Mutation of a critical residue of fascin eliminates the protein’s actin-bundling activity but maintains its positive role in filopodia formation
Abstract
Fascin is an evolutionarily conserved actin-binding protein that plays a key role in forming filopodia. It is widely thought that this function involves fascin directly bundling actin filaments, which is controlled by an N-terminal regulatory serine residue. In this paper, by studying cellular processes in Drosophila melanogaster that require fascin activity, we identify a regulatory residue within the C-terminal region of the protein (S289). Unexpectedly, although mutation (S289A) of this residue disrupted the actin-bundling capacity of fascin, fascin S289A fully rescued filopodia formation in fascin mutant flies. Live imaging of migrating macrophages in vivo revealed that this mutation restricted the localization of fascin to the distal ends of filopodia. The corresponding mutation of human fascin (S274) similarly affected its interaction with actin and altered filopodia dynamics within carcinoma cells. These data reveal an evolutionarily conserved role for this regulatory region and unveil a function for fascin, uncoupled from actin bundling, at the distal end of filopodia.
Introduction
Filopodia are dynamic membrane extensions generated by bundles of F-actin that are involved in numerous biological processes (Wood and Martin, 2002; Mattila and Lappalainen, 2008). However, the molecular mechanisms behind filopodia formation are still not well understood. One of the most widely studied proteins critical for the formation of filopodia is the actin-binding protein fascin (Vignjevic et al., 2006). In the current model of filopodia formation, called convergent elongation, initiation begins with the coalescing of branched actin filaments through a filopodial tip complex; subsequently, fascin is thought to convert these clustered filaments into a stable, bundled filopodia (Gupton and Gertler, 2007; Faix et al., 2009). This role of fascin in filopodia promotes cell migration in vitro (Adams, 2004) and during development (Zanet et al., 2009), and high fascin expression is correlated with invasion and metastasis of various tumors (Hashimoto et al., 2005). Despite this clinical relevance, the molecular mechanisms regulating fascin activity are not well understood. Fascin is thought to contain two actin-binding sites at either ends of the protein; the N-terminal site is best characterized and inhibited by phosphorylation of a conserved serine (S39 in humans) by PKC (Ono et al., 1997; Hashimoto et al., 2007). Phosphorylation of this residue in cells in vitro affects cell migration, filopodia formation, and as shown more recently, invadopodia dynamics (Vignjevic et al., 2006; Hashimoto et al., 2007; Li et al., 2010). However, the roles and regulation of fascin in vivo are not well understood.
The function of Drosophila melanogaster fascin, also known as singed, has been well described in adult stages in which it is necessary for bristle formation and female fertility (Cant et al., 1994). In addition, during embryogenesis, macrophages (hemocytes) express high levels of fascin, which is essential for their developmental migration (Zanet et al., 2009). Drosophila macrophage dispersal is therefore a good system to study fascin function and regulation during cell migration, in a model amenable to analysis of cytoskeletal dynamics in vivo (Stramer et al., 2005, 2010; Wood et al., 2006; Siekhaus et al., 2010). Unexpectedly, although phosphorylation of S52 (the equivalent of S39 in human fascin [hfascin]) blocks fascin activity in adult bristles, it does not impair fertility or macrophage dispersal. Furthermore, the unphosphorylatable and phosphomimetic mutations at S52 can equally compensate for the lack of endogenous fascin and fully rescue macrophage filopodia and transient actin bundles during oogenesis (Zanet et al., 2009), supporting the existence of overlooked regulatory mechanisms.
We therefore investigated other residues of the fascin protein that could influence its activity. In Drosophila, a previous genetic screen identified an S289 point mutation that affects fascin function in adult flies (Cant and Cooley, 1996). As this serine is conserved between species and the three human paralogues (Sedeh et al., 2010; Jansen et al., 2011), we investigated the putative regulatory role of this residue during Drosophila development, by replacing endogenous fascin with the unphosphorylatable (S289A) and the phosphomimetic (S289D) mutant forms. These functional assays reveal that, in contrast to fascin S289D, fascin S289A is capable of rescuing fascin mutant flies. However, the S289A mutation impairs fascin localization and induces its stable association to the distal end of filopodial filaments. Intriguingly, despite its ability to fully rescue filopodia formation, this mutation completely disrupts the actin-bundling activity of fascin as confirmed by both in vitro and in vivo assays. Furthermore, we show that this serine residue in hfascin is also playing a key role in filopodial dynamics within carcinoma cells. These data highlight a critical evolutionarily conserved function for this serine residue of fascin and unveil a role for fascin in filopodia uncoupled from its well-characterized actin-bundling activity.
Results and discussion
S289 of fascin is essential for Drosophila macrophage migration and filopodia in vivo
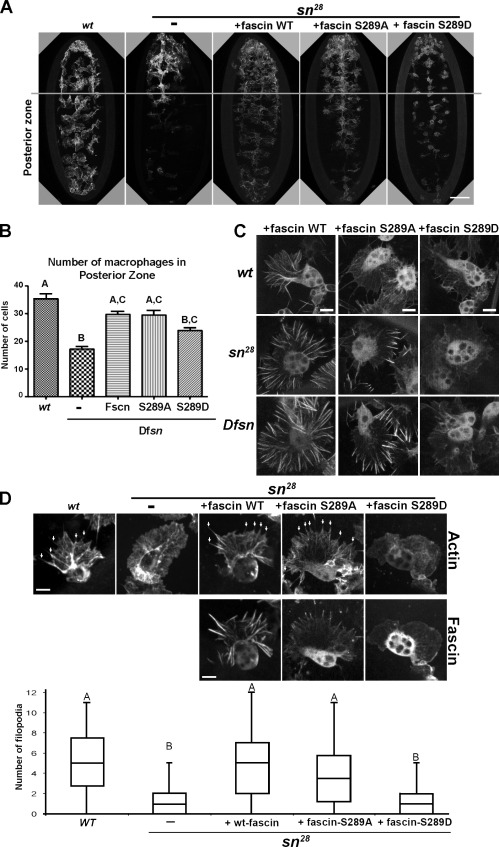
We first generated transgenic flies allowing tissue-specific expression of GFP-tagged fascin (Brand and Perrimon, 1993) containing point mutations at serine S289 (S289A/D). We assayed their ability to rescue macrophage migration in mutant embryos that carry a small deficiency uncovering the fascin locus (Dfsn), which represents a null fascin allele (Zanet et al., 2009). To quantify the migratory behavior, we counted the number of macrophages that dispersed from the head to the posterior 2/3 of the embryo along the ventral surface (Fig. 1 A). In mutant embryos, macrophage migration was reduced, and this defect was rescued by specific expression within macrophages of wild-type or fascin S289A; in contrast, fascin S289D was less efficient at rescuing migration (Fig. 1, A and B). We subsequently investigated the subcellular localization of GFP-tagged fascin variants in macrophages by time-lapse imaging. In the absence of the endogenous protein, fascin S289A showed a striking localization to the periphery of lamellae, whereas S289D remained cytoplasmic (Fig. 1 C and Video 1). Finally, we investigated the ability of the fascin mutant transgenes to rescue filopodia formation in fascin mutant embryos. Despite its altered localization, fascin S289A was able to rescue filopodia number, whereas fascin S289D could not (Fig. 1 D). These experiments reveal that S289 is a regulatory residue that modulates fascin activity during cell migration in vivo.
Figure 1.
Drosophila fascin S289 regulates embryonic macrophage migration. (A) Comparison of macrophage dispersal on the ventral surface of stage 15 embryos in wild-type (wt) and deficiency (Dfsn) mutants expressing GFP-fascin transgenes. The gray line demarcates the anterior one third of the embryo. (B) Bar graph revealing the mean number of macrophages that migrated from the anterior region of the embryo (n = 10 embryos). Letters highlight no significant difference by one-way ANOVA (P < 0.05). Error bars show means ± SD. (C) Wild-type GFP-fascin and GFP-fascin S289A/D were expressed specifically in macrophages in wild-type and singed mutant embryos (sn28 and Dfsn). In the fascin mutants, fascin S289A localized to short filaments at the distal region of lamellae, whereas S289D remained cytoplasmic. (D) Filopodia (arrows) were quantified in wild type, the singed mutant, and singed mutants expressing fascin transgenes using a fluorescent actin probe. Letters highlight no significant difference by one-way ANOVA (P < 0.05). Boxes define the 25th and 75th percentile. The bands in the middle are the medians. The whiskers represent the 1.5 interquartile range. Bars: (A) 50 µm; (C and D) 5 µm. See also Video 1.
Fascin S289A specifically localizes to the distal end of filopodia and rescues microspike formation within lamellae
To further characterize the influence of S289 on fascin localization, GFP-fascin S289A/D were coexpressed with wild-type fascin tagged with mCherry within migrating macrophages. Although fascin S289D remained cytoplasmic, fascin S289A was highly enriched within the distal end of the protrusion when compared with wild-type fascin (Fig. 2, A and B; and Video 2). Interestingly, in human cells, it has been shown that after initial filopodial extension, fascin bundles become incorporated into lamellae through retrograde actin flow (Nemethova et al., 2008). We observed a similar retrograde movement of wild-type fascin bundles in Drosophila macrophages, whereas fascin S289A was lost from the actin filament before incorporation into lamellae (Video 3). The enhanced localization to the distal end of filopodia was maintained throughout its lifetime (Fig. 2 B and Videos 2 and 3). This is in contrast to the reported tip localization of the hfascin S39E mutation in B16F1 melanoma cells, in which binding to the distal ends of filopodia was only transient (Vignjevic et al., 2006).
Figure 2.
Fascin S289A localizes to the distal end of filopodia and rescues filopodia formation. (A) Wild-type mCherry-fascin (purple) and GFP-fascin S289A/D (green) were coexpressed specifically in macrophages of sn28 mutant embryos. Fascin S289A localized to distal ends of filopodia extending beyond the leading edge (arrows) and to nascent filopodial bundles (asterisks). (B) Line scan analysis of both mCherry-fascin (purple) and GFP-fascin S289A (green) within a dynamic filopodia. (C) LifeAct-GFP was expressed in macrophages in wild-type (wt) and sn28 mutant embryos. (D) LifeAct-GFP and mCherry-fascin or mCherry-fascin S289A were specifically expressed in macrophages in sn28 mutant embryos. (E) High magnification of filopodia in D and line scan analysis show an enrichment of fascin at the distal end of the filopodia (arrows). Note that fascin S289A enrichment was enhanced. Bars, 5 µm. See also Videos 2, 3, 4, and 5.
We subsequently examined whether, in the absence of wild-type protein, fascin S289A could also rescue actin microspike formation within lamellae as visualized by coexpression with an F-actin probe LifeAct-GFP (Riedl et al., 2008). In the absence of fascin, macrophages lack most actin-rich filopodia and microspikes within lamellae (Fig. 2 C), and we found that S289A rescued these defects to the same extent as the wild-type protein (Fig. 2 D and Videos 4 and 5). Interestingly, line scan analysis revealed that the peak of fascin S289A accumulation was immediately distal to the F-actin bundle in filopodia (Fig. 2 E). Although weaker and more transient, wild-type fascin was also detected ahead of the F-actin signal, supporting the idea that the localization of the S289A mutant represents a normally occurring distribution of fascin at the very distal end of the filopodia (Fig. 2 E and Videos 4 and 5). These data show that a critical aspect of fascin function in Drosophila macrophages resides at the distal end of the filopodial filament and that this subcellular distribution can be inhibited by a phosphomimetic mutation at S289 of the fascin protein.
The conserved residue in hfascin (S274) controls filopodia dynamics in carcinoma cells
To further explore this novel regulatory mechanism, we introduced equivalent phosphomutations in the human protein at the conserved serine residue (S274) and examined their effects in a carcinoma cell line in which fascin expression was stably silenced by small hairpin RNA (shRNA; Fig. S1 A). Depletion of endogenous fascin significantly inhibited the ability of this cell line to generate filopodia (not depicted), and reexpression of monomeric RFP (mRFP)–tagged hfascin rescued filopodia production (Fig. S1 E). hfascin S274A–expressing cells exhibited significantly fewer and shorter filopodia compared with those expressing wild-type fascin (Fig. 3, A–C). Live imaging revealed that S274A strongly localized to filopodia at early stages of formation (Fig. 3 B); however, these filopodia failed to extend and showed a shorter lifetime, suggesting that hfascin S274A cannot fully rescue the formation of the F-actin bundle in this cell type (Figs. 3 D and S1 F and Video 6). Expression of hfascin S274D also was unable to rescue filopodia number (Fig. 3 C). Furthermore, filopodia, although normal in length, were significantly less stable and quickly collapsed (Figs. 3 D and S1 and Video 6). These data show that the S274 residue in hfascin is critical for regulating filopodial dynamics in human carcinoma cells.
Figure 3.
Mutation of the conserved serine residue of hfascin affects filopodia dynamics and impairs its actin-bundling in vitro. (A) Similar mutations were generated in mRFP-tagged human fascin (hfascin; purple) and expressed in MDA-MB-231 cells that were depleted of endogenous fascin. Double-headed arrows highlight filopodia examined by line scan analysis in B. (B) High magnification of filopodia highlighted in A and line scan analysis show different distribution of hfascin transgenes within filopodia compared with actin (LifeAct-GFP). (A and B) Bars, 1 µm. (C) Quantification of mean filopodia length and number during transgene expression. *, P < 0.05; **, P < 0.01. See also Video 6. Error bars show means ± SD. (D) Time course of filopodia growth over 5 min (from the kymograph in Fig. S1 F) reveals that phosphomutations on residue S274 alter filopodial dynamics. (E and F) Actin-bundling (E) and actin-binding (F) cosedimentation assays show that S274A/D mutations decreased actin-bundling/binding properties of fascin in vitro. In the bundling assay, note the reduction of actin in the pellet of fascin S39D and S289A/D (red squares). In the actin-binding assay, note the reduction of these fascin mutants in the pellet (red squares). SN, supernatant; P, pellet. (G) Transmission electron micrographs reveal that S274 mutations prevent the formation of organized bundles. mag., magnification; WT, wild type.
This conserved serine residue can be phosphorylated and regulates the actin-binding/bundling capacity of fascin
The different effects of these phosphomutations suggested that this residue of fascin might be phosphorylated. To test this, we first quantified the level of phosphorylation of wild-type hfascin versus hfascin S274A in human carcinoma cells, by using the Pro-Q Diamond reagent, and found that this mutation decreased phosphorylation levels by ∼20% (Fig. S1 C). As a control, we also examined the effect of mutation of the known phosphorylation site of the protein (S39) and found an equivalent reduction in phosphorylation levels, which was enhanced in the double mutation (S39A and S274A; Fig. S1 C; Ono et al., 1997). An identical reduction in phosphorylation levels was revealed by examining the metabolic incorporation of radioactive orthophosphate by the different mutant forms of fascin expressed in cancer cells (Fig. S1 B). Interestingly, the S289A mutation in Drosophila fascin led to a similar reduction in phosphorylation levels when expressed in embryos (Fig. S1 D), highlighting the evolutionary conservation of this regulatory mechanism.
As fascin is thought to directly bundle actin filaments during filopodia formation, we wanted to determine how the S274 mutations affect the actin-interacting properties of the protein. We performed in vitro actin-binding/bundling assays on the different mutant forms of fascin using a cosedimentation assay with F-actin. We first validated our assay by using S39A and S39D mutants, which enhance and decrease the bundling/binding properties of the protein, respectively (Fig. 3, E and F; Ono et al., 1997; Vignjevic et al., 2006). This cosedimentation assay showed that both S274A and D mutations reduced the ability of fascin to bundle and bind F-actin (Fig. 3, E and F). We also performed EM on these in vitro fascin-actin bundles to analyze the morphology of these actin structures (Jansen et al., 2011). This revealed that the S274A mutation severely affected actin filament bundling (Fig. 3 G). In contrast, S274D was capable of inducing some bundling; however, the morphology of these bundles showed no regular organization when compared with wild-type fascin (Fig. 3 G). This analysis demonstrates that S274, along with S39, modulates the actin-interacting capacity of the protein. Interestingly, these data are consistent with the recent characterization of a second major actin-binding domain of fascin located in the region surrounding S274 (Jansen et al., 2011).
Fascin S289A rescues nurse cell actin bundles despite an impaired actin-binding capacity
The intriguing observation that the fascin S289A mutant was capable of rescuing macrophage migration with an impaired ability to interact with actin suggested a new function that is independent of actin bundling. To further examine this possibility in vivo, we analyzed the effect of this new mutation in Drosophila nurse cells. These cells display a dense fascin-dependent filopodial-like actin network necessary to anchor nuclei in the center of the cell, and in the absence of fascin, the actin network is completely lost, resulting in female sterility (Cant et al., 1994). When expressed in nurse cells of fascin mutant females, wild-type GFP-fascin restored the assembly of actin bundles where it colocalized with actin cables (Fig. 4 A). As observed in macrophages, fascin S289D failed to rescue actin bundle formation in nurse cells and prevented localization to the residual actin filaments at the cell membrane (Fig. 4 A). In contrast, the S289A mutation was able to rescue the formation of actin bundles. Surprisingly fascin S289A did not localize along the length of these rescued F-actin bundles (Fig. 4 A). Higher magnification images revealed that fascin S289A instead localized specifically to puncta at the distal ends of actin bundles along the cell membrane (Fig. 4 A), where barbed ends of actin filaments have been reported to localize (Guild et al., 1997; Gates et al., 2009). Furthermore, aside from rescuing nurse cell actin bundles, fascin S289A (unlike fascin S289D) restored female sterility, demonstrating that this mutant form is fully functional during this developmental process.
Figure 4.
Drosophila fascin S289A can rescue fascin function in nurse cells without interacting with actin. (A) Expression of wild-type (wt) fascin in sn28 mutant flies rescued the formation of actin bundles and fertility. Note that wild-type fascin (green) colocalizes with actin bundles (purple). The S289D mutation failed to rescue fertility, bundle assembly, or sterility. In contrast, fascin S289A rescued fertility but did not colocalize with actin bundles. Note that in the high magnification image (bottom), fascin S289A localized to the barbed ends of actin filaments at the cell membrane (arrows). Boxes represent the regions magnified at the bottom. (B) FRET/FLIM analysis of GFP-fascin transgenes with LifeAct-TagRFP in nurse cells. Lifetime of the GFP is represented by a color code from no interaction (blue) to high interaction (red). (C) Quantification of FRET efficiency between Drosophila fascin transgenes and F-actin. Both S289A/D significantly disrupted the ability of fascin to interact with F-actin. n = 6; ***, P < 0.001. Error bars show means ± SD. Bars, 5 µm.
We next wanted to determine whether modification of S289 modulates the actin-interacting capacity of fascin in vivo. We developed a novel fluorescence resonance energy transfer (FRET)/fluorescence lifetime imaging microscopy (FLIM)–based assay to quantify the interaction of fascin with F-actin in intact human SW480 carcinoma cells as previously used to test the interaction between hfascin and PKC (Parsons and Adams, 2008). We first measured interactions of Drosophila fascin S52A/E (equivalent of S39 in human) with actin; as expected, we found that S52A led to an increase in the interaction with actin over wild type, whereas fascin S52E was severely reduced (Fig. S2, A–C). This demonstrates that the Drosophila protein behaves similarly to the human form in a mammalian cell line as well as validating that the FRET/FLIM assay reports on the capacity of Drosophila fascin to bind to actin bundles. We then extended this approach to nurse cells, which can be fixed without altering the actin network, thus allowing us to perform FRET/FLIM in vivo. Similar to Drosophila fascin in mammalian cells, the S52A/E mutations showed comparable actin-interacting properties when expressed in vivo (Fig. S2, D and E). We next examined the effects of the S289 mutations and discovered that S289D significantly decreased the ability of fascin to interact with F-actin in nurse cells (Fig. 4, B and C). The interaction between fascin and F-actin was also completely abrogated by the S289A mutation (Fig. 4, B and C), despite its ability to fully rescue actin bundles.
Conclusion
Here, we reveal an evolutionarily conserved residue of fascin (S274/289) capable of regulating its activity and localization within filopodia. Using functional tests in Drosophila, we find that the S → A mutant, despite a restricted localization to the distal end of filopodia and an impaired actin-bundling capacity, is capable of rescuing both cell migration and sterility in fascin mutant flies. Furthermore, in vitro assays indicate that the S → A mutation directly inhibits the actin-bundling activity of fascin, challenging the current view that the only role of fascin in filopodia is restricted to the bundling of preformed actin filaments. Instead, our results collectively suggest that fascin plays a key role at the distal end of filopodia to promote the assembly and growth of filopodial filaments.
There are several proteins known to accumulate at the tip of the filopodia where they mediate early stages (Faix et al., 2009) and late stages (Breitsprecher et al., 2011) of filament dynamics. It is intriguing that previous work on the S39E mutant of hfascin demonstrated enhanced filopodial tip localization, leading the authors to speculate that a tip complex member may be involved in fascin recruitment by a mechanism independent of actin bundling (Vignjevic et al., 2006). However, the localization of fascin S289A does not exhibit a tip-specific localization pattern similar to the other members of the tip complex, such as Ena (Tucker et al., 2011), suggesting that fascin does not belong to this complex. The distal enrichment of S289A may therefore reflect an actin bundle architecture that is not homogenous along the length of the filament. Indeed, there are reported differences in the composition of actin-binding proteins along the length of filopodia (Geraldo et al., 2008; Schäfer et al., 2010) as well as biochemical differences in the actin filament itself (Dent and Gertler, 2003).
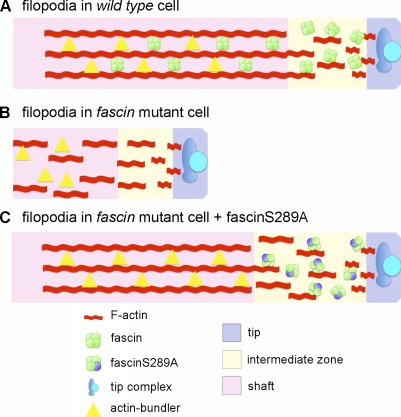
We therefore propose an additional role for fascin in filopodia at the distal end of the filament that occurs immediately after the tip complex (Fig. 5); this intermediate zone, unveiled by fascin S289A, may involve a complex that helps stabilize the subsequent bundling process through an unknown actin-independent mechanism. Interestingly, activated β1 integrin displays a similar tapered localization pattern at the distal end of filopodial filaments in growth cones (Galbraith et al., 2007), and it is possible that fascin may also be part of an adhesion-dependent complex, which has been described within filopodia (Schäfer et al., 2010). To date, our experiments to detect phosphorylation of this residue (and S39) in vivo suggest a very small level of phosphorylation, possibly caused by an extremely low percentage of phosphorylated fascin protein in the cell. However, the distinct effects of phosphomutations on fascin distribution suggest that a regulatory cycle could occur within the intermediate zone and be required for fascin localization to this region. Nevertheless, we cannot exclude the possibility that this residue is in a critical folded region of the protein with point mutations (or posttranslational modifications) subtly affecting an intramolecular interaction that alters both localization and actin bundling, as previously suggested (Cant and Cooley, 1996).
Figure 5.
A model proposing a new function for fascin within the distal end of growing filopodia. (A) During filopodia initiation, filaments of actin are coalesced by proteins belonging to the tip complex. Fascin is then loaded onto the filopodia within a region we term the “intermediate zone,” which may be regulated by a cycle of fascin phosphorylation and dephosphorylation. Subsequently, fascin, or another bundling factor, can help stabilize the actin bundle within the shaft of the filopodia. (B) In the absence of fascin, the tip complex may initiate filopodia; however, the process is aborted without the presence of fascin within the intermediate zone. (C) The S289A mutation stabilizes fascin localization to the distal end of the filopodia. Despite an inability of this mutant to bundle actin itself, an unknown role of fascin within the intermediate zone allows filopodia to elongate if another actin-bundling factor is present.
Fascin clearly can play a direct role in bundling F-actin; however, this function is not absolutely required, as the S289A mutation was capable of rescuing Drosophila macrophages and nurse cells without localizing along the length of the actin bundle. In nurse cells for example, another actin-bundling protein, such as the villin-like protein quail (Mahajan-Miklos and Cooley, 1994; Cant et al., 1998), may replace fascin’s bundling role. In contrast, in the carcinoma cell line tested here, hfascin S274A is insufficient to rescue filopodia formation, suggesting that fascin is the primary actin-bundling protein within this cell type.
In conclusion, our results have revealed a specific role for fascin within the distal ends of filopodia, which appears to be uncoupled from its actin-bundling activity. The enhanced affinity of the fascin S289A mutant for this region may allow its use as a tool to decipher these unknown initial stages of filopodia formation.
Materials and methods
Fly stocks
We used the following lines obtained from the Bloomington Drosophila Stock Center and the community: sn28, DfsnC128, upstream activation sequence (UAS)-GFP-Moesin (Dutta et al., 2002), UAS-mCherry-Moesin (Millard and Martin, 2008), UAS-GFP-fascin (Zanet et al., 2009), and nanos-Gal4VP16. The percentage of the rescue of fertility was calculated by counting the number of pupae generated by one female sn28/sn28 expressing the different GFP-fascin transgenes in nurse cells under the nanos-Gal4 driver in comparison with a wild-type fly.
Plasmids and transgenesis
UAS-fascin lines were generated by inserting a full-length singed cDNA (RH62992) into the pUASp vector. GFP-fascin and mCherry-fascin fusions were obtained by fusing GFP or mCherry sequences to the N terminus of full-length singed cDNA, and site-directed mutagenesis on serine S289 was performed by PCR. The LifeAct-GFP and LifeAct-TagRFP lines were generated by inserting the cDNA sequence of the LifeAct (Riedl et al., 2008) tagged with the GFP or the TagRFP (Evrogen) in the C terminus of the pUASp vector.
Singed (sn)-Gal4 lines were obtained by cloning the genomic region (X chromosome position: 7,867,099–7,867,592) of the singed gene, which controls Drosophila macrophage expression of fascin, upstream of the Gal4 sequence. Genomic DNA was amplified by PCR by using the following primers: 5′-CTCCATTAAATTGAAGAACCTC-3′ and 5′-CAGTACGTTGGGCATTCC-3′. This fragment was subcloned in the pGEM-T Easy (Promega) vector then cloned in the pPTGal4 plasmid to obtain the driver singed-Gal4. Transgenic flies were generated by P element insertion. For each individual construct, we established and tested a minimum of three independent lines.
For hfascin knockdown, the target shRNA, 5′-GCCTGAAGAAGAAGCAGAT-3′, was cloned into the pLLT3.7 vector (gift from J. Adams, University of Bristol, Bristol, England, UK). shRNA-resistant hfascin cDNA (in which the shRNA target sequence was changed to 5′-GCCTGAAAAAAAAACAGAT-3′ [bold letters indicate changes]), and its wild-type and mutant forms were generated and tagged in the N-terminal site with EGFP (pEGFP vector; Takara Bio Inc.) or mRFP into pcDNA3.1 (Invitrogen). GFP-LifeAct and Ruby-LifeAct constructs were a gift from R. Wedlich-Soldner (Max Planck Institute, Munich, Germany; Riedl et al., 2008). For recombinant fascin purification, the cDNA encoding hfascin and mutants were cloned between NotI and XhoI restriction sites in the pET-30a(+) vector (EMD).
Cell culture and transfections
MDA-MB-231 (human breast carcinoma) and SW480 (human colon carcinoma) cells were cultured in DME supplemented with 10% FBS. For fascin localization and filopodia formation analysis, MDA-MB-231 fascin knockdown cells were established by lentiviral infection. These cells were transiently cotransfected with the mRFP-tagged shRNA-resistant fascin constructs and GFP-LifeAct using Lipofectamine 2000 reagent (Invitrogen). After transfection, cells were plated on 10 µg/ml fibronectin-coated coverslips (Millipore) and, 24 h later, sealed in imaging chambers and visualized as described in Live imaging.
Live imaging
UAS transgenic constructs encoding fluorescent proteins (GFP or mCherry) were expressed in macrophages using the singed-Gal4 driver line. Live embryos were mounted as previously described (Wood et al., 2006). In brief, stage 15 embryos were dechorionated in bleach and mounted under coverslips on hydrophobic Lumox dishes (Sarstedt) in Voltalef oil. Embryos were then imaged using a confocal microscope (TCS SP5 [Leica] or UltraVIEW VoX [PerkinElmer] spinning disk with a 63× N.A. 1.4 objective). For colocalization experiments of GFP- and mCherry-tagged proteins, images were acquired sequentially in the red and green channels; inverting the order of acquisition had no effect on the localization. For filopodia formation analysis in human carcinoma cells, MDA-MB-231 cells were visualized using a confocal microscope (A1R; Nikon). Filopodia number and length were measured from three independent experiments using ImageJ software (National Institutes of Health), and statistical significance of differences between groups was assessed by analysis of variance (ANOVA).
Immunoprecipitation and Pro-Q Diamond staining
Cells were transfected with the different hfascin constructs (GFP-fascin wild type, GFP-fascin S39A, GFP-fascin S274A, and GFP-fascin S39AS274A) and then lysed in radioimmunoprecipitation assay buffer supplemented with antiphosphatase inhibitor cocktails (EMD). Drosophila fascin (UAS-mCherry-fascin and UAS-mCherry-fascin S289A) was expressed in embryos with the ubiquitous daughterless-Gal4 driver. Wild-type and mutant fascins were immunoprecipitated with GFP-Trap or RFP-Trap beads (Chromotek). Proteins were blotted on polyvinylidene fluoride membrane and stained with Pro-Q Diamond (Invitrogen). Total fascin protein amounts were detected on the same blot with Coomassie or the antifascin antibody (sn7C; Developmental Studies Hybridoma Bank). Pro-Q Diamond signal and protein levels were quantified with ImageJ.
For metabolic incorporation of radioactive orthophosphate, cells were transfected with the different hfascin constructs (GFP-fascin S39A, GFP-fascin S274A, and GFP-fascin S39AS274A). Cells were starved for 4 h in DME depleted in phosphate and containing 0.5% dialyzed FBS (Invitrogen). This media were subsequently replaced by DME depleted in phosphate containing 10% dialyzed FBS and 0.5 mCi/ml of radiolabeled P32-orthophosphate (PerkinElmer) for 1 h. Cells were lysed and immunoprecipitated as described in the previous paragraph. Proteins were separated in an SDS-PAGE gel and analyzed using a phosphoimager (FLA-3000; Fujifilm).
FRET/FLIM
The LifeAct-TagRFP and the GFP-fascin (wild-type and mutated forms) were coexpressed in the fly germline using the driver nanos-Gal4VP16. Ovaries were dissected in PBS and fixed in 4% paraformaldehyde for 25 min. Ovaries were rinsed in PBS and quenched for nonspecific fluorescence in PBS containing sodium borohydride (Sigma-Aldrich) at 1 mg/ml for 20 min. Ovaries were extensively rinsed in PBS/0.1% Tween 20 and then mounted in Vectashield mounting medium (Vector Laboratories). For FRET/FLIM cell culture experiments, wild-type MDA-MB-231 and human colon carcinoma SW480 cells were transiently cotransfected with GFP-tagged fascin (wild type and mutants) and the LifeAct-Ruby construct. Time domain FLIM was performed with a multiphoton microscope system (with TE2000 microscope; Nikon) described in detail previously (Parsons and Adams, 2008). In brief, cells were fixed in 4% paraformaldehyde and permeabilized in 0.2% (wt/vol) Triton X-100 in PBS. After quenching with 1 mg/ml sodium borohydride in PBS for 10 min at RT, cells were washed in PBS and mounted in Mowiol containing 2.5% Dabco (Sigma-Aldrich). Fluorescence lifetime imaging capability was provided by time-correlated single-photon counting electronics (SPC-700; Becker & Hickl). A 40× objective was used throughout (Plan Fluor NA 1.3; CFI 60; Nikon), and data were collected at 500 ± 20 nm through a band pass filter (35–5040; Coherent, Inc.). Acquisition times of the order of 300 s at a low 890-nm excitation power were used to achieve sufficient photon statistics for fitting, while avoiding either pulse pile up or significant photobleaching. Histogram data are plotted as mean FRET efficiency from >10 cells per sample over three independent experiments or from six egg chambers per sample. ANOVA was used to test statistical significance between different populations of data. Lifetime images of example cells are presented using a pseudocolor scale, whereby blue depicts normal GFP lifetime (no FRET) and red depicts lower GFP lifetime (areas of FRET).
Protein purification
Recombinant fascin expression was induced in the BL21 Escherichia coli strain with 1 mM IPTG (Sigma-Aldrich) for 5 h at 25°C. The bacterial pellet was resuspended in 300 mM NaCl, 10 mM imidazole, and 50 mM Na2HPO4, pH 8, sonicated, cleared, and purified using Ni–nitriloacetic acid beads (QIAGEN). Proteins were dialyzed against a buffer containing 150 mM NaCl, 10 mM imidazole, and 50 mM Na2HPO4, pH 8, at 4°C. Actin from rabbit muscle was purchased from Sigma-Aldrich.
Actin-binding/bundling assay
30 µM actin was polymerized in the following buffer for 2 h at 4°C: 50 mM KCl, 2 mM MgCl2, and 1 mM ATP. F-actin was then pelleted for 1.5 h at 50,000 g at 4°C and dialyzed overnight at 4°C in G buffer (2 mM Tris, pH 7.4, 0.2 mM CaCl2, 0.2 mM ATP, and 0.5 mM DTT). The different forms of purified fascin proteins were first centrifuged at 80,000 g for 30 min to remove potential aggregates, and actin was centrifuged at 13,000 rpm five times at RT. Fascin and actin at a final concentration of 15 µM each were incubated for 1 h at RT in KME buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7). Samples were centrifuged for 30 min at two different speeds, 13,000 g (for bundling experiments) and 80,000 g (for binding experiments). Equal volumes of supernatant and pellet were analyzed by 8% SDS-PAGE, and proteins were stained with the Imperial Protein Stain (Thermo Fisher Scientific).
EM analysis of fascin and actin bundles
For the ultrastructural analysis of fascin-actin bundles, actin-binding experiments were initially performed as described in the previous section. Before centrifugation steps, samples were diluted 1:10 in KME buffer and prepared as previously described (Jansen et al., 2011). In brief, 3 µl of the diluted sample was left to adsorb onto Formvar- and carbon-coated 200-mesh copper grids for 30 s, excess solution was removed, and the sample was negatively stained with uranyl acetate 1% for 1 min. The remaining uranyl acetate solution was removed, and the grids were dried at RT. The EM study was performed in a transmission electron microscope (F20; Tecnai) at 25,000× magnification with an accelerating voltage of 200 kV.
Online supplemental material
Fig. S1 shows blots of the knockdown of fascin in MDA-MB-231 and the reexpression of fascin transgenes. Fig. S2 shows FRET/FLIM data of different forms of fascin interacting with actin in human cells and Drosophila nurse cells. Video 1 shows time-lapse imaging of wild-type fascin, fascin S289A, and fascin S289D in migrating macrophages in embryos. Video 2 shows coexpression of wild-type mCherry-fascin and GFP-fascinS289A in macrophages in sn28 mutant embryos. Video 3 shows high magnification time-lapse sequence of a macrophage coexpressing wild-type mCherry-fascin and GFP-fascinS289A in sn28 mutant embryos. Videos 4 and 5 show time-lapse imaging of mCherry-fascin and mCherry-fascinS289A, respectively, and LifeAct-GFP in migrating macrophages. Video 6 shows time-lapse imaging of filopodia from human cells which express the different transgenes of hfascin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110135/DC1.
Acknowledgments
We would like to thank François Payre, Phillip Gordon-Weeks, and Tanya Shaw for helpful discussions and Dan Worth for his technical advice in the EM experiments.
This work was funded by a Biotechnology and Biological Sciences Research Council project grant to B. Stramer and J. Zanet. M. Parsons is funded by the Royal Society (University Research Fellowship), A. Jayo is funded by the Basque Government, Spain (Research Staff Training Fellowship), and S. Plaza is funded by Association pour la Recherche sur le Cancer and SFI20101201669.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- fluorescence resonance energy transfer
- hfascin
- human fascin
- mRFP
- monomeric RFP
- shRNA
- small hairpin RNA
- UAS
- upstream activation sequence
References
- Adams J.C. 2004. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16:590–596 10.1016/j.ceb.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D., Koestler S.A., Chizhov I., Nemethova M., Mueller J., Goode B.L., Small J.V., Rottner K., Faix J. 2011. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J. Cell Sci. 124:3305–3318 10.1242/jcs.086934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K., Cooley L. 1996. Single amino acid mutations in Drosophila fascin disrupt actin bundling function in vivo. Genetics. 143:249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K., Knowles B.A., Mooseker M.S., Cooley L. 1994. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 125:369–380 10.1083/jcb.125.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K., Knowles B.A., Mahajan-Miklos S., Heintzelman M., Cooley L. 1998. Drosophila fascin mutants are rescued by overexpression of the villin-like protein, quail. J. Cell Sci. 111:213–221 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Gertler F.B. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 40:209–227 10.1016/S0896-6273(03)00633-0 [DOI] [PubMed] [Google Scholar]
- Dutta D., Bloor J.W., Ruiz-Gomez M., VijayRaghavan K., Kiehart D.P. 2002. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis. 34:146–151 10.1002/gene.10113 [DOI] [PubMed] [Google Scholar]
- Faix J., Breitsprecher D., Stradal T.E., Rottner K. 2009. Filopodia: Complex models for simple rods. Int. J. Biochem. Cell Biol. 41:1656–1664 10.1016/j.biocel.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Galbraith C.G., Yamada K.M., Galbraith J.A. 2007. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 315:992–995 10.1126/science.1137904 [DOI] [PubMed] [Google Scholar]
- Gates J., Nowotarski S.H., Yin H., Mahaffey J.P., Bridges T., Herrera C., Homem C.C., Janody F., Montell D.J., Peifer M. 2009. Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev. Biol. 333:90–107 10.1016/j.ydbio.2009.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S., Khanzada U.K., Parsons M., Chilton J.K., Gordon-Weeks P.R. 2008. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell Biol. 10:1181–1189 10.1038/ncb1778 [DOI] [PubMed] [Google Scholar]
- Guild G.M., Connelly P.S., Shaw M.K., Tilney L.G. 1997. Actin filament cables in Drosophila nurse cells are composed of modules that slide passively past one another during dumping. J. Cell Biol. 138:783–797 10.1083/jcb.138.4.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S.L., Gertler F.B. 2007. Filopodia: the fingers that do the walking. Sci. STKE. 2007:re5 10.1126/stke.4002007re5 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Skacel M., Adams J.C. 2005. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int. J. Biochem. Cell Biol. 37:1787–1804 10.1016/j.biocel.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Parsons M., Adams J.C. 2007. Dual actin-bundling and protein kinase C-binding activities of fascin regulate carcinoma cell migration downstream of Rac and contribute to metastasis. Mol. Biol. Cell. 18:4591–4602 10.1091/mbc.E07-02-0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., Dominguez R. 2011. Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286:30087–30096 10.1074/jbc.M111.251439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Dawson J.C., Forero-Vargas M., Spence H.J., Yu X., König I., Anderson K., Machesky L.M. 2010. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20:339–345 10.1016/j.cub.2009.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Cooley L. 1994. The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell. 78:291–301 10.1016/0092-8674(94)90298-4 [DOI] [PubMed] [Google Scholar]
- Mattila P.K., Lappalainen P. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- Millard T.H., Martin P. 2008. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development. 135:621–626 10.1242/dev.014001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemethova M., Auinger S., Small J.V. 2008. Building the actin cytoskeleton: filopodia contribute to the construction of contractile bundles in the lamella. J. Cell Biol. 180:1233–1244 10.1083/jcb.200709134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Yamakita Y., Yamashiro S., Matsudaira P.T., Gnarra J.R., Obinata T., Matsumura F. 1997. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272:2527–2533 10.1074/jbc.272.4.2527 [DOI] [PubMed] [Google Scholar]
- Parsons M., Adams J.C. 2008. Rac regulates the interaction of fascin with protein kinase C in cell migration. J. Cell Sci. 121:2805–2813 10.1242/jcs.022509 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer C., Born S., Möhl C., Houben S., Kirchgessner N., Merkel R., Hoffmann B. 2010. The key feature for early migratory processes: Dependence of adhesion, actin bundles, force generation and transmission on filopodia. Cell Adh Migr. 4:215–225 10.4161/cam.4.2.10745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedeh R.S., Fedorov A.A., Fedorov E.V., Ono S., Matsumura F., Almo S.C., Bathe M. 2010. Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein. J. Mol. Biol. 400:589–604 10.1016/j.jmb.2010.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekhaus D., Haesemeyer M., Moffitt O., Lehmann R. 2010. RhoL controls invasion and Rap1 localization during immune cell transmigration in Drosophila. Nat. Cell Biol. 12:605–610 10.1038/ncb2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Wood W., Galko M.J., Redd M.J., Jacinto A., Parkhurst S.M., Martin P. 2005. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168:567–573 10.1083/jcb.200405120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Moreira S., Millard T., Evans I., Huang C.Y., Sabet O., Milner M., Dunn G., Martin P., Wood W. 2010. Clasp-mediated microtubule bundling regulates persistent motility and contact repulsion in Drosophila macrophages in vivo. J. Cell Biol. 189:681–689 10.1083/jcb.200912134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P.K., Evans I.R., Wood W. 2011. Ena drives invasive macrophage migration in Drosophila embryos. Dis Model Mech. 4:126–134 10.1242/dmm.005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G.G. 2006. Role of fascin in filopodial protrusion. J. Cell Biol. 174:863–875 10.1083/jcb.200603013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., Martin P. 2002. Structures in focus—filopodia. Int. J. Biochem. Cell Biol. 34:726–730 10.1016/S1357-2725(01)00172-8 [DOI] [PubMed] [Google Scholar]
- Wood W., Faria C., Jacinto A. 2006. Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. J. Cell Biol. 173:405–416 10.1083/jcb.200508161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Stramer B., Millard T., Martin P., Payre F., Plaza S. 2009. Fascin is required for blood cell migration during Drosophila embryogenesis. Development. 136:2557–2565 10.1242/dev.036517 [DOI] [PubMed] [Google Scholar]