Abstract
Structures of horse liver alcohol dehydrogenase complexed with NAD+ and unreactive substrate analogues, 2,2,2-trifluoroethanol or 2,3,4,5,6-pentafluorobenzyl alcohol, were determined at 100 K at 1.12 or 1.14 Å resolution, providing estimates of atomic positions with overall errors of about 0.02 Å, geometry of ligand binding, descriptions of alternative conformations of amino acid residues and waters, and evidence of a strained nicotinamide ring. The four independent subunits from the two homodimeric structures differ only slightly in the peptide backbone conformation. Alternative conformations for amino acid side chains were identified for 50 of the 748 residues in each complex, and Leu-57 and Leu-116 adapt different conformations to accommodate the different alcohols at the active site. Each fluoroalcohol occupies one position, and the fluorines of the alcohols are well resolved. These structures closely resemble the expected Michaelis complexes with the pro-R hydrogens of the methylene carbons of the alcohols directed toward the re-face of C4N of the nicotinamide rings with a C to C distance of 3.40 Å. The oxygens of the alcohols are ligated to the catalytic zinc at a distance expected for a zinc alkoxide (1.96 Å) and participate in a low-barrier hydrogen bond (2.52 Å) with the hydroxyl group of Ser-48 in a proton relay system. As determined by X-ray refinement with no restraints on bond distances and planarity, the nicotinamide rings in the two complexes are slightly puckered (quasi-boat conformation, with torsion angles of 5.9° for C4N and 4.8° for N1N relative to the plane of the other atoms) and bond distances that are somewhat different as compared to those found for NAD(P)+. It appears that the nicotinamide ring is strained toward the transition state on the path to alcohol oxidation.
Horse liver alcohol dehydrogenase (ADH, EC1.1.1.1) is a very well-studied enzyme.1 The kinetics and catalytic mechanism have been described.2 Three-dimensional structures have been determined for a variety of complexes with native and mutated enzymes.3,4 ADH is the first enzyme for which a conformational change involving rotation of domains upon binding a substrate was described;5 this conformational change is intimately related to catalysis.6 Kinetic isotope effects provide evidence for quantum mechanical hydrogen tunneling during catalysis,7 and temperature-independent isotope effects are consistent with a role for vibrationally assisted tunneling.8–11 Computational studies, based on three-dimensional structures of ADH, have addressed the energetics of proton and hydride transfer steps and the roles of protein motions in catalysis.12–18 Nevertheless, the connection between protein dynamics and tunneling is not yet established.19 Furthermore, some atomic resolution X-ray studies of ADH raise questions about the chemistry of interconversion of ternary complexes during catalysis.20,21
Atomic resolution structures22 provide a wealth of detail that is critical for understanding enzyme function. For such studies, it is important to crystallize the enzyme with the actual substrates or ligands that are sterically and chemically similar to the substrates. For alcohol dehydrogenase, complexes with NAD+ and fluoroalcohols are especially informative because the fluorines are sterically similar to hydrogens, are readily visible in electron density maps, and produce an unreactive substrate analogue due to the electron withdrawal, so that the nicotinamide ring should remain oxidized. The alcohols chosen for this study, 2,3,4,5,6-pentafluorobenzyl alcohol (PFB) and 2,2,2-trifluoroethanol (TFE) are potent competitive inhibitors of alcohol oxidation by ADH (Ki values of 0.52 and 8.4 μM, respectively). A structure with wild-type enzyme complexed with PFB was previously determined at 2.1 Å resolution at 4 °C.23 PFB is a good analog of benzyl alcohol, a good substrate that has been used extensively for kinetic studies and evaluation of quantum mechanical tunneling. TFE is an analog of the natural substrate, ethanol, and no high resolution structure with wild-type enzyme has been reported, although there are moderate resolution structures with mutated ADHs that are center pieces for understanding the requirements for hydrogen tunneling.19 With cryo-crystallography and synchrotron radiation, the desired atomic resolution structures could be determined.
EXPERIMENTAL PROCEDURES
Crystallization
Wild-type (natural) crystalline horse liver alcohol dehydrogenase (EE isoenzyme) and LiNAD+ were purchased from Roche Molecular Biochemicals. The fluoroalcohols (98–99%) were purchased from Aldrich and used without further purification. 2-Methyl-2,4-pentanediol was obtained from Kodak and treated with activated charcoal before use. The crystals of wild-type ADH complexed with NAD+ and the fluoroalcohols were prepared as described by the general procedure used previously.23 The enzyme was first recrystallized from 10 mM sodium phosphate buffer, pH 7.0, at 4 °C with 10% ethanol; the crystals were redissolved in buffer with some added KCl, and the solution was dialyzed extensively against 50 mM ammonium N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonate buffer, 0.25 mM EDTA, pH 6.7 (measured at 25 °C, pH 7.0 at 4 °C). The solution was clarified by centrifugation. About 1 mL of 10 mg/mL (A280 = 0.455/cm for 1 mg/mL) enzyme was dialyzed in washed 1/4-inch diameter tubing at 4 °C against 10 mL of the same buffer with 1 mM NAD+ and 10 mM pentafluorobenzyl alcohol or 100 mM trifluoroethanol for about 1 hr, and then the concentration of 2-methyl-2,4-pentanediol was raised by dialysis over some days to 12% when crystals formed. After that, the concentration of the diol was raised slowly to a final of 25%, which was sufficient for cyroprotection at 100 K.
X-ray Crystallography
The crystals (about 0.3 – 0.4 mm thick) for both complexes were mounted on fiber loops (Hampton Research) and flash cooled by plunging them into liquid N2. X-ray data were collected at 100 K. The data for the pentafluorobenzyl alcohol complex were collected March 9, 2006, on the GM/CA-CAT 23ID beamline with a MAR300CCD detector at the Advanced Photon Source with the X-ray wavelength at 0.9537 Å, a 0.025 by 0.075 mm beam, at 133 mm distance with 8 sec exposures and 0.2° oscillations over 360° total and then for a low-resolution pass at 300 mm with 2 sec exposures with 0.2° oscillations over 360° total. The data for the trifluoroethanol complex were collected June 24, 2009, on the SBC 19ID beamline with an ADSC315r Quantum detector at APS with the X-ray wavelength at 0.9184 Å with a 0.05 by 0.05 mm beam, at 130 mm distance, with 4 sec exposures with 0.5° oscillations for 760 images covering 380° total. Data were processed with d*TREK.24 Both structures were solved by molecular replacement using the coordinates for the refined wild-type ADH–NAD+–2,3,4,5,6-pentafluorobenzyl alcohol complex (1HLD.pdb) as a model.23 The structures were refined by cycles of restrained refinement with REFMAC525) and model building with the program O.26 Model bias was avoided during the initial refinement by not including the fluoroalcohol in the model, and omit maps were used where structural features were in doubt. Initially, the nicotinamide ring in the NAD+ molecule dictionary was set to be planar, but as test of the identity of the coenzyme, the monomer dictionary used by REFMAC was modified to remove the restraints on planarity and relax the restraints on bond distances (from 0.02 to 0.10 Å) for the nicotinamide ring. This modified coenzyme is named NAJ in our coordinate files in order to distinguish it from the NAD (with tight restraints) listed in other structures. SHELXL27 was also used was used for refinement with no restraints for the coenzyme. After rounds of blocked, diagonal least squares refinement, a blocked, full-matrix refinement was used in order to determine bond distances and their errors for important structural features.
During the early stages of refinement, hydrogens were included in riding positions, and isotropic temperature factors were refined. The hydrogens contribute to scattering, decreasing the R values (by about 1%), but not sufficiently to be refined independently of the parent atom. Hydrogens in methyl, amino, and hydroxyl groups, where the torsion angle is not known, were given zero occupancy. Alternative conformations of amino acids and waters were assigned on the basis of indisputable visual evidence: divergent electron densities at +1 σ level above the average in 2|Fo| − |Fc| maps, divergent lobes, and consistent plus/minus densities at +3.5/−3.5 σ in |Fo| − |Fc| maps. Occupancies were adjusted to make the temperature factors similar for the alternative positions. An arbitrary cutoff for B-values for water molecules was not applied, but waters were included when there was density above +1 σ in 2|Fo| − |Fc| maps and the distances to other atoms were compatible with hydrogen bonds. Single waters were fitted in most cases, but bilobate (“dumbbell”) densities were fitted with two alternative atoms when the distances were between about 1.6 to 2.4 Å. Some waters were placed in ellipsoidal densities, which were assumed to be characterized by anisotropic refinement. Extensive clusters of waters were observed. Partial waters were included to fit alternative conformations of amino acid residues. In the final stages, refinement with anisotropic temperature factors for all atoms with a restraint of 10 for sphericity was used. The final models were well-described by the electron density maps, but side chains for some residues on the surface of the protein (such as lysine and glutamic acid residues) were not completely within density, and many peaks of density, probably due to additional water or methylpentanediol molecules, were not modeled because of lack of contacts with the protein. The amino terminal acetyl groups found for the enzyme isolated from horse liver1 were not modeled, for lack of density. The structures were checked with PROCHECK.28 All residues had favored or allowed backbone conformational angles, except that Cys-174 residues (ligated to the catalytic zincs) were in the generously allowed region. EXCEL was used for statistical analyses.
RESULTS AND DISCUSSION
Refinement and model building
Table 1 summarizes the X-ray data collection and refinement. The homodimeric molecule in the asymmetric unit has a total of 748 amino acid residues, 4 zinc atoms, 2 NAD+ molecules, 4 methylpentanediols, 2 fluoroalcohols, and 1040 water molecules (in the complex with pentafluorobenzyl alcohol), and 1025 waters (in the complex with trifluoroethanol). Because of the alternative conformations, the number of fitted atoms is larger than the actual number in the structures.
Table 1.
X-ray Data and Refinement Statistics for Horse Liver Alcohol Dehydrogenase Complexed with NAD+ and Fluoroalcohols
| 2,3,4,5,6-pentafluoro-benzyl alcohol | 2,2,2-trifluoroethanol | |
|---|---|---|
| PDB entry name | 4DWV | 4DXH |
| space group | P1 | P1 |
| homodimeric molecules per unit cell | 1 | 1 |
| cell dimensions, Å | 44.29, 51.44, 92.49 | 44.25, 51.16, 92.53 |
| cell angles, deg | 91.72, 103.1, 110.1 | 91.91, 103.0, 109.9 |
| resolution range, Å | 20.0–1.14 | 20–1.12 |
| measured reflections: total, unique | 1203625, 253394 | 1073887, 267668 |
| completeness, % (outer shell) | 93.7 (84.5) | 94.2 (91.4) |
| Rmeas, % (outer shell)a | 6.5 (30.8) | 5.4 (42.2) |
| mean <I>/σ<I> (outer shell) | 12.9 (3.2) | 9.7 (2.1) |
| Rvalue, Rfree, test %b | 12.4, 14.4, 1.0 | 12.7, 14.7, 0.5 |
| Mean B-value, Wilson, REFMAC, Å2 | 11.4, 14.9 | 9.6, 13.3 |
| rsmd for bond distances, Åc | 0.013 | 0.011 |
| rmsd for bond angles, degc | 1.67 | 1.64 |
| estimated errors in coordinates, Å | 0.020 | 0.019 |
| number of atoms fitted | 6996 | 6973 |
The asymmetric unit is a dimer, made up of two identical chemical subunits, but differing slightly in conformation. The structures of the complexes with PFB and TFE are essentially identical, superimposing all alpha carbons with an rmsd of 0.09 Å. Superpositioning only the coenzyme binding domains (residues 176–318) of subunit A onto subunit B for the structure with pentafluorobenzyl alcohol gives an rmsd of 0.08 Å, whereas superpositioning of all residues is accomplished with an rmsd of 0.17 Å. The alpha carbons of the superpositioned residues are almost identical, suggesting that the conformations of both subunits are very similar, but subunit B has a slightly more open conformation, giving rise to differences of up to 1 Å in the catalytic domain as compared to subunit A. The similarity in the subunits suggests that both active sites can be catalytically active and provides no evidence for cooperativity in catalysis.
The structure of ADH crystallized with NAD+ and PFB previously reported at a resolution of 2.1 Å, determined at 4 °C,23 and the present one superimpose, using the coenzyme binding domains (residues 176–318) with an rmsd of 0.31 Å (0.37 for all alpha carbons), but there is a small expansion of about 0.7 Å in overall size at the 177 °C higher temperature. Previous studies that compared a series of structures of other proteins determined at 80–98 to 295–320 K also noted an expansion.29–31 Most features are the same in both ADH structures, but some side chains have somewhat different positions. The additional data (250,801 unique reflections compared to 37,512) lead to a greatly improved structure, with many alternative conformations, more accurate estimates of non-covalent bond distances and anisotropic atomic displacement parameters. A structure of horse liver ADH with the A317C substitution (located near the carboxyamido group of the nicotinamide ring) was solved at 1.2 Å resolution and is almost identical to the present structure (3OQ6.pdb).32 The current study analyzes the details of the atomic resolution structures.
Active Site Architecture
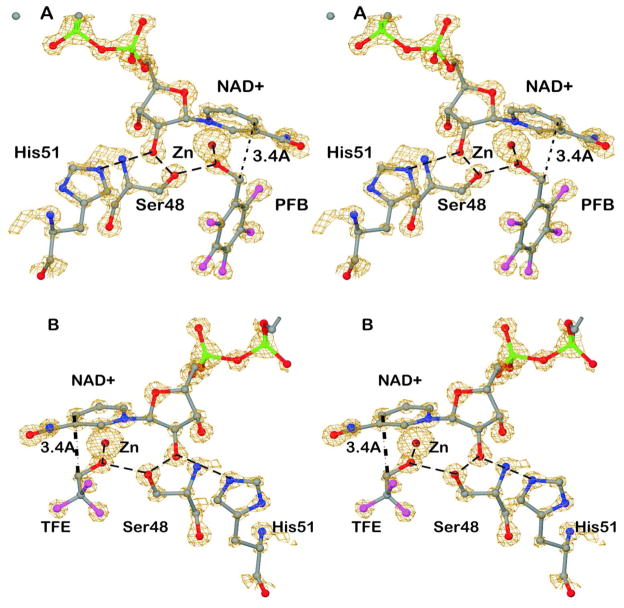
The X-ray data provided electron density maps at atomic resolution. Figure 1 shows the binding of the NAD+ and fluoroalcohols in maps that are contoured to show how the electron density envelopes increase with increasing atomic number. The positions of the atoms are well-defined, and the fluorine atoms stand out. The plane of the pentafluorobenzyl group is clear. The trifluoromethyl group has one position, suggesting a lack of free rotation about the C1-C2 bond. In each structure, the hydrogen-bonded network connecting the oxygen of the alcohol to His-51, which can act as the base in catalysis, is shown in dashed lines.23,33,34 The distance between C4N of the nicotinamide ring and the methylene carbon of the alcohol is 3.4 Å in all four subunits, illustrating a ground-state complex that mimics the expected Michaelis complex.
Figure 1.
Electron density maps for complexes of ADH with NAD+ and the fluoroalcohols. Interactions in the proton relay system shown in dashed lines. The dotted line with label 3.4 Å shows the distance between C4N of the nicotinamide ring and the methylene carbon of the alcohol. The stereoviews are rotated about 180° in order to see different views. (A) Binding of pentafluorobenzyl alcohol. Electron density is at about 4 σ above the mean. (B) Binding of trifluoroethanol. Electron density is at about 4.9 σ above the mean.
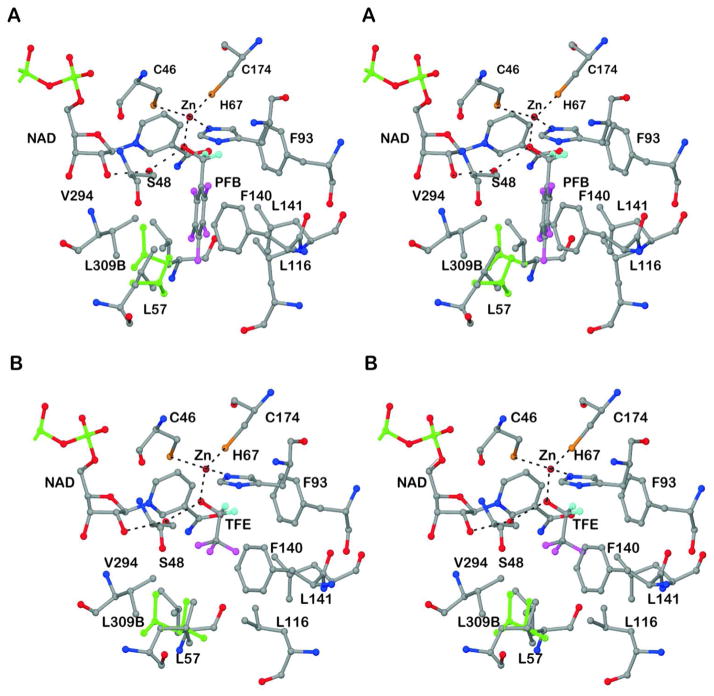
Figure 2 illustrates the interactions of the two different fluoroalcohols in the substrate binding sites. The hydrogen atoms riding on the methylene carbons are included, to indicate that the pro-R hydrogen is directed toward the position expected for direct transfer of hydride to C4N (re-face) of the nicotinamide ring.35 The binding site is hydrophobic, consistent with the observations that this enzyme is very selective for non-polar, primary alcohols. Ethanol is a major natural substrate, but longer chain alcohols, benzyl alcohols and some secondary alcohols are also good substrates.36 The conformations of amino acid residues in the binding site are adaptable, allowing different inhibitors to bind.37,38 In the structures with the fluoroalcohols, residues Leu-57 and Leu-116 in the substrate binding pocket adopt different conformations (Figure 2), apparently optimizing hydrophobic interactions. The atomic resolution provides a more complete picture, as Leu-309 (from the other subunit) has alternative conformations in the structures with both alcohols, and Leu-A57 has different and also alternative conformations in the structures with PFB and TFE. Leu-B57 and leucine residues A116 and B116 have different, but single, conformations in the structures with PFB and TFE; the alternative position for Leu-116 in the structure with TFE makes a cavity that accommodates a water that is hydrogen bonded to the carbonyl oxygen of Phe-93. (However, no water molecules are near the zinc or close to the nicotinamide ring.) Leu-309 has alternative conformations in both active sites of the two structures. The differing conformations represent energetically accessible states that can affect substrate specificity and the dynamics that lead to catalysis.
Figure 2.
Interactions of alcohols with active site residues. The coordination of the catalytic zinc ion and the hydrogen bonds of the alcohol in the proton relay system are shown as dotted lines. The riding hydrogens of the methylene carbons are shown in cyan and the other atoms have usual atomic coloring, except that the alternative conformations of the side chains of Leu-57 and Leu-309B (other subunit) are shown in green. Note that the conformations of Leu-57 and Leu-116 differ in the two structures. (A) Pentafluorobenzyl alcohol. (B) Trifluoroethanol.
Since the van der Waals radii for H and F are similar (1.30 – 1.38 and 1.20 Å, respectively,39,40 the fluoroalcohols are expected to be good steric analogs of the corresponding substrates, but the electronegative fluorines may also participate in weak C-H···F hydrogen bonds and affect binding.41,42 The pentafluorobenzyl group binds in one position, and its rotation would be hindered by steric interactions with Leu-57, Leu-116, and Ser-48. Its position might also be stabilized by the weak interactions (3.2 to 3.3 Å F to C distance) of F4 and F5 with the CD1 and CD2 methyl groups of Leu-57 and of F6 with the CB methylene of Ser-48. Trifluoroethanol binds with a staggered conformation of the O relative to the fluorines, and the fluorines are about 3.2 Å from the carbons of CE1 of Phe-93, the CG2 methyl group of Val-294, and the CB of Ser-48. A 60 degree rotation of the trifluoromethyl group produces an eclipsed conformation, and the contact with Phe-93 is lost. These structures may offer some evidence for weak interactions between the fluorines and the protein.
The enzyme catalyzes the transfer the pro-R hydrogen of ethanol, and the structure with trifluoroethanol provides a basis for explaining this stereospecificity. If the methyl group is rotated about the torsion angle for the oxygen and the methylene C (note this C is labeled as C2 in the PDB file) so that the pro-S hydrogen would point toward C4N of the nicotinamide ring, the methyl group would clash with the benzene ring of Phe-93. However, the reaction may only be highly stereoselective, as about 10% of the pro-S hydrogen from 1-octanol can be transferred.43 This lack of specificity is an indication of structural flexibility and is also consistent with the observation that 2-propanol is a (poor) substrate.
Alternative Conformations
Fitting alternative conformations in structures determined by X-ray crystallography is required to get the best structures (lowest R-factors and B-factors), to identify potential chemical heterogeneity, and to describe conformational flexibility that reflects overall protein dynamics. The rapid cooling of the crystal in liquid nitrogen traps various conformational states that can exist at 37 °C, although the thermodynamics of conformational changes may shift the equilibrium among states.44,45 In the present study, four different subunits were modeled, providing an opportunity to confirm assignments and to identify consistent conformations. As illustrated in Figure 2, leucine residues 57 and 309 have alternative conformations in each structure, and Leu-116 has different conformations depending upon which alcohol is bound. Altogether, alternative conformations were modeled for 50 amino acid residues of the 748 in the dimeric molecule of each complex (Table 2). About half of these residues are found in both subunits, indicating similar environments. Since the asymmetric unit is the dimeric molecule, the subunits can be different, but the structures of the subunits are almost identical. Four residues differ in their alternative conformations between the two structures. We can expect that other alternative conformations are present, but not yet identified, since fitting of electron density maps is never finished.
Table 2.
Alternative Amino Acid Conformations and Associated Partial Waters
Amino acid residues are numbered from 1 to 374, and waters from 401 upward, in subunits A and B. “AB” signifies both subunits. Associated partial water molecules are given in parentheses.
| Arg | B120a |
| Asp | A161, AB273, AB252 |
| Cys | A46b, AB282 |
| Gln | A251 (A880, A1029), B251 (B746, B819), A299 (A700, A831, A1028, MRD A379), B299 (B864) |
| Glu | A107 (C=O, A729), B107 (C=O, B741), B167, A239 (A998), B239 (B930), AB252, A256 (A724, A901, B793), B256 (B812), B353 (B703), B366 (side chain and C=O, B690) |
| His | A138 (A829, A914b) |
| Leu | A57, A61, AB309 |
| Lys | A39, A185a, AB188, B212 (B778), A228 (A664), B228 (B907), AB247 (C=O), B315, AB330, A338, B338b |
| Met | AB40; AB336 |
| Ser | A117, AB280, AB302 |
| Val | AB184 |
| Waters | (alternative, doubled) A488, A514, A517, A615, A623, A646, A664 (by A228 Lys), A729 (by A107), A764, A857a, A862, A880 (by A251), A915, A926, A958, A980, A986, B444, B544, B630, B762, B774, B775, B876, B904, B907 (by B228 Lys), B909, B910, B911, B912, B927a |
Only in TFE structure
Only in PFB structure
As expected, the longer-chain amino acids (glutamine, glutamate, and lysine) on the surface have some propensity for occupying different positions, typically in energetically reasonable conformations. In several cases, two conformations were identified and more may be present, but the electron densities were not sufficient to model every position. Some of the glutamate positions result from rotation of 120° around the CA-CB bond, putting the carboxylates in very different places. The epsilon amino group of lysine often seems to occupy alternative positions. Some alternative positions are unusual. The minor (4%) position for Cys-A46 in the structure with PFB may result from the loss of some catalytic zinc, and thus would represent artifactual, chemical heterogeneity. Such heterogeneity was not observed in the positions of the other three Cys-46 residues. Backbone carbonyl groups for Glu-107 and Lys-247 exhibit some “wobble” that was enclosed in an enlarged electron density, but the side chain positions were not affected. Glu-B366 has the whole residue in alternative positions. The origin of these alternative conformations is not obvious.
Several water molecules with partial occupancies interact with the partially occupied amino acid side chains. The occupancies of these waters were adjusted to reflect the occupancies of the side chains, rather than to let the temperature factors account for the partial occupancies. Several water molecules (30 in common for the two structures) also have alternative positions, which were modeled as doubled atoms.
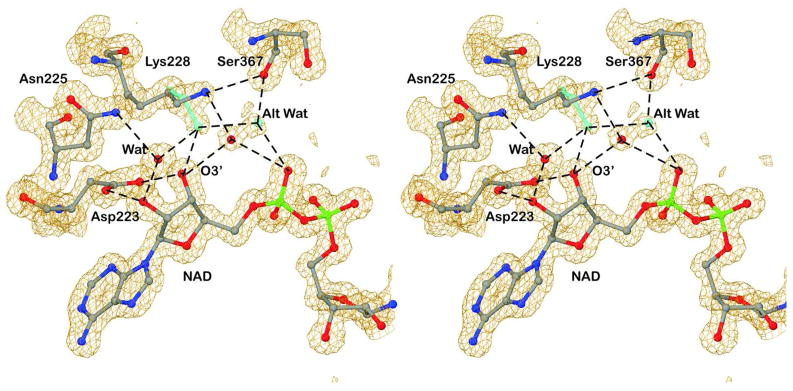
An interesting example of alternative positions for a side chain and a water is that of Lys-228, which interacts with the adenosine monophosphate portion of NAD (Figure 3). The epsilon amino (NE) group can interact indirectly via a water molecule with O3′ of the adenosine ribose and a phosphate oxygen or directly with O3′ and indirectly via the water in the alternative position with the phosphate oxygen. The occupancies range from about 50:50 in the B subunit to 80:20 in the A subunit. It appears that the varied conformations may modulate binding interactions by increasing the fluidity of the site. The role of Lys-228 has been studied. The K228R substitution moderately increases steady-state kinetic constants (e.g., Kd values for NAD+ and NADH increase by 4 to 7-fold), and the guanidino group of Arg-228 displaces the water and is accommodated with small, local changes.34,46 Larger, chemical modifications of Lys-228 substantially affect coenzyme binding and can result in large increases in catalytic activity.6,47
Figure 3.
Alternative conformation for Lys-228 and its accompanying water. The green stick model and water represent the alternative positions. Hydrogen bonds are represented by the dashed lines. The electron density map is contoured at about 1 σ above the mean.
Recent studies suggest that enzymes exist in an ensemble of different conformational states, including alternative conformations of side chains, and these states have different catalytic activities.48,49 If each of the 50 alternative amino acid conformations identified in this study are independent, then the ensemble could include 1015 different conformations! Although amino acid residues throughout the protein may contribute to the dynamics of the scaffold, we suggest that the different conformations of leucine residues 57 and 309 may be most directly relevant for catalysis. Indeed, the L57F substitution unmasked hydrogen tunneling, and increased catalytic efficiency on benzyl alcohol by 2.8-fold.7,50 Perhaps the phenylalanine residue limits conformational flexibility and helps to position the substrate for reaction. The contributions of many other residues remain to be investigated.
Geometry of Ligand Binding
The high resolution structures provide significant details of the arrangement of ligands and bond distances that are relevant for the mechanism (Table 3). As analogues of the Michaelis complexes, the distances between C4N of NAD+ and what would be the reactive carbon (-CH2-) of the alcohol are 3.40 ± 0.04 Å for the structures with PFB and TFE, when the distances in the four subunits of the two structures are averaged. (The individual values from refinements with SHELXL are given for each structure, and the values for the four fluoroalcohols from the refinements with REFMAC were averaged for comparison to other structures refined similarly.) The distance of 3.40 Å is a typical van der Waals distance, and the geometry is appropriate for direct transfer of a hydride ion from the alcohol to C4N of NAD+. The complexes appear to be ready to form a transition state, without significant rotation of the single bonds around the methylene carbon.
Table 3.
Distances (in Å units) for Ligand Binding in Complexes of ADH
| Structure | NAD C4N - Lig “C”a | Zn - Lig O | Ser48 OG - Lig O |
|---|---|---|---|
| PFB b, A subunit | 3.36 ± 0.02 | 1.97 ± 0.01 | 2.48 ± 0.01 |
| B subunit | 3.36 ± 0.03 | 1.98 ± 0.01 | 2.52 ± 0.03 |
| TFE b, A subunit | 3.43 ± 0.03 | 1.97 ± 0.01 | 2.48 ± 0.01 |
| B subunit | 3.42 ± 0.06 | 1.98 ± 0.01 | 2.47 ± 0.10 |
| FAlc c, ave of 4 | 3.40 ± 0.03* | 1.96 ± 0.02* | 2.52 ± 0.02* |
| 1P1R, ave of 4d | 3.48 ± 0.09 | 2.15 ± 0.04* | 2.72 ± 0.03* |
| 3BTO, ave of 4e | 3.56 ± 0.03* | 2.10 ± 0.04* | 2.67 ± 0.04* |
The ligands (Lig) and the atoms corresponding to the reactive carbon (“C”) are pentafluorobenzyl alcohol (C7), trifluoroethanol (C1, labeled as C2 for “ETF” in the PDB file), N-1-methylhexylformamide (C1), 3-butylthiolane 1-oxide (S6).
The distances and errors calculated with SHELXL.
The distances for the four values for the fluoroalcohols from the REFMAC refinements were averaged and compared to the averages of the four distances for the complexes with methylhexylformamide and 3-butylthiolane 1-oxide, and the * indicates that the differences relative to the complexes with fluoroalcohols were significant at p <0.01.
X-ray data for NADH and (R)-N-1-methylhexylformamide [CH3(CH2)4CH(CH3)NHCHO], Ki = 15 μM, complexed with ADH at 1.57 Å resolution; average of 4 subunits.66
X-ray data for NADH and (1S,3S)-3-butylthiolane 1-oxide, Ki = 0.72 μM, complexed with ADH at 1.66 Å resolution; average of 4 subunits.37
Two moderately high resolution (2.0 Å) structures have been determined for mutated horse liver ADHs complexed with NAD+ and TFE: F93W enzyme (1AXE.pdb51) and F93W/V203A enzyme (1A71.pdb52). The unit cell for the structure determined at 277 K (1AXE) is slightly larger than the cell determined at 100 K, and the coenzyme binding domains can be superimposed with an rmsd of 0.26 Å. Since one or two amino acids are substituted in the active site, some differences can be expected in ligand binding relative to wild-type or each mutated enzyme. It appears the nicotinamide ring and the TFE are positioned differently in the mutated enzymes. The distance between C1 of TFE and C4N of NAD+ averages 3.16 Å in the mutated F93W ADH, whereas the C1-C4N distance is 3.54 Å in the doubly mutated F93W/V203A ADH. The difference is attributed to the V203A substitution and is considered to be a critical factor in the extent of quantum mechanical tunneling during hydride transfer.19,52 In the wild-type enzyme the distance is 3.43 ± 0.03 Å. In the complex with NAD+ and TFE with (non-zinc) ADH from Drosophila melanogaster, the distance between C4N and C1 of TFE is 3.29 Å (1SBY.pdb).
The distances in the NAD+-alcohol complexes can be compared to those in structures that are analogues of the NADH-aldehyde complex in order to characterize the changes that occur during the catalytic reaction. The distances between C4N of NADH and the atom that corresponds to the reducible C of the carbonyl group in the structures with methylhexylformamide (1P1R.pdb) or 3-butylthiolane 1-oxide (3BTO.pdb) average 3.53 or 3.57 Å (Table 3). This would be a reasonable distance for the ground state structure for the Michaelis complex of NADH and a carbonyl compound. The complex with the formamide may be the closest model of the Michaelis complex with an aldehyde, whereas the complex with the thiolane oxide may resemble the complex with cyclohexanone, which is a good substrate. The difference in distances between the structures with the alcohols and the 3-butylthiolane 1-oxide is significant, even though the resolutions of the structures with NADH are not as high as those with NAD+ and the fluoroalcohols.
The average distance in the complexes between the O of the fluoroalcohols and the catalytic zinc is 1.96 Å, which would be consistent with the binding of an alkoxide. For comparison, the distances between the zinc and the doubly-bonded O atom in the complexes with NADH are 2.15 or 2.10 Å, which differ significantly from the distances for the fluoroalcohols. Transient kinetic studies of proton release during binding of trifluoroethanol suggest that the pK of the ternary complex is 4.6,53 and the crystals were prepared at pH 7, making it reasonable to assume an alkoxide is present. Comparable pH dependence studies have not been done with pentafluorobenzyl alcohol, but electron-withdrawal by the five fluorine atoms is expected to facilitate deprotonation of the alcohol. Molecular dynamics simulations provide distances for the Zn-O of about 2.0 Å for the complex with NAD+ and benzyl alcohol, increasing to about 2.2 Å in the complex with NADH and benzaldehyde.14,54
Microscopic pK values for the ionization of substrate alcohols are difficult to assign, as the alcohols participate in the hydrogen-bonded network linked to His-51,33 but apparent pK values derived from the pH dependence of hydride transfer gives a pK value of 6.4 for ethanol and benzyl alcohol oxidations at 25 °C.55,56 The pK for water bound to the enzyme-NAD+ complex is estimated to be 7.3,53 and it is reasonable to assume that the pK values of the bound alcohols would be lower than 7.3, yielding the alkoxides in the complexes.
The average distance between the O of the fluoroalcohol and the hydroxyl group of Ser-48 is 2.48 ± 0.01 Å (SHELXL refinement), shorter than expected for a typical hydrogen bond and in the range expected for a low-barrier hydrogen bond, which may be important for catalysis in enzymes.57 Thus, the fluoroalcohol and the hydroxyl group share the proton, and each O would have alkoxide character. The structural results corroborate the studies on solvent isotope effects for benzyl alcohol oxidation, which showed an inverse effect of 2-fold (faster in D2O than in H2O), consistent with a partial charge on the substrate oxygen of − 0.3 in the transition state.50,58,59 The corresponding distance in the complexes with aldehyde analogs are 2.69 ± 0.04 Å, significantly longer than in the alcohol complexes and typical of a normal hydrogen bond.
High resolution structures (1.45 to 1.65 Å) of human liver alcohol dehydrogenase isoenzymes complexed with formamides show geometry similar to those for the horse enzyme, with average distances of 3.85 Å between the carbonyl carbon of the formamide and C4N of NAD in ADH1B1 and 3.43 in ADH1C2 (1U3U.pdb, 1U3V.pdb, 1U3W.pdb).60 Likewise, the average distance between the O of the formamide and the catalytic zinc is 2.22 Å, and the distance of the O to the hydroxyl group of Ser/Thr-48 is 2.61 Å. However, in these structures, the coenzyme was refined as NAD+ and the nicotinamide rings are planar. Perhaps these structures resemble “abortive” complexes.
Identification of the Coenzyme
The horse liver ADH was crystallized with NAD+ and the fluoroalcohol, and it could be assumed that the crystals would contain the oxidized nicotinamide ring since the fluoroalcohols are not substrates and they would also strongly inhibit oxidation of methylpentanediol and adventitious alcohols in the solutions. However, microspectrophotometry led to the suggestion that crystals prepared with NAD+ and trifluoroethanol61 contained 100% of the coenzyme as NADH.62 We believe the formation of NADH in the crystals was an artifact due to washing of the crystals before spectrometry with a buffer containing only 30% methylpentanediol, which would reduce the NAD+ when the inhibiting trifluoroethanol was removed. We found that mixing stoichiometric concentrations of enzyme and NAD+ with excess methylpentanediol led to substantial formation of NADH.63 Note that our crystals were directly flash cooled in liquid N2 so that the NAD+ and fluoroalcohol were still present. Nevertheless, the long time for crystallization (some weeks at 4 °C) and the possibility of radiation-induced reduction or damage of the nicotinamide ring requires that oxidation state of the coenzyme be assessed. The atomic resolution data allow the structure of the coenzyme to be determined when we use a procedure appropriate for refinement of structures of enzymes crystallized with NAD+ or NADH.
As described in Experimental Procedures, we used a modified dictionary for NAD in which the restraints on bond distances and ring planarity for the nicotinamide ring were relaxed, so that refinements could fit either NAD+ or NADH. This molecule is named NAJ in our PDB entries, and it is based on the highest resolution structure of NAD+ available.64 The results in Table 4 summarize the bond distances in the nicotinamide rings in various structures with oxidized and reduced nicotinamide rings. The first four lines in Table 4 present the results for the refinements with SHELXL. The bond distances in the nicotinamide rings for the A and B subunits are the same within errors. The fifth line has the weighted averaged values from refinement of the PFB and TFE complexes, and the propagated errors are about 0.01 Å for each distance. On the sixth line are the averages of the four distances from refinement with REFMAC. The bond distances determined by both refinement procedures are the same within the standard errors of about 0.02 Å, the same as the overall estimated coordinate error.
Table 4.
Bond distances (Å units) in Nicotinamide Rings
| Structure | N1-C2 | C2-C3 | C3-C4 | C4-C5 | C5-C6 | C6-N1 | average deviation |
|---|---|---|---|---|---|---|---|
| 1 PFB, A subunita | 1.365 ± 0.011 | 1.362 ± 0.012 | 1.441 ± 0.013 | 1.415 ± 0.014 | 1.358 ± 0.013 | 1.420 ± 0.012 | 0.012 |
| 2 PFB, B subunita | 1.375 ± 0.024 | 1.353 ± 0.033 | 1.443 ± 0.018 | 1.394 ± 0.034 | 1.352 ± 0.034 | 1.391 ± 0.015 | 0.026 |
| 3 TFE, A subunita | 1.378 ± 0.013 | 1.365 ± 0.014 | 1.416 ± 0.017 | 1.371 ± 0.016 | 1.371 ± 0.014 | 1.398 ± 0.014 | 0.015 |
| 4 TFE, B subunita | 1.384 ± 0.087 | 1.354 ± 0.128 | 1.408 ± 0.026 | 1.376 ± 0.123 | 1.365 ± 0.123 | 1.389 ± 0.026 | 0.086 |
| 5 Weighted average of 4 subunitsa | 1.371 ± 0.008 | 1.363 ± 0.009 | 1.432 ± 0.009 | 1.396 ± 0.010 | 1.363 ± 0.009 | 1.404 ± 0.007 | 0.009 |
| 6 REFMAC, averageb | 1.37 | 1.35 | 1.43 | 1.38 | 1.36 | 1.38 | 0.018 |
| 7 NAJc | 1.346 | 1.391 | 1.398 | 1.384 | 1.379 | 1.347 | 0.005 |
| 8 LiNAD+d | 1.39 | 1.37 | 1.40 | 1.42 | 1.37 | 1.35 | 0.02 |
| 9 NAD(P)+e | 1.36 | 1.38 | 1.40 | 1.39 | 1.38 | 1.37 | 0.014 |
| 10 dihydronicotinamidef | 1.38 | 1.32 | 1.51 | 1.53 | 1.32 | 1.43 | 0.012 |
| 11 NADH, as NAJg | 1.40 | 1.38 | 1.50 | 1.46 | 1.36 | 1.46 | 0.034 |
| 12 NADH in ADHh | 1.40 | 1.35 | 1.48 | 1.47 | 1.41 | 1.40 | 0.031 |
| 13 NADPHi | 1.38 | 1.38 | 1.49 | 1.48 | 1.38 | 1.42 | 0.026 |
Bond distances and errors from refinement with SHELXL with no restraints on the bond distances and planarity for the coenzyme.
Average bond distances (of 4 subunits) from refinement of PFB and TFE structures with REFMAC with relaxed restraints (0.10 Å) on distances and no restraints on planarity of the nicotinamide ring. (Note that protons were removed on N1A and phosphate AO2 to represent the coenzyme at neutral pH).
Neutron diffraction at 0.65 Å resolution with estimated errors of 0.005 Å.64
X-ray data at 1.09 Å with estimated average errors of 0.02 Å.89 The distances are within 0.03 Å of those found for N-1-(2,6-dichlorobenzyl)-3-carbamidopyridinium iodide.90
Five structures of enzymes complexed with NAD+ determined at 1.0 – 1.2 Å resolution (PDB entries: 1ZJZ, 1SBY, 1T2D, 3JY0, 2O23) and five structures with NADP+ determined at 0.66 to 1.0 Å resolution (PDB entries: 1US0, 3BCJ, 1PWM, 2J8T, 1ZK4). These values are within 0.01 Å of the target values in the dictionary from REFMAC where the restraints usually are set at 0.02 Å.
N-Benzyl-1,4-dihydronicotinamide.91 The nicotinamide ring is planar.
Average of eight subunits in complexes of ADH with NADH, refined as NAJ, and (R)-N-1-methylhexylformamide (1.57 Å resolution, 1P1R.pdb66) or (1S,3S)-3-butylthiolane 1-oxide (1.66 Å resolution, 3BTO.pdb37).
Four structures (1.0 – 1.2 Å, average of 8 subunits) of zinc or cadmium horse liver ADH complexed with NADH and isobutyramide, dimethyl sulfoxide, or hydroxide partly adducted to the nicotinamide ring (PDB files: 1HET, 1HEU, 2JHG, 1JHF).20,21
Five structures of NADPH in enzymes determined at 1.09 to 1.35 Å resolution; all nicotinamide rings were essentially planar (PDB files: 3DJJ, 1LQU, 1YNQ, 1HEJ, 1KMS).
For comparison, Table 4 also gives the bond distances determined for the oxidized coenzyme (NAJ and LiNAD, lines 7 and 8) and averages for NAD(P)+ in structures of ten enzymes solved at resolution better than 1.2 Å (line 9). Lines 10 – 13 contain experimental values for the reduced nicotinamide ring of a model compound as well as those from four structures of ADH with NADH or five structures with NADPH determined at high resolution (1.1 to 1.35 Å). The bond distances in the nicotinamide ring in the structures with the fluoroalcohols (line 5) most closely fit the distances determined for the oxidized coenzyme (line 9) rather than those for NAD(P)H (lines 10 – 13). In particular, the bond distances for C3–C4 and C4–C5 in the complexes with the fluoroalcohols are shorter by 0.06 to 0.08 Å than those for the reduced nicotinamide rings. When the averaged values (4 subunits) for the structures with pentafluorobenzyl alcohol and trifluoroethanol (line 5) are compared with those (8 subunits) for the structures of ADH complexed with NADH and carbonyl analogues (line 11, refined as NAJ), the differences are significant at p < 0.01, and as compared to the values for NADPH (line 13), the differences are significant at p < 0.002.
However, close examination shows some small, but probably mechanistically significant differences in bond distances in the complexes. The C3–C4 bond distances for the nicotinamide rings in the complexes with fluoroalcohols are longer (compare lines 5 and 9) than those for NAD(P)+ structures by about 0.03 Å (p < 0.03), whereas the C4–C5 distance is not distinguishable from NAD(P)+. Moreover, the C2–C3 bond distance is about 0.02 Å shorter than the bonds in either NAD(P)+ or NADPH (p < 0.02). The C5–C6 distance is about 0.02 Å shorter than the bond in either NAD(P)+ (p < 0.01) or NADPH (p < 0.1). The bond distances for N1-C2 and C6-N1 are in-between (with differences of 0.01 – 0.03 Å) those for NAD(P)+ (p <0.02) and NADPH (p < 0.3). It does not appear that some mixture of NAD+ and NADH is present in the complexes with the fluoroalcohols because the C4-C5 bond distance is the same as in the structures with oxidized nicotinamide rings. The electron density maps show no difference densities at 2 σ above the average in the |Fo| − |Fc| maps. It was the plus/minus difference densities observed near C4N in the structures with NADH (1P1R.pdb) and NAD+-pyrazole (1N8K.pdb) that led us to conclude that the nicotinamide rings were puckered and that the restraints in the refinement needed to be relaxed. As observed in Figure 1, the electron densities around C4N are similar to those for the other atoms in the ring, but the B-factors for C4N are slightly higher (about 12%) than the average of the other five atoms in the ring. This indicates that the atoms are fitted well in density, but perhaps there is some loss of electron density or an increase in motion at C4N. There is no evidence that the X-rays alter the state of oxidation of the coenzyme, as structures with either coenzyme are obtained. The observed structures are consistent with the hypothesis that the nicotinamide ring becomes slightly puckered with small alterations in bond distances and develops a partial positive charge on C4N in the transition state.65
Puckered Nicotinamide Rings
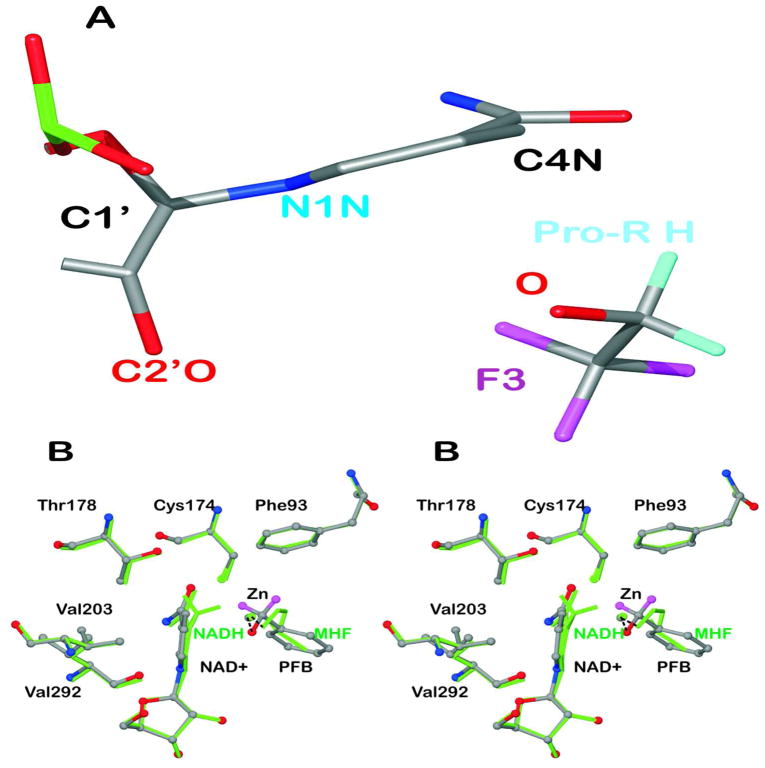
The structures with NAD+ and fluoroalcohols have slightly puckered nicotinamide rings (quasi-boat conformation) with a torsion angle of 5.9 ± 1.5° (αC420) for displacement of C4N in the plane with C3-C4-C5 from the plane of atoms C2-C3-C6 and an angle of 4.8 ± 1.6° (αN1) for displacement of N1N in the plane with N1-C2-C6 from the plane with C2-C3-C6 as determined from the refinements of the four subunits with SHELXL (Figure 4A). These calculations account for the errors in atomic positions and a slight twist of the nicotinamide ring, but the torsion angle defined by atoms C2-C3-C4-C5 is the same within error. Nicotinamide rings of NADH have more obvious boat conformations, as demanded by the electron density maps and described for the first time for the complex of ADH with NADH and methylhexylformamide66 (Figure 4B). The puckered rings (αC4 = 18°, αN1 = 12° for 1P1R.pdb) place C4N relatively closer to the carbonyl carbon of the substrate analogue, which may facilitate hydride transfer in reactive complexes.14,54,67
Figure 4.
Puckering of the nicotinamide rings. (A) The structure of the complex with NAD+ and TFE (subunit A) is shown with atoms C2N and C3N aligned on top of atoms C5N and C6N so that the displacements of N1N and C4N from the plane are visible. (B) Comparison of the structure of the complex with NAD+ and pentafluorobenzyl alcohol to that with NADH and methylhexylformamide. The “A” subunits of the structures (amino acid residues A1 to A374) were superimposed, with an rmsd of 0.25 Å. The complex with NAD+ and pentafluorobenzyl alcohol (with H atoms in magenta but F atoms removed for clarity) are shown in atom coloring with ball and stick representation. The complex with NADH (with H atoms on C4N) and methylhexylformamide (“MHF”, with atoms C3-C6 removed for clarity) is shown in green. The residues making close contacts are labeled. The distances are given in Table 5.
Previous structural analyses of enzymes that bind NAD(P) could have found puckered, oxidized nicotinamide rings, but atomic resolution is required, and details of the refinement procedures and the state of oxidation are often not clear. Unfortunately, the default REFMAC dictionaries for oxidized and reduced NAD(P) enforce planarity for the nicotinamide rings, although bond distances are longer for the reduced ring. A systematic study of 340 Protein Data Bank entries for structures containing NAD(P) solved at better than 2.0 Å concluded that the structures with an oxidized nicotinamide ring, and even some with a reduced nicotinamide ring, were planar.68 A small subset had distorted nicotinamide rings, and all of those with severe puckering (14 of them with torsion angles > 10°) had close electrostatic interactions with oxygen atoms.68 Some of these close contacts may be due to covalent adducts, as was described for NAD+-pyrazole complexes with ADH, where the bond distances in the nicotinamide ring are close to those for NADH (refined as NAJ, 1N92.pdb, 1N8K.pdb).69
We surveyed the Protein Data Bank for structures of enzymes with NAD(P)+ determined at atomic resolution, which should yield good bond distances and perhaps evidence of ring puckering and close contacts. Five structures with NAD+ have planar nicotinamide rings, and four of five structures with NADP+ have slightly puckered rings, and all have bond distances (Table 4, line 9) fitting those for NAJ and LiNAD (lines 7 and 8). All of the nicotinamide rings, except those described in 1ZJZ.pdb, 3JYO.pdb and 1ZK4.pdb, appear to have close contacts to oxygen atoms of the protein or a ligand. Good evidence for puckered, oxidized nicotinamide rings is sparse, except in the structures of ADH with fluorinated alcohols (including also 3OQ6.pdb).
Structures with NAD(P)H may be expected to have puckered nicotinamide rings, as for two structures of ADH (Table 4, line 11). However, several structures with NADPH (Table 4, line 13) all have essentially planar rings, as does N-benzyldihydronicotinamide (line 10). Some structures have been reported with NADH, but the identification of the coenzyme and the refinement procedures are often not clear. A slightly puckered nicotinamide ring was refined for the abortive NADH/phenylalanine complex with L-phenylalanine dehydrogenase (1C1D.pdb, 1.25 Å), with bond distances most similar to reduced nicotinamide rings.70 Somewhat puckered nicotinamide rings were described for the abortive NADH/UDP-glucose complex of UDP-galactose 4-epimerase (1EK6.pdb, 1.5 Å71) and for the NADH complex with aminoadipate semialdehyde dehydrogenase (2J6L.pdb, 1.3 Å), but the bond distances in the nicotinamide rings are closer to those for NAD+ than for NADH. The abortive NADH/R-phenylethanol complex with the R-specific alcohol dehydrogenase (1ZJY.pdb, 1.05 Å) shows a planar nicotinamide ring with bond distances almost identical to those for NAD+.72 Some of these structures have close contacts to the nicotinamide rings, but their significance is not clear.
In contrast, atomic resolution (1.0 – 1.2 Å) studies on several complexes of horse liver alcohol dehydrogenase with NADH, using refinements with relaxed restraints on the coenzyme, identified substantially puckered nicotinamide rings and bond distances that fit reduced nicotinamide rings (Table 4, line 12).20,21 However, the interpretations of the electron density maps are complicated because of the evidence for covalent adducts with the nicotinamide rings, which produce a mixture of NADH forms in each active site. In one structure (1HET.pdb), 60% of the molecules apparently have a hydrated nicotinamide ring (“hydroxide” adduct, but the covalent bond distance is somewhat long at 1.96 Å), and the other 40% have the water in an alternative position that is typically occupied by the oxygen of the substrate analogs ligated to the catalytic zinc. The hydroxide adduct is suggested to activate the nicotinamide ring for hydride transfer and to participate as a base in alcohol oxidation in a proposed central complex with a pentacoordinated zinc.20 However, there is no good chemical rationale for this mechanism, and there is little room to accommodate five ligands to the zinc without some structurally significant rearrangements.33 Moreover, in another structure (2JHG.pdb), with NADH and isobutyramide (an inhibitory analog of a carbonyl substrate), the extra water is totally excluded, but adducts (with a bond distance of 1.95 Å) have apparently formed between the amino group of the amide and C4N of the nicotinamide with partial occupancy.21 Hydroxide adducts with NADH were also proposed for complexes of ADH in which the catalytic zinc was replaced with Cd2+.20,21 Because of the heterogeneity of the complexes with adducts of NADH in each active site, it is not clear from these studies which structures are relevant for the catalytic mechanism.
Strained Nicotinamide Rings
Considering the slight puckering of the nicotinamide ring observed in the structures with fluoroalcohols (REFMAC or SHELXL refinements) and the altered bond distances as compared to oxidized nicotinamide rings, we suggest that the complexes represent a strained, oxidized ring part way on the path to the transition state. Although the statistical analyses are consistent with altered bond distances, the overall pattern of changes is illuminating as they are very similar to the differences calculated for the transition state structure as compared to ground state NAD+ and NADH for formate dehydrogenase,65 as discussed later.
Examination of the environment of the nicotinamide ring suggests that some close contacts may be responsible for the ring puckering. Figure 4B can illustrate the ground state structures for the substrates and products. The nicotinamide ring in the complexes formed with NAD+ and fluoroalcohols is slightly puckered (Figure 4A) and becomes more obviously puckered in complexes with NADH and carbonyl compounds. We suggest that close contacts with the protein may stress the nicotinamide ring in the oxidized state, and the stress is relieved in the reduced ring, which is obviously puckered as could be expected from the tetrahedral bonding at C4N. Table 5 lists these contacts along with the corresponding distances in the structure with NADH and methylhexylformamide. The distances between atoms of the reduced ring to Cys174 SG, Thr178 CG2, and Val292 O are significantly longer (0.05 to 0.20 Å) than those distances in the NAD+-fluoroalcohol complexes. The puckering of the reduced nicotinamide ring observed in the structures with methylhexylformamide and 3-butylthiolane oxide (1P1R and 3BTO) moves C4N about 0.25 Å closer, as compared to the position with NAD+, to the atom of the carbonyl analog that would accept the hydride.
Table 5.
Close Interactions of the Nicotinamide Rings in Complexes with ADH
| Contact to NAD(H) | NAD+-Fluoroalcohol | NADH, 1P1R |
|---|---|---|
| Cys174 SG - C4N | 3.52 ± 0.01a | 3.62 ± 0.04 |
| Thr178 OG1 - C4N | 3.42 ± 0.02 | 3.52 ± 0.04 |
| Thr178 CG2 - C4N | 3.39 ± 0.02a | 3.59 ± 0.06a |
| Val292 O - C2N | 3.03 ± 0.01a | 3.08 ± 0.02a |
| Val203 CG2 - C6N | 3.64 ± 0.05 | 3.70 ± 0.05 |
| Lig378 O - C5N | 3.04 ± 0.03a | 3.21 ± 0.04a |
| Val292 O - N7N | 2.95 ± 0.02 | 2.98 ± 0.02 |
| Ala317 O - N7N | 2.98 ± 0.02 | 2.97 ± 0.02 |
| Phe319 NH - O7N | 2.85 ± 0.03 | 2.80 ± 0.04 |
The differences between the average distances in the four subunits of complexes with the fluoroalcohols and methylhexylformamide are significant at p < 0.02.
If the contacts observed between the enzyme and the oxidized coenzyme are contributing to the ring strain, substitution of the amino acid residues should affect catalytic activity. Indeed, the T178S substitution (loss of the CG2 methyl group contact) decreases the observed rate constant for hydride transfer with benzyl alcohol from 24 s−1 to 2.9 s−1 without affecting coenzyme binding, whereas the T178V substitution decreased affinity for NADH and NAD+ by 4 and 8-fold, respectively, without affecting hydride transfer.69 The V203A substitution decreased the observed rate constant for hydride transfer to 1.5 s−1. It is more difficult to probe the role of the carbonyl O of Val-292, but the V292A, S, or T substitutions decreased affinity for coenzymes 30 – 60-fold and the rate constant for hydride transfer to 5 – 9 s−1. The isomerization of the V292S enzyme-NAD+ complex (open conformation to closed) is hindered,6,9 but the V292T enzyme forms a closed complex with the NAD+-pyrazole adduct, which has a severely puckered nicotinamide ring that could resemble the transition state.69 In these V292S or T enzymes, a new water molecule that makes hydrogen bonds with the new hydroxyl group is inserted, but how this affects the conformational equilibrium is not clear. By comparing the structures of the wild-type and V292T enzymes complexed with NAD+ and pyrazole (1N92.pdb and 1N8K.pdb), the distances between C4N of the nicotinamide ring and Thr-178 OG1 or CG2 are about 0.07 Å longer in the mutated as compared to wild-type enzyme, but the distances between Val-292 O and C2N are not affected. Although the alterations in rate constants and contact distances due to the amino acid substitutions are not very large, the experimental evidence for structural explanations is consistent with the suggestion that binding to the protein causes distortion of the nicotinamide ring in the ground state.
Mechanistic Conclusions
The atomic resolution structures of the enzyme with NAD+ and alcohols represent two different complexes that resemble the expected Michaelis complexes. We suggest that hydride would be transferred directly between the coenzyme and the substrate in an environment with a tetrahedral zinc after the proton from the alcohol is relayed through the hydrogen-bonded network to His-51 (Figure 1).33,34 These structures complement those with NADH and analogues of the aldehyde substrate, particularly the one with methylhexylformamide, which resembles the Michaelis complex expected for the reverse reaction (Figure 4B). The geometry of the binding defines the distances for proton and hydride ion transfers. Each fluoroalcohol is formally deprotonated, but the alkoxide forms a low-barrier hydrogen bond to the Ser-48 OG hydroxyl group. Calculations show that the interaction of the alkoxide with zinc decreases the energy barrier for hydride transfer.73 The distances between the O of the carbonyl analogues and Ser-48 OG and the zinc are significantly longer in the complexes mimicking the ADH-NADH-aldehyde complexes, consistent with calculations.14 The interactions of the alcohols with the amino acid side chains determine that the pro-R hydrogen of the alcohol would be most directly transferred to the re-face of the nicotinamide ring (Figure 2).
We do not think that any evidence supports a role for a water to activate the coenzyme in a central complex that would be poised to transfer hydrogen.20 A central, pentacoordinated complex with both the substrate oxygen and a water bound to the catalytic zinc was first proposed to explain NMR results,74 but the idea was abandoned when the structure of a ternary complex showed no such coordination.33 The various structures with water and substrate analogs in the same active site may be transient species that occur during replacement of the water ligated to the zinc by the oxygen of the substrate.21 The exchange of water and substrate may occur via a pentacoordinate species or by a double displacement mechanism involving Glu-68.53
The potential significance of the strained ground state for catalysis may be illustrated by comparing the equilibrium constant for the overall reaction (NAD+ + alcohol == NADH + aldehyde) to the constant when the substrates are bound to the enzyme. At pH 7, the equilibrium constant (Keq) for the oxidation of benzyl alcohol is 3.8 × 10−4 (favoring alcohol), whereas bound to the enzyme, the “on-enzyme” equilibrium constant (Kint) is about 0.25.63 Thus, it appears that the enzyme shifts the equilibrium position in favor of alcohol oxidation by a factor of 650, or 3.8 kcal/mole. Some of the intrinsic binding energy for NAD+ may distort the nicotinamide ring, accounting for some of shift. The positively-charged nicotinamide ring also seems to shift the pK of the water bound to the catalytic zinc in the binary enzyme-NAD+ complex by 2.2 units (3.0 kcal/mole).53 If a similar shift occurs for alcohol bound to the zinc in the ternary complex, which would facilitate formation of the alkoxide, the distortion and pK shift could account for the shift in the equilibrium constant. At pH 7, the dissociation constant for NAD+ is 120 μM and that for NADH is 0.41 μM.63 If the intrinsic binding energy of NAD+ is the same as that of NADH, some 3.4 kcal/mole is available to shift the on-enzyme equilibrium. (Some intrinsic binding energy is also used when the enzyme changes conformation, which may be about the same for both coenzymes.) The magnitudes of these energy changes are reasonable, but are not definitively assigned to particular structural changes.
The on-enzyme equilibrium can also be affected by interactions of the enzyme with NADH and the aldehyde. Raman spectroscopy shows that the carbonyl group of a formamide is stabilized by 5.5 kcal/mole upon binding to ADH.75 In contrast, a Raman study of the homologous lactate and malate dehydrogenases led to the suggestion that the binding NADH did not involve stabilization of the pro-R hydrogen in the binary complexes.76 Further experimental and theoretical studies on ADH should determine the energetics of each step in the enzyme reaction.
As described in reviews of the role of strain in enzyme catalysis, the enzyme must bind, in principle, the transition state relatively more tightly than it binds the substrates and products.77,78 If binding of substrates and the transition state were equally tight, the chemical reaction would occur no more rapidly in the presence of the enzyme than in the absence because the difference in energy between substrates and the transition state would be unchanged. Binding of the substrates may be less tight because some potential binding energy is used to destabilize the substrates, raising the energy of the ground states and differentially lowering the barrier for the chemical step. Since the enzyme is not a rigid, or static, scaffold, the enzyme is also strained by the binding of substrates, and strain by itself could not account for all of the catalytic efficiency.
Computational analyses on the reaction of dehydrogenases have suggested that the nicotinamide ring is puckered in the transition state for hydride transfer, which would favorably position C4N of the nicotinamide ring for transfer of hydride from an alcohol to the re-face of the ring leading to a pseudo-axial orientation for the pro-R hydrogen in the reduced nicotinamide ring.14,65,67,79–82 Distortion of the oxidized nicotinamide ring might also stabilize some carbonium ion character on C4N and facilitate the chemistry.83 (The 15N isotope effect of 1.0 for N1N-labeled coenzyme on the reaction does not disprove the hypothesis, because bond order at N1N may not change significantly.84) Calculations for formate dehydrogenase show that deformation of the oxidized nicotinamide ring, to produce the quasi-boat structure of the proposed transition state, with puckering angles αC4 of 15° and αN1 of 5°, is estimated to require 16 kcal/mole, whereas similar puckering of the reduced nicotinamide ring requires only 1.5 kcal/mole and has a relatively flat free-energy profile over a 20 to 30° range.67,85 The energy profile for deforming the oxidized nicotinamide ring has not been calculated.
Further calculations for the formate dehydrogenase reaction (gas phase or acetonitrile) characterized a transition state structure for the coenzyme.65 Relative to the oxidized ring in NAD+, bond distances for N1-C2 and C6-N1 increased in the transition state by 0.02 – 0.03 Å and by 0.03 – 0.05 Å in the reduced ring. The distances for C3-C4 and C4-C5 increased by 0.04 – 0.06 Å in the transition state and by 0.11 – 0.14 Å in the final reduced ring of NADH. The distances for C2-C3 and C5-C6 decreased in the transition state by 0.02 – 0.03 Å and decreased further to 0.04 – 0.05 Å in the reduced state. The changes in bond distances for the calculated transition states are very similar to those that are listed in Table 4 for NAD+ in the complexes with the fluoroalcohols as compared to free NAD+, except that the C3-C4 and C4-C5 distances are 0.03 – 0.05 Å shorter in the complexes as compared to the calculations. (We also note that the calculated changes in C2-C3 and C5-C6 distances in the reduced ring more closely match those in N-benzyldihydronicotinamide rather than in NAD(P)H as observed in the various enzyme complexes.) The αC4 puckering angle in the calculated transition state was 9° as compared to about 6° in the structures with fluoroalcohols. Of course, the crystallized enzyme complexes are not at the transition state, but the observed structures may define a strained state on the pathway to the transition state.
High-level, hybrid calculations that include all protein atoms and consider the quantum mechanical hydrogen tunneling provide important insights into the nature of the transition state. These calculations are validated by reproducing the free energy barrier for the transition state (15.5 kcal/mole for a rate constant for hydride transfer from benzyl alcohol of 38 s−1)58 and the primary and secondary kinetic isotope effects that are evidence of the tunneling.14,16,17 There are substantial motions in reaching the transition state, as the donor-acceptor distance between the reacting carbons decreases to about 2.6–2.7 Å. Many features of the enzyme contribute to the dynamics, but it was concluded that hydrophobic interactions, including those listed in Table 5 above, do not appreciably change the potential energy surface along the reaction path, whereas several polar residues (including Ser-48 and Asp-49) are important.16,17 On the other hand, it appears that the steric interactions between Val-203 and Thr-178 with the nicotinamide ring affect the equilibrium free energy via promoting motions that change the donor-acceptor distance.14,19 It appears that the complexes described here with the substrate analogs are beautifully “preorganized” 19 for hydride transfer, but what motions (reorganization) might be correlated to the chemical step need to be determined yet. Molecular dynamics and normal mode analyses suggest that motions of domains (related to the global conformational change) could “push” the substrates together.13,86 Equilibrium, vibrational motions might also be sufficient, however, as temperature factors of 10 Å2 (observed in these structures, albeit at 100 K) could correspond to motions of 0.35 Å, leading to a decrease in the donor-acceptor distance from 3.4 to 2.7 Å. However, a recent, empirically-parameterized model suggests that the “tunneling ready state” could have a donor acceptor distance of 3.2 Å.87 The atomic resolution X-ray structures presented here resemble such a state and describe the scaffold and ensembles that can be relevant for further studies on the dynamics and catalysis.
Acknowledgments
Funding
This work was supported by the United States Public Health Service, National Institutes of Health, grants GM078446 and AA00279 to B.V.P.
We thank Dr. Eric N. Brown for preliminary studies on the complex with trifluoroethanol, Dr. Lokesh Gakhar for assistance with the X-ray crystallography, and The University of Iowa Protein Crystallography Facility for instrumentation. Synchrotron data were collected at the Advanced Photon Source at Argonne National Laboratory. Results were obtained on beamline 23ID in the GM/CA-CAT, which has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-Eng-38. Results were also derived from work performed on beamline 19ID in the Structural Biology Center, with special assistance from Dr. Stephan Ginell. Argonne National Laboratory is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
ABBREVIATIONS
- ADH
alcohol dehydrogenase
- PFB
2,3,4,5,6-pentafluorobenzyl alcohol
- TFE
2,2,2-trifluoroethanol
- rmsd
root-mean-square deviation
Footnotes
The X-ray coordinates and structure factors have been deposited in the RCSB Protein Data Bank with the entry names 4DWV for the enzyme complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol and 4DXH for enzyme complexed with NAD+ and 2,2,2-trifluoroethanol.
The authors declare no competing interests.
References
- 1.Brändén CI, Jörnvall H, Eklund H, Furugren B. Alcohol Dehydrogenases. The Enzymes. (3) 1975;11:103–190. [Google Scholar]
- 2.Pettersson G. Liver alcohol dehydrogenase. CRC Crit Rev Biochem. 1987;21:349–389. [PubMed] [Google Scholar]
- 3.Eklund H, Brändén C-I. Biological Macromolecules and Assemblies: Volume 3-Active Sites of Enzymes. Wiley; 1987. Alcohol Dehydrogenase; pp. 73–141. [Google Scholar]
- 4.Eklund H, Ramaswamy S. Medium- and short-chain dehydrogenase/reductase gene and protein families: Three-dimensional structures of MDR alcohol dehydrogenases. Cell Mol Life Sci. 2008;65:3907–3917. doi: 10.1007/s00018-008-8589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brändén CI, Eklund H. Coenzyme-induced conformational changes and substrate binding in liver alcohol dehydrogenase. Ciba Found Symp. 1978;60:63–80. doi: 10.1002/9780470720424.ch5. [DOI] [PubMed] [Google Scholar]
- 6.Plapp BV. Conformational changes and catalysis by alcohol dehydrogenase. Arch Biochem Biophys. 2010;493:3–12. doi: 10.1016/j.abb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahnson BJ, Park DH, Kim K, Plapp BV, Klinman JP. Unmasking of hydrogen tunneling in the horse liver alcohol dehydrogenase reaction by site-directed mutagenesis. Biochemistry. 1993;32:5503–5507. doi: 10.1021/bi00072a003. [DOI] [PubMed] [Google Scholar]
- 8.Kohen A, Cannio R, Bartolucci S, Klinman JP. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature. 1999;399:496–499. doi: 10.1038/20981. [DOI] [PubMed] [Google Scholar]
- 9.Rubach JK, Ramaswamy S, Plapp BV. Contributions of valine-292 in the nicotinamide binding site of liver alcohol dehydrogenase and dynamics to catalysis. Biochemistry. 2001;40:12686–12694. doi: 10.1021/bi011540r. [DOI] [PubMed] [Google Scholar]
- 10.Tsai S, Klinman JP. Probes of hydrogen tunneling with horse liver alcohol dehydrogenase at subzero temperatures. Biochemistry. 2001;40:2303–2311. doi: 10.1021/bi002075l. [DOI] [PubMed] [Google Scholar]
- 11.Mincer JS, Schwartz SD. Rate-promoting vibrations and coupled hydrogen-electron transfer reactions in the condensed phase: A model for enzymatic catalysis. J Chem Phys. 2004;120:7755–7760. doi: 10.1063/1.1690239. [DOI] [PubMed] [Google Scholar]
- 12.Caratzoulas S, Mincer JS, Schwartz SD. Identification of a protein-promoting vibration in the reaction catalyzed by horse liver alcohol dehydrogenase. J Am Chem Soc. 2002;124:3270–3276. doi: 10.1021/ja017146y. [DOI] [PubMed] [Google Scholar]
- 13.Luo J, Bruice TC. Low-frequency normal modes in horse liver alcohol dehydrogenase and motions of residues involved in the enzymatic reaction. Biophys Chem. 2007;126:80–85. doi: 10.1016/j.bpc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Billeter SR, Webb SP, Agarwal PK, Iordanov T, Hammes-Schiffer S. Hydride transfer in liver alcohol dehydrogenase: quantum dynamics, kinetic isotope effects, and role of enzyme motion. J Am Chem Soc. 2001;123:11262–11272. doi: 10.1021/ja011384b. [DOI] [PubMed] [Google Scholar]
- 15.Hammes-Schiffer S. Impact of enzyme motion on activity. Biochemistry. 2002;41:13335–13343. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- 16.Cui Q, Elstner M, Karplus M. A theoretical analysis of the proton and hydride transfer in liver alcohol dehydrogenase (LADH) J Phys Chem B. 2002;106:2721–2740. [Google Scholar]
- 17.Alhambra C, Corchado J, Sanchez ML, Garcia-Viloca M, Gao J, Truhlar DG. Canonical variational theory for enzyme kinetics with the protein mean force and multidimensional quantum mechanical tunneling dynamics. Theory and application to liver alcohol dehydrogenase. J Phys Chem B. 2001;105:11326–11340. [Google Scholar]
- 18.Villà J, Warshel A. Energetics and dynamics of enzymatic reactions. J Phys Chem B. 2001;105:7887–7907. [Google Scholar]
- 19.Nagel ZD, Klinman JP. Update 1 of: Tunneling and dynamics in enzymatic hydride transfer. Chem Rev. 2010;110:PR41–67. doi: 10.1021/cr1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijers R, Morris RJ, Adolph HW, Merli A, Lamzin VS, Cedergren-Zeppezauer ES. On the enzymatic activation of NADH. J Biol Chem. 2001;276:9316–9321. doi: 10.1074/jbc.M010870200. [DOI] [PubMed] [Google Scholar]
- 21.Meijers R, Adolph HW, Dauter Z, Wilson KS, Lamzin VS, Cedergren-Zeppezauer ES. Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase. Biochemistry. 2007;46:5446–5454. doi: 10.1021/bi6023594. [DOI] [PubMed] [Google Scholar]
- 22.Dauter Z. Protein structures at atomic resolution. Methods Enzymol. 2003;368:288–337. doi: 10.1016/S0076-6879(03)68016-X. [DOI] [PubMed] [Google Scholar]
- 23.Ramaswamy S, Eklund H, Plapp BV. Structures of horse liver alcohol dehydrogenase complexed with NAD+ and substituted benzyl alcohols. Biochemistry. 1994;33:5230–5237. doi: 10.1021/bi00183a028. [DOI] [PubMed] [Google Scholar]
- 24.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D, Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 25.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D, Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick GM. A short history of SHELX. Acta Crystallogr A, Found Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:286–290. [Google Scholar]
- 29.Frauenfelder H, Hartmann H, Karplus M, Kuntz ID, Jr, Kuriyan J, Parak F, Petsko GA, Ringe D, Tilton RF, Jr, Connolly ML, Max N. Thermal expansion of a protein. Biochemistry. 1987;26:254–261. doi: 10.1021/bi00375a035. [DOI] [PubMed] [Google Scholar]
- 30.Tilton RF, Jr, Dewan JC, Petsko GA. Effects of temperature on protein structure and dynamics: X-ray crystallographic studies of the protein ribonuclease-A at nine different temperatures from 98 to 320 K. Biochemistry. 1992;31:2469–2481. doi: 10.1021/bi00124a006. [DOI] [PubMed] [Google Scholar]
- 31.Kurinov IV, Harrison RW. The influence of temperature on lysozyme crystals. Structure and dynamics of protein and water. Acta Crystallogr D, Biol Crystallogr. 1995;51:98–109. doi: 10.1107/S0907444994009261. [DOI] [PubMed] [Google Scholar]
- 32.Herdendorf TJ, Plapp BV. Origins of the high catalytic activity of human alcohol dehydrogenase 4 studied with horse liver A317C alcohol dehydrogenase. Chem-Biol Interact. 2011;191:42–47. doi: 10.1016/j.cbi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eklund H, Plapp BV, Samama JP, Brändén CI. Binding of substrate in a ternary complex of horse liver alcohol dehydrogenase. J Biol Chem. 1982;257:14349–14358. [PubMed] [Google Scholar]
- 34.LeBrun LA, Park DH, Ramaswamy S, Plapp BV. Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase. Biochemistry. 2004;43:3014–3026. doi: 10.1021/bi036103m. [DOI] [PubMed] [Google Scholar]
- 35.Levy HR, Vennesland B. The stereospecificity of enzymatic hydrogen transfer from diphosphopyridine nucleotide. J Biol Chem. 1957;228:85–96. [PubMed] [Google Scholar]
- 36.Green DW, Sun HW, Plapp BV. Inversion of the substrate specificity of yeast alcohol dehydrogenase. J Biol Chem. 1993;268:7792–7798. [PubMed] [Google Scholar]
- 37.Cho H, Ramaswamy S, Plapp BV. Flexibility of liver alcohol dehydrogenase in stereoselective binding of 3-butylthiolane 1-oxides. Biochemistry. 1997;36:382–389. doi: 10.1021/bi9624604. [DOI] [PubMed] [Google Scholar]
- 38.Ramaswamy S, Scholze M, Plapp BV. Binding of formamides to liver alcohol dehydrogenase. Biochemistry. 1997;36:3522–3527. doi: 10.1021/bi962491z. [DOI] [PubMed] [Google Scholar]
- 39.Bondi A. van der Waals volumes and radii. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 40.Nyburg SC, Faerman CH. A revision of van der Waals atomic radii for molecular crystals: N, O, F, S, Cl, Se, Br and I bonded to carbon. Acta Crystallogr. 1985;B41:274–279. [Google Scholar]
- 41.Thalladi VR, Weiss HC, Blaser D, Boese R, Nangia A, Desiraju GR. C-H···F interactions in the crystal structures of some fluorobenzenes. J Am Chem Soc. 1998;120:8702–8710. [Google Scholar]
- 42.Kim CY, Chang JS, Doyon JB, Baird TT, Jr, Fierke CA, Jain A, Christianson DW. Contribution of fluorine to protein-ligand affinity in the binding of fluoroaromatic inhibitors to carbonic anhydrase II. J Am Chem Soc. 2000;122:12125–12134. [Google Scholar]
- 43.Shapiro S, Arunachalam T, Caspi E. Equilibration of 1-octanol with alcohol dehydrogenase. Evidence for horse liver alcohol dehydrogenase responsibility for exchange of the 1-pro-S hydrogen atom. J Am Chem Soc. 1983;105:1642–1646. [Google Scholar]
- 44.Halle B. Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc Natl Acad Sci USA. 2004;101:4793–4798. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeBrun LA, Plapp BV. Control of coenzyme binding to horse liver alcohol dehydrogenase. Biochemistry. 1999;38:12387–12393. doi: 10.1021/bi991306p. [DOI] [PubMed] [Google Scholar]
- 47.Plapp BV. Enhancement of the activity of horse liver alcohol dehydrogenase by modification of amino groups at the active sites. J Biol Chem. 1970;245:1727–1735. [PubMed] [Google Scholar]
- 48.Hammes GG, Benkovic SJ, Hammes-Schiffer S. Flexibility, diversity, and cooperativity: Pillars of enzyme catalysis. Biochemistry. 2011;50:10422–10430. doi: 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagel ZD, Dong M, Bahnson BJ, Klinman JP. Impaired protein conformational landscapes as revealed in anomalous Arrhenius prefactors. Proc Natl Acad Sci USA. 2011;108:10520–10525. doi: 10.1073/pnas.1104989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plapp BV. Catalysis by alcohol dehydrogenases. In: Kohen A, Limbach HH, editors. Isotope Effects in Chemistry and Biology. CRC Press, Taylor and Francis; Boca Raton: 2006. pp. 811–835. [Google Scholar]
- 51.Bahnson BJ, Colby TD, Chin JK, Goldstein BM, Klinman JP. A link between protein structure and enzyme catalyzed hydrogen tunneling. Proc Natl Acad Sci USA. 1997;94:12797–12802. doi: 10.1073/pnas.94.24.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colby TD, Bahnson BJ, Chin JK, Klinman JP, Goldstein BM. Active site modifications in a double mutant of liver alcohol dehydrogenase: Structural studies of two enzyme-ligand complexes. Biochemistry. 1998;37:9295–9304. doi: 10.1021/bi973184b. [DOI] [PubMed] [Google Scholar]
- 53.Kovaleva EG, Plapp BV. Deprotonation of the horse liver alcohol dehydrogenase-NAD+ complex controls formation of the ternary complexes. Biochemistry. 2005;44:12797–12808. doi: 10.1021/bi050865v. [DOI] [PubMed] [Google Scholar]
- 54.Luo J, Bruice TC. Dynamic structures of horse liver alcohol dehydrogenase (HLADH): results of molecular dynamics simulations of HLADH-NAD+-PhCH2OH, HLADH-NAD+-PhCH2O−, and HLADH-NADH-PhCHO. J Am Chem Soc. 2001;123:11952–11959. doi: 10.1021/ja0109747. [DOI] [PubMed] [Google Scholar]
- 55.Brooks RL, Shore JD, Gutfreund H. The effects of pH and temperature on hydrogen transfer in the liver alcohol dehydrogenase mechanism. J Biol Chem. 1972;247:2382–2383. [PubMed] [Google Scholar]
- 56.Kvassman J, Pettersson G. Effect of pH on the process of ternary-complex interconversion in the liver-alcohol-dehydrogenase reaction. Eur J Biochem. 1978;87:417–427. doi: 10.1111/j.1432-1033.1978.tb12391.x. [DOI] [PubMed] [Google Scholar]
- 57.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 58.Sekhar VC, Plapp BV. Rate constants for a mechanism including intermediates in the interconversion of ternary complexes by horse liver alcohol dehydrogenase. Biochemistry. 1990;29:4289–4295. doi: 10.1021/bi00470a005. [DOI] [PubMed] [Google Scholar]
- 59.Ramaswamy S, Park DH, Plapp BV. Substitutions in a flexible loop of horse liver alcohol dehydrogenase hinder the conformational change and unmask hydrogen transfer. Biochemistry. 1999;38:13951–13959. doi: 10.1021/bi991731i. [DOI] [PubMed] [Google Scholar]
- 60.Gibbons BJ, Hurley TD. Structure of three class I human alcohol dehydrogenases complexed with isoenzyme specific formamide inhibitors. Biochemistry. 2004;43:12555–12562. doi: 10.1021/bi0489107. [DOI] [PubMed] [Google Scholar]
- 61.Plapp BV, Eklund H, Brändén CI. Crystallography of liver alcohol dehydrogenase complexed with substrates. J Mol Biol. 1978;122:23–32. doi: 10.1016/0022-2836(78)90105-5. [DOI] [PubMed] [Google Scholar]
- 62.Bignetti E, Rossi GL, Zeppezauer E. Microspectrophotometric measurements on single crystals of coenzyme containing complexes of horse liver alcohol dehydrogenase. FEBS Lett. 1979;100:17–22. doi: 10.1016/0014-5793(79)81122-9. [DOI] [PubMed] [Google Scholar]
- 63.Shearer GL, Kim K, Lee KM, Wang CK, Plapp BV. Alternative pathways and reactions of benzyl alcohol and benzaldehyde with horse liver alcohol dehydrogenase. Biochemistry. 1993;32:11186–11194. doi: 10.1021/bi00092a031. [DOI] [PubMed] [Google Scholar]
- 64.Guillot B, Lecomte C, Cousson A, Scherf C, Jelsch C. High-resolution neutron structure of nicotinamide adenine dinucleotide. Acta Crystallogr D, Biol Crystallogr. 2001;57:981–989. doi: 10.1107/s0907444901007120. [DOI] [PubMed] [Google Scholar]
- 65.Schiøtt B, Zheng YJ, Bruice TC. Theoretical investigation of the hydride transfer from formate to NAD+ and the implications for the catalytic mechanism of formate dehydrogenase. J Am Chem Soc. 1998;120:7192–7200. [Google Scholar]
- 66.Venkataramaiah TH, Plapp BV. Formamides mimic aldehydes and inhibit liver alcohol dehydrogenases and ethanol metabolism. J Biol Chem. 2003;278:36699–36706. doi: 10.1074/jbc.M305419200. [DOI] [PubMed] [Google Scholar]
- 67.Almarsson Ö, Bruice TC. Evaluation of the factors influencing reactivity and stereospecificity in NAD(P)H dependent dehydrogenase enzymes. J Am Chem Soc. 1993;115:2125–2138. [Google Scholar]
- 68.Meijers R, Cedergren-Zeppezauer E. A variety of electrostatic interactions and adducts can activate NAD(P) cofactors for hydride transfer. Chem-Biol Interact. 2009;178:24–28. doi: 10.1016/j.cbi.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 69.Rubach JK, Plapp BV. Amino acid residues in the nicotinamide binding site contribute to catalysis by horse liver alcohol dehydrogenase. Biochemistry. 2003;42:2907–2915. doi: 10.1021/bi0272656. [DOI] [PubMed] [Google Scholar]
- 70.Brunhuber NMW, Thoden JB, Blanchard JS, Vanhooke JL. Rhodococcus L-phenylalanine dehydrogenase: Kinetics, mechanism, and structural basis for catalytic specifity. Biochemistry. 2000;39:9174–9187. doi: 10.1021/bi000494c. [DOI] [PubMed] [Google Scholar]
- 71.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 72.Schlieben NH, Niefind K, Muller J, Riebel B, Hummel W, Schomburg D. Atomic resolution structures of R-specific alcohol dehydrogenase from Lactobacillus brevis provide the structural bases of its substrate and cosubstrate specificity. J Mol Biol. 2005;349:801–813. doi: 10.1016/j.jmb.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 73.Agarwal PK, Webb SP, Hammes-Schiffer S. Computational studies of the mechanism for proton and hydride transfer in liver alcohol dehydrogenase. J Am Chem Soc. 2000;122:4803–4812. doi: 10.1021/ja011384b. [DOI] [PubMed] [Google Scholar]
- 74.Dworschack RT, Plapp BV. pH, isotope, and substituent effects on the interconversion of aromatic substrates catalyzed by hydroxybutyrimidylated liver alcohol dehydrogenase. Biochemistry. 1977;16:2716–2725. doi: 10.1021/bi00631a020. [DOI] [PubMed] [Google Scholar]
- 75.Deng H, Schindler JF, Berst KB, Plapp BV, Callender R. A Raman spectroscopic characterization of bonding in the complex of horse liver alcohol dehydrogenase with NADH and N-cyclohexylformamide. Biochemistry. 1998;37:14267–14278. doi: 10.1021/bi981477e. [DOI] [PubMed] [Google Scholar]
- 76.Deng H, Zheng J, Sloan D, Burgner J, Callender R. A vibrational analysis of the catalytically important C4-H bonds of NADH bound to lactate or malate dehydrogenase - ground state effects. Biochemistry. 1992;31:5085–5092. doi: 10.1021/bi00136a022. [DOI] [PubMed] [Google Scholar]
- 77.Jencks WP. Catalysis in Chemistry and Enzymology. Dover; New York: 1986. [Google Scholar]
- 78.Hackney DD. Binding energy and catalysis. In: Sigman DS, Boyer PD, editors. The Enzymes. 3. Academic Press; San Diego: 1990. pp. 1–36. [Google Scholar]
- 79.Tapia O, Cardenas R, Andres J, Colonna-Cesari F. Transition structure for hydride transfer to pyridinium cation from methanolate - Modeling of LADH catalyzed reaction. J Am Chem Soc. 1988;110:4046–4047. [Google Scholar]
- 80.Wu YD, Houk KN. Theoretical evaluation of conformational preferences of NAD+ and NADH - An approach to understanding the stereospecificity of NAD+ NADH-dependent dehydrogenases. J Am Chem Soc. 1991;113:2353–2358. [Google Scholar]
- 81.Almarsson Ö, Karaman R, Bruice TC. Kinetic importance of conformations of nicotinamide adenine dinucleotide in the reactions of dehydrogenase enzymes. J Am Chem Soc. 1992;114:8702–8704. [Google Scholar]
- 82.Webb SP, Agarwal PK, Hammes-Schiffer S. Combining electronic structure methods with the calculation of hydrogen vibrational wavefunctions: Application to hydride transfer in liver alcohol dehydrogenase. J Phys Chem B. 2000;104:8884–8894. [Google Scholar]
- 83.Cook PF, Oppenheimer NJ, Cleland WW. Secondary deuterium and nitrogen-15 isotope effects in enzyme- catalyzed reactions. Chemical mechanism of liver alcohol dehydrogenase. Biochemistry. 1981;20:1817–1825. doi: 10.1021/bi00510a016. [DOI] [PubMed] [Google Scholar]
- 84.Rotberg NS, Cleland WW. Secondary 15N isotope effects on the reactions catalyzed by alcohol and formate dehydrogenases. Biochemistry. 1991;30:4068–4071. doi: 10.1021/bi00230a035. [DOI] [PubMed] [Google Scholar]
- 85.Young L, Post CB. Free-energy calculations involving internal coordinate constraints to determine puckering of a 6-membered ring molecule. J Am Chem Soc. 1993;115:1964–1970. [Google Scholar]
- 86.Luo J, Bruice TC. Anticorrelated motions as a driving force in enzyme catalysis: The dehydrogenase reaction. Proc Natl Acad Sci USA. 2004;101:13152–13156. doi: 10.1073/pnas.0405502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roston D, Kohen A. Elusive transition state of alcohol dehydrogenase unveiled. P Natl Acad Sci USA. 2010;107:9572–9577. doi: 10.1073/pnas.1000931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brunger AT. Free R-value - A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 89.Reddy BS, Saenger W, Mühlegger K, Weimann G. Crystal and molecular structure of the lithium salt of nicotinamide adenine dinucleotide dihydrate (NAD+, DPN+, Cozymase, Codehydrase I) J Am Chem Soc. 1981;103:907–914. [Google Scholar]
- 90.Brändén CI, Lindqvist I, Zeppezauer M. Model compounds for NAD-reactions: I. The crystal structure of N-1-(2,6-dichlorobenzyl)-3-carbamidopyridinium iodide monohydrate. Arkiv för Kemi. 1968;30:41–50. [Google Scholar]
- 91.Karle IL. Crystal structure of N-benzyl-1,4-dihydronicoticotinamide. Acta Crystallogr. 1961;14:497–502. [Google Scholar]