. 2004. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 279:26475–26480 10.1074/jbc.M313973200 [DOI] [PubMed] [Google Scholar]
- Mbalaviele G., Chen H., Boyce B.F., Mundy G.R., Yoneda T. 1995. The role of cadherin in the generation of multinucleated osteoclasts from mononuclear precursors in murine marrow. J. Clin. Invest. 95:2757–2765 10.1172/JCI117979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.A., Diaz B., Bromann P.A., Tsai J.H., Kawakami Y., Maurer J., Stewart R.A., Izpisúa-Belmonte J.C., Courtneidge S.A. 2011. A Src-Tks5 pathway is required for neural crest cell migration during embryonic development. PLoS ONE. 6:e22499 10.1371/journal.pone.0022499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi-Koga T., Takayanagi H. 2009. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol. Rev. 231:241–256 10.1111/j.1600-065X.2009.00821.x [DOI] [PubMed] [Google Scholar]
- Nygren J.M., Liuba K., Breitbach M., Stott S., Thorén L., Roell W., Geisen C., Sasse P., Kirik D., Björklund A., et al. 2008. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat. Cell Biol. 10:584–592 10.1038/ncb1721 [DOI] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., Takenawa T. 2008. Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 182:157–169 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M., Kozuka-Hata H., Tasaki S., Semba K., Hattori S., Sugano S., Inoue J., Yamamoto T. 2009. Temporal perturbation of tyrosine phosphoproteome dynamics reveals the system-wide regulatory networks. Mol. Cell. Proteomics. 8:226–231 [DOI] [PubMed] [Google Scholar]
- Pawelek J.M., Chakraborty A.K. 2008a. The cancer cell—leukocyte fusion theory of metastasis. Adv. Cancer Res. 101:397–444 10.1016/S0065-230X(08)00410-7 [DOI] [PubMed] [Google Scholar]
- Pawelek J.M., Chakraborty A.K. 2008b. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat. Rev. Cancer. 8:377–386 10.1038/nrc2371 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring C., Ginsberg M.H., Haling J., Pendergast A.M. 2011. Abl-interactor-1 (Abi1) has a role in cardiovascular and placental development and is a binding partner of the alpha4 integrin. Proc. Natl. Acad. Sci. USA. 108:149–154 10.1073/pnas.1012316108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saginario C., Sterling H., Beckers C., Kobayashi R., Solimena M., Ullu E., Vignery A. 1998. MFR, a putative receptor mediating the fusion of macrophages. Mol. Cell. Biol. 18:6213–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltel F., Chabadel A., Bonnelye E., Jurdic P. 2008. Actin cytoskeletal organisation in osteoclasts: a model to decipher transmigration and matrix degradation. Eur. J. Cell Biol. 87:459–468 10.1016/j.ejcb.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F., Jr, Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., Courtneidge S.A. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 7:155–165 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Sens K.L., Zhang S., Jin P., Duan R., Zhang G., Luo F., Parachini L., Chen E.H. 2010. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J. Cell Biol. 191:1013–1027 10.1083/jcb.201006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P., Montgomery C., Geske R., Bradley A. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 64:693–702 10.1016/0092-8674(91)90499-O [DOI] [PubMed] [Google Scholar]
- Stylli S.S., Stacey T.T., Verhagen A.M., Xu S.S., Pass I., Courtneidge S.A., Lock P. 2009. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 122:2727–2740 10.1242/jcs.046680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., et al. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell. 3:889–901 10.1016/S1534-5807(02)00369-6 [DOI] [PubMed] [Google Scholar]
- Tang Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T.R., Peng X., Hu J., et al. 2009. TGF-β1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 15:757–765 10.1038/nm.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S., Faccio R., Chandrasekar I., Ross F.P., Cooper J.A. 2006. Cortactin has an essential and specific role in osteoclast actin assembly. Mol. Biol. Cell. 17:2882–2895 10.1091/mbc.E06-03-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi S. 2006. A complex of Wiskott-Aldrich syndrome protein with mammalian verprolins plays an important role in monocyte chemotaxis. J. Immunol. 176:6576–6585 [DOI] [PubMed] [Google Scholar]
- Wickner W., Schekman R. 2008. Membrane fusion. Nat. Struct. Mol. Biol. 15:658–664 10.1038/nsmb.1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd-Katz S.E., Brunner M.C., Mirlas N., Geiger B. 2011. Analysis of the signaling pathways regulating Src-dependent remodeling of the actin cytoskeleton. Eur. J. Cell Biol. 90:143–156 10.1016/j.ejcb.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., et al. 2005. DC-STAMP is essential for cell–cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 202:345–351 10.1084/jem.20050645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Birnbaum M.J., MacKay C.A., Mason-Savas A., Thompson B., Odgren P.R. 2008. Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation. J. Cell. Physiol. 215:497–505 10.1002/jcp.21331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Patrick Ross F. 2007. Mechanisms of osteoclastic secretion. Ann. NY Acad. Sci. 1116:238–244 10.1196/annals.1402.058 [DOI] [PubMed] [Google Scholar]