Abstract
EBF proteins have diverse functions in the development of multiple lineages, including neurons, B cells and adipocytes. During Drosophila muscle development EBF proteins are expressed in muscle progenitors and are required for muscle cell differentiation, but there is no known function of EBF proteins in vertebrate muscle development. In this study, we examine the expression of ebf genes in Xenopus muscle tissue and show that EBF activity is necessary for aspects of Xenopus skeletal muscle development, including somite organization, migration of hypaxial muscle anlagen toward the ventral abdomen, and development of jaw muscle. From a microarray screen, we have identified multiple candidate targets of EBF activity with known roles in muscle development. The candidate targets we have verified are MYOD, MYF5, M-Cadherin and SEB-4. In vivo overexpression of the ebf2 and ebf3 genes leads to ectopic expression of these candidate targets, and knockdown of EBF activity causes downregulation of the endogenous expression of the candidate targets. Furthermore, we found that MYOD and MYF5 are likely to be direct targets. Finally we show that MYOD can upregulate the expression of ebf genes, indicating the presence of a positive feedback loop between EBF and MYOD that we find to be important for maintenance of MYOD expression in Xenopus. These results suggest that EBF activity is important for both stabilizing commitment and driving aspects of differentiation in Xenopus muscle cells.
Keywords: Early B cell factor (EBF), skeletal muscle, myogenic regulatory factor, transcription, Xenopus
Introduction
The processes of cell commitment and cell differentiation are important aspects of the development of muscle tissue. The group of transcription factors known as myogenic regulatory factors (MRFs), which includes the proteins MYOD and MYF5, is critical in driving both commitment and differentiation in muscle tissue, as seen by the complete lack of muscle cells in Myod, Myf5 double knockout mice (Rudnicki et al., 1993). Additional transcription factors also regulate aspects of muscle development, often as part of the MRF regulatory network. In Drosophila, the Early B cell factor (EBF, also known as COE (collier/olfactory/EBF)) family member Collier plays a role in muscle development, but the roles of EBF proteins in vertebrate muscle development have not been explored.
EBF family members are transcription factors involved in development in several different cell lineages, including neurons, B cells, adipocytes and muscle cells (reviewed in (Dubois and Vincent, 2001; Liberg et al., 2002; Lukin et al., 2008)). These proteins contain a zinc finger DNA binding domain and an atypical helix-loop-helix dimerization domain (Hagman et al., 1993; Hagman et al., 1995; Wang and Reed, 1993; Wang et al., 1997). There are four family members in mammals (EBF1, EBF2, EBF3 and O/E4), two known members in Xenopus (EBF2 and EBF3), and one in zebrafish (ZCOE2) (Bally-Cuif et al., 1998; Dubois et al., 1998; Garel et al., 1997; Hagman et al., 1993; Malgaretti et al., 1997; Pozzoli et al., 2001; Wang and Reed, 1993; Wang et al., 1997). Among invertebrates, the proteins Collier in Drosophila and UNC3 (CeO/E) in C. elegans belong to the EBF family (Crozatier et al., 1996; Prasad et al., 1998).
In Drosophila, the collier gene is expressed in progenitors for several muscles, and is required for myoblast fusion (Crozatier and Vincent, 1999). The expression of collier in Drosophila is driven by both Collier itself and by the MYOD ortholog Nautilus, and this upregulation is synergistic when the two genes are present together (Dubois et al., 2007). In Xenopus and mouse, Northern blot analysis and RNase protection assays show that Ebf2 and Ebf3 are expressed in adult muscle (Dubois et al., 1998; Garel et al., 1997; Malgaretti et al., 1997) Furthermore, EBF proteins are known to bind to the negative regulatory element of the glut4 gene in muscle (Dowell and Cooke, 2002), which allows for insulin-mediated glucose uptake in multiple tissue types (Kahn, 1998). However, the exact expression patterns, transcriptional targets, and functions of EBF genes in the development of vertebrate muscle are not understood.
The process of muscle development has been intensively investigated in multiple vertebrate models, including Xenopus. During vertebrate development, early mesoderm tissue forms somites, which contain myotome cells that will become myoblasts and give rise to muscle tissue. In Xenopus, cells in presomitic mesoderm undergo an early rotational event that gives rise to somites (reviewed in (Elinson, 2007)). Somites contain two separate muscle cell lineages. The region of the somite called the dermamyotome contains a dorsal lip, near the neural tube, with cells that will form epaxial muscles (muscles of the deep back), and a ventral lip, far from the neural tube, with cells that will form hypaxial muscles (muscles of the body wall and limbs) (Gros et al., 2005; Mariani et al., 2001). In Xenopus the hypaxial cells bud off from the somite and migrate ventrally along the body wall before completing the processes of muscle development (Martin and Harland, 2001; Martin and Harland, 2006). Next, myoblasts localize to their correct positions and exit the cell cycle. In most species, myoblasts then align with neighboring myoblasts, undergo fusion, and continue differentiation as multinucleated muscle fibers. However, in Xenopus, muscle cells generated before metamorphosis utilize amitotic rounds of nuclear division to generate multi-nucleated muscle cells (Boudjelida and Muntz, 1987; Kielbowna, 1966).
Across many species, MYOD and MYF5 are expressed from the somite stage. In Xenopus, MYOD and MYF5 are expressed even in presomitic mesoderm (Dosch et al., 1997; Hopwood et al., 1989; Hopwood et al., 1991), and when MYF5 function is blocked by morpholinos, normal development of both the presomitic mesoderm region and of somites is disrupted (Keren et al., 2005). MYOD and MYF5 are also expressed in Xenopus migrating hypaxial cells (Martin and Harland, 2001).
To identify transcriptional targets for EBF factors, we performed a microarray screen of Xenopus animal cap tissue with active EBF3 protein (Green and Vetter, 2011), and unexpectedly found that several muscle-related genes were among the most strongly up-regulated targets, suggesting a role for EBF factors in regulating vertebrate muscle differentiation. Here, we demonstrate the sufficiency and requirement of EBF2 and EBF3 for in vivo expression of the muscle-related genes myod, myf5, seb-4 (also called rbm24), and m-cadherin, identified in our microarray screen. We also describe the expression patterns of ebf2 and ebf3 in the tissues that give rise to Xenopus skeletal muscle, and show a requirement for EBF2 and EBF3 activity in normal muscle development. Finally, we provide evidence that myf5 and myod are direct targets of EBF activity and that MYOD can drive expression of ebf2 and ebf3, in vivo. Our results suggest several new functions of EBF proteins in vertebrate muscle development, and provide evidence in vertebrates of a reciprocal transcriptional relationship between EBF proteins and MYOD.
Materials and methods
Microinjection of RNA and morpholinos
The following constructs were used as DNA templates to make capped RNA: pCS2+Noggin (Lamb et al., 1993), pCS2+hGR-MT-Xebf2, pCS2+hGR-MT-Xebf3, pCS2+NLS-DN-EBF, p64T-MyoD-GR (Kolm and Sive, 1995), and pCS2+nβgal (Chitnis et al., 1995). Capped RNA was generated in vitro using the Message mMachine kit (Ambion). Antisense morpholino oligonucleotides (MOs) were designed by Gene Tools, and directed against a region at or near the translational start site of ebf2 (5’-GCGCTTTGTCTCTCAAGGCAGTTCC-3’) and ebf3 (5’-GTATATTTTCCTGAATCCCAAACAT-3’).
For testing sufficiency of EBF and MYOD to drive target gene expression, a volume of 4nl containing RNA was injected into one blastomere of two cell stage embryos. The following amounts of capped RNA were used for injection: hGR-Xebf2 (0.5ng for overexpression and 50pg for coinjection with MOs), hGR-Xebf3 (0.5ng), MyoD-hGR (0.5ng for overexpression and 25pg for coinjection with MOs)), and nuclear β-galactosidase (nβgal, 30pg). For morpholino experiments, each of two vegetal blastomeres of 8-cell stage embryos were injected with EBF2 MO (Gene Tools, 7.5ng or 10ng), EBF3 MO (Gene Tools, 7.5ng or 10ng), and capped mRNA encoding nβgal (20pg). Capped mRNA encoding NLS-DN-EBF (1ng) was also injected into two vegetal blastomeres of 8-cell stage embryos. In all microinjections, nβgal capped mRNA was co-injected with other capped mRNA or morpholinos into embryos as a tracer. Embryos were then grown and staged (Nieuwkoop and Faber, 1994). Embryos, which were injected with mRNA encoding hGR-XEBF2, hGR-XEBF3 and MYOD-hGR, were treated with 30µM DEX from the gastrula stage (stage 11/11.5) to the neurula stage (stage 14/15), which is 6–8 hours at room temperature. Alternatively, embryos injected with hGR-XEBF3 mRNA were treated with 30µM DEX from the blastula stage (stage 9) for 4.5 hours (to the gastrula stage (stage 10.5~11)) at 23°C. All embryos were then fixed with 4% paraformaldehyde (PFA) in PBS for 30 minutes. After washing the embryos 3 times with PBS, X-gal staining was performed as described (Turner and Weintraub, 1994). Further fixation was done for one hour at room temperature or overnight at 4°C.
Reverse transcriptase PCR
For reverse transcriptase PCR (RT-PCR) experiments, 1 ng hGR-XEBF3 mRNA and/or 0.2 ng Noggin mRNA were injected into Xenopus embryos at the one-cell stage, and animal caps were isolated at stage 9. If the embryos were injected with hGR-XEBF3 mRNA, animal caps from the embryos were divided into two groups, with one group being treated with DEX for 4.5 hours, (during which time sibling embryos reached stage 11–11.5), and the other used as a non-treated control. For another control, animal caps were isolated from uninjected sibling embryos. Total RNA was isolated from the animal caps with Trizol (Invitrogen), genomic DNA was removed with the RNeasy mini kit (Qiagen), and then the total RNA was extracted with phenol/chloroform. To make cDNA from isolated total RNA from animal caps, SuperScript III RT (Invitrogen) was used according to the manufacturer’s instructions. MacVector Software was used to design the gene specific primers (Supplemental Table 1). PCR was performed as previously described (Hutcheson and Vetter, 2001; Pozzoli et al., 2001). RT-PCR for brachyury was performed to ensure there was no mesoderm contamination in animal caps. We also confirmed that muscle actin was not expressed in uninjected or untreated caps, further confirming the lack of mesoderm contamination (Supplemental Figure 1). The primer sequences for muscle actin were obtained from Xenbase (http://www.xenbase.org/other/static/methods/RT-PCR.jsp).
Whole mount in situ hybridization
The following constructs were used to generate antisense RNA probes: pBS-Xebf2 (Pozzoli et al., 2001), pBS-Xebf3 (Pozzoli et al., 2001), pSP73-XMyoD (Hopwood et al., 1989), pBS-XMyf5 (Gawantka et al., 1998; Hopwood et al., 1991), M-Cadherin (IMAGE ID 5440166, ATCC), XSEB-4 (IMAGE ID 4970239, ATCC), Actin alpha (IMAGE ID 5542285, ATCC), and Tnnc1 (IMAGE ID 4407474, ATCC). Antisense RNA probe was generated in vitro using SP6, T7 or T3 RNA polymerase (Ambion) and labeled with digoxigenin-11-UTP (Roche). Whole mount in situ hybridization was performed on the fixed and X-gal stained embryos as described (Harland, 1991; Kanekar et al., 1997).
Immunostaining
For whole mount immunostaining, pigmented embryos were bleached with 1% hydrogen peroxide and 5% formamide in 0.5X SSC solution under fluorescent light for about 1hour. The bleached embryos were fixed again with 4% PFA. 12/101 antibody hybridoma supernatant was used to stain differentiated skeletal muscle (Developmental Studies Hybridoma Bank, (Kintner and Brockes, 1984)). After washing embryos three times (1 hour per wash) with PBS at 4°C, embryos were incubated with blocking solution containing 1% triton X-100 and 10% heat inactivated goat serum in PBS for 3 to 5 hours at RT. 12/101 antibody and β-galactosidase antibody were diluted in the blocking solution (1:300) and incubated for 2 to 4 days at 4°C. The embryos were then washed three times (1 hour per wash) with the blocking solution, and the alexa 488 conjugated goat anti-mouse IgG secondary antibody (Invitrogen) and alexa 568 conjugated goat anti-rabbit IgG secondary antibody (Invitrogen) were diluted in the blocking solution (1:1000) and incubated with embryos for 2 days at 4°C. The embryos were washed with PBS three times and then photographed.
Real-time quantitative PCR
For real-time quantitative PCR (RT-QPCR) experiments, 1 ng hGR-XEBF3 mRNA was injected into Xenopus embryos at the one-cell stage and animal caps were isolated at stage 9. The animal caps were divided into four groups. The control group received no treatment (−C−D). The second group was treated with 30 µM DEX alone for 3 hours (−C+D), and the third group was treated with 5µg/ml cycloheximide (CHX) alone for 3.5 hours (+C−D). Finally, the fourth group was treated with 5 µg/ml CHX for 30 minutes and then 30 µM DEX was added for 3 hours (+C+D). Total RNA was purified from animal caps as described above.
The Superscript III Platinum two-step RT-QPCR kit and SYBR Green (Invitrogen) were used to make cDNA and to generate the PCR solution, and RT-QPCR was performed on a 7900HT Real Time PCR system (Applied Biosystems). The relative gene expression levels were determined by normalizing the expression level of each gene to the expression level of histone H4. Finally, expression levels were normalized by setting the expression level in the condition of −C+D to 100.
Results
EBF3 drives expression of multiple muscle development genes in explanted Xenopus animal caps
To identify transcriptional targets of EBF3, we performed a microarray screen on Xenopus animal cap explants, comparing animal cap explants with and without active EBF3 protein ((Green and Vetter, 2011) and see GEO database: GSE25734 and GSE27084). A hormone-inducible fusion protein (hGR-XEBF3) was used to allow regulation of EBF activity using the hormone dexamethasone (DEX). In the absence of DEX, EBF3 remains inactive, but adding DEX to the explants induces EBF3 activity (Kolm and Sive, 1995). Originally the aim of the microarray was to identify neuronal-specific targets of EBF activity, so we used Noggin to neuralize the animal caps (Lamb et al., 1993). Nonetheless, we found a number of non-neuronal genes to be upregulated by EBF3 activity in the microarray screen ((Green and Vetter, 2011) and GEO: GSE25734). We have performed an additional microarray screen without Noggin, and obtained similar results ((Green and Vetter, 2011) and GEO: GSE27084). We found that genes involved in muscle development were among the most strongly upregulated genes on the array, with myod being the second most strongly upregulated target of all genes. The candidate targets with expected neuronal functions are described elsewhere (Green and Vetter, 2011).
We found a variety of candidate targets that have known functions or expression in muscle tissue (Green and Vetter, 2011), and we performed additional analysis on six of these: myod (80-fold upregulated, Genbank accession number BC073672), muscle-cadherin (m-cadherin, also called as cadherin 15, 39-fold, CF288050), actin, alpha skeletal muscle (actin alpha, also called as acta1, 34-fold, BC046739), seb-4 (also called as rbm24, 16-fold, BC072812), cardiac troponin c (tnnc1, 9-fold, BC082829), and myf5 (6-fold, AJ009303). These results are the first to show myod transcriptionally regulated by an EBF family member, and suggest a potentially critical role of EBF proteins in Xenopus muscle development. To confirm our microarray results, we performed RT-PCR and found that each candidate target gene listed above was upregulated in animal cap explants in the presence of active hGR-XEBF3 (Figure 1). To ensure that Noggin is not responsible for the expression of the candidate targets, we performed RT-PCR controls showing that myod can be upregulated by Xebf3 in the absence of Noggin, and that Noggin alone does not induce myod expression (Supplemental Figure 1).
Figure 1. Confirmation of candidate EBF targets by RT-PCR.
hGR-Xebf3 mRNA and Noggin mRNA were injected into embryos at the single-cell stage. At the blastula stage, the animal caps were dissected then divided into two groups and either treated with DEX or left as untreated controls. Following a 4.5 hour incubation, total RNA was isolated and RT-PCR performed. The column labeled –RT is a negative control in which reverse transcriptase was omitted at the cDNA synthesis step. Total embryo (TE) cDNA from stage 12 (for myf5, brachyury and histone h4) or stage 27 (for the remaining genes) was used as a positive RT-PCR control. brachyury was analyzed to ensure there was no mesoderm contamination in animal caps. histone h4 was used as a loading control. All tested genes (except the loading and contamination controls) were upregulated in the presence of DEX.
EBF2 and EBF3 are expressed in developing muscle tissue
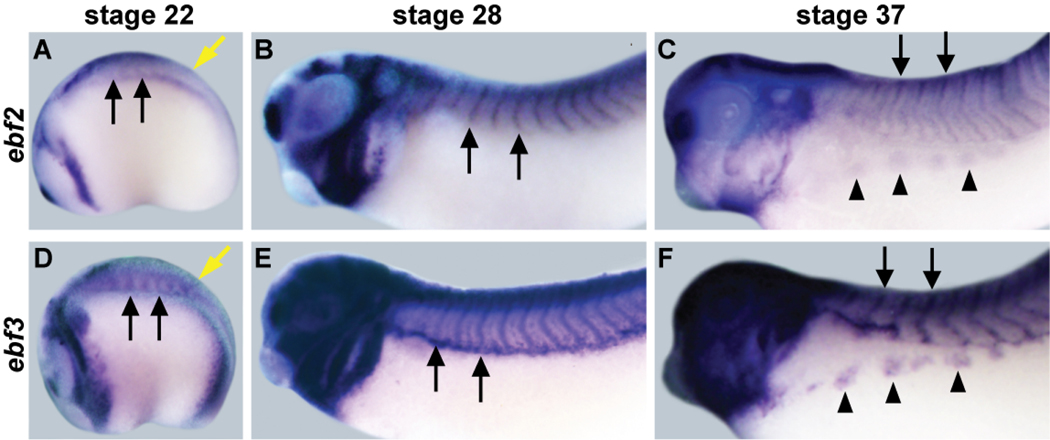
EBF2 is another known EBF family member in Xenopus, and the function of EBF2 is known to be similar to that of EBF3, so EBF2 was also included in the remaining experiments of this study (Dubois et al., 1998; Pozzoli et al., 2001). Previous studies in Xenopus have focused mainly on neuronal expression of the ebf2 and ebf3 genes (Dubois et al., 1998; Pozzoli et al., 2001), but it has been reported that ebf2 is expressed in Xenopus muscle by Northern blot (Dubois et al., 1998) and there is also apparent expression of ebf3 in somites of stage 28 and stage 32 Xenopus embryos (Pozzoli et al., 2001). Since we have found that EBF activity could drive the expression of our muscle specific candidate target genes in animal cap explants, we performed whole mount in situ hybridization (WM-ISH) to obtain a more detailed picture of both ebf2 and ebf3 expression in somites and developing muscle tissue (Figure 2). At stage 22, there is expression of ebf2 and ebf3 in presomitic mesoderm tissue (Figure 2A, D; yellow arrows). Expression of ebf2 and ebf3 is detectable in somites as well (Figure 2A, D; black arrows), and this somite expression becomes very clear at stage 28 (Figure 2B, E; black arrows). The somites will give rise to structures including dorsal epaxial muscle and ventral hypaxial muscle. At stage 37, ebf2 and ebf3 maintain somitic expression, and maintain expression in the migrating hypaxial muscle tissue (Figure 2C, F; arrowheads). These expression patterns are consistent with a role for ebf2 and ebf3 in regulating skeletal muscle development.
Figure 2. Expression patterns of ebf2 and ebf3 in Xenopus muscle.
ebf2 (A-C) and ebf3 (D-F) are expressed in multiple developing neural and muscle tissues. At stage 22, ebf2 and ebf3 are expressed in pre-somitic mesoderm (yellow arrows). At all three stages, ebf2 and ebf3 are expressed in the developing somites (black arrows). At stage 37, they are also expressed in the migrating hypaxial muscle anlagen (arrowheads).
EBF2 and EBF3 are involved in Xenopus muscle development
In order to determine if EBF2 and EBF3 have a functional requirement in Xenopus muscle development, we assessed development after inhibiting the function of EBF2 and EBF3 using two different approaches. First, antisense morpholinos (MO) were used to block translation of ebf2 and ebf3 (EBF2 MO and EBF3 MO). Controls demonstrating the specificity of these morpholinos are discussed below, and in a previous report (Green and Vetter, 2011). Second, a truncated dominant negative EBF (NLS-DN-EBF) construct was used, which blocks the function of endogenous EBF proteins by forming non-functional dimers that do not bind DNA (Green and Vetter, 2011), and (Dubois et al., 1998; Hagman et al., 1993; Hagman et al., 1995)). In this construct, the putative nuclear localization signal (NLS) (Wang and Reed, 1993) of EBF3 was deleted along with the DNA binding domain, and replaced with the NLS of SV40 large T antigen. Either the MOs or NLS-DN-EBF mRNA were injected into two vegetal cells of eight-cell stage embryos, which make minimal contributions to neuronal tissue where EBF factors are also known to be required (Dubois et al., 1998; Green and Vetter, 2011; Pozzoli et al., 2001). We then examined the expression pattern of the skeletal muscle marker myod (Dosch et al., 1997; Hopwood et al., 1989; Martin and Harland, 2001) at stage 39/40 (Figure 3, A-H) by using WM-ISH.
Figure 3. Defective skeletal muscle development after knockdown of EBF2 and EBF3.
Two vegetal cells of eight-cell stage embryos were injected with control MO or EBF2 MO (2 MO) and EBF3 MO (3 MO), either alone or together. β-gal mRNA was coinjected as a marker of the injected side (light blue). At stage 39/40, myod expression was examined (A-H), and 12/101 antibody was used as a marker of skeletal muscle tissue (I-L). The left column (A, C, E, G, I, and K) shows the uninjected control side of the embryos. The right column (B, D, F, H, J, and L) shows the injected side, and (B and F) in some embryos there is more light blue staining in the pronephros, the functional larval kidney, which largely develops from the two vegetal cells that we targeted (Moody, 1987). All panels show lateral views. After injection of 2 MO or 3 MO, myod expression patterns show that the chevron shape of somites is abnormal (black arrows), the hypaxial muscle anlagen are smaller, and the migration distance is reduced (black arrowheads), compared to the uninjected side. The expression of myod in jaw muscle is also reduced (yellow arrows). When 2 MO and 3 MO were coinjected (H), these defects were more severe than 2 MO or 3 MO alone (D and F). Control MO has no effect (B). 12/101 antibody staining shows that when 2 MO and 3 MO were coinjected, somite segmentation is not complete, and the chevron shape of somites is abnormal (white arrows). Also jaw muscle differentiation is reduced (yellow arrow) and abdominal hypaxial muscle differentiation is strongly reduced (white arrowheads), while control MO shows a mild defect of only hypaxial muscle differentiation (J). To visualize the injected side after immunostaining, β-galactosidase antibody (not shown) was used for coimmunostaining along with 12/101 antibody.
After MO knockdown of EBF2 and EBF3, the migrating hypaxial muscle anlagen were reduced, and the remaining anlagen migrated a shorter distance (Figure 3D, F, H; black arrowheads) than on the uninjected side. The chevron shape of somites was more irregular (Figure 3D, F, H; black arrows) than the uninjected side, and myod expression levels in jaw muscle were downregulated (Figure 3D, F, H; yellow arrows). These defects of muscle development were present following single knockdown of either EBF2 or EBF3, and were more severe after double knockdown of both. There were no visible defects after injection of control MO (Figure 3, A and B). Since myod is one of our candidate targets of EBF activity, we verified our findings by labeling embryos with an antibody against the differentiated skeletal muscle marker 12/101 ((Kintner and Brockes, 1984) and Figure 3, I-L). At stage 39/40, skeletal muscle tissue staining positively with 12/101 antibody can be seen in somites, jaw, and abdomen. Control MO slightly delays hypaxial muscle differentiation, but does not affect overall muscle differentiation (Figure 3, I and K). However when EBF2 MO and EBF3 MO were coinjected, the region of skeletal muscle tissue was reduced in jaw (Figure 3L; yellow arrows) and abdomen (Figure 3L, white arrowheads) compared to the uninjected side (Figure 3K). In the somite region, the segmentation between somites was not clear, and the chevron shape was abnormal (Figure 3L, white arrows). We found similar defects of skeletal muscle differentiation after injection of NLS-DN-EBF (data not shown). These defects of muscle development after knockdown of EBF2 and EBF3 give us good evidence that EBF proteins are required for normal Xenopus skeletal muscle development.
Classes and expression patterns of candidate target genes
Since the loss-of-function experiments demonstrated that EBF activity is required for Xenopus muscle development, we wished to further analyze the candidate EBF targets with possible roles in muscle development. First, they were classified based on their known functions in Xenopus and other species. MYOD and MYF5 are basic helix-loop-helix (bHLH) transcription factors and also are myogenic regulatory factors (MRF) (Braun et al., 1989; Davis et al., 1987; Hopwood et al., 1989; Hopwood et al., 1991); M-Cadherin is a cell membrane protein (Donalies et al., 1991); skeletal muscle alpha actin composes the core of the thin filament of the sarcomere in muscle; SEB-4 is an RNA binding protein (Fetka et al., 2000); and TNNC1 (cardiac troponin C) is a member of the EF-hand Ca2+ binding protein family (Yuasa et al., 1998).
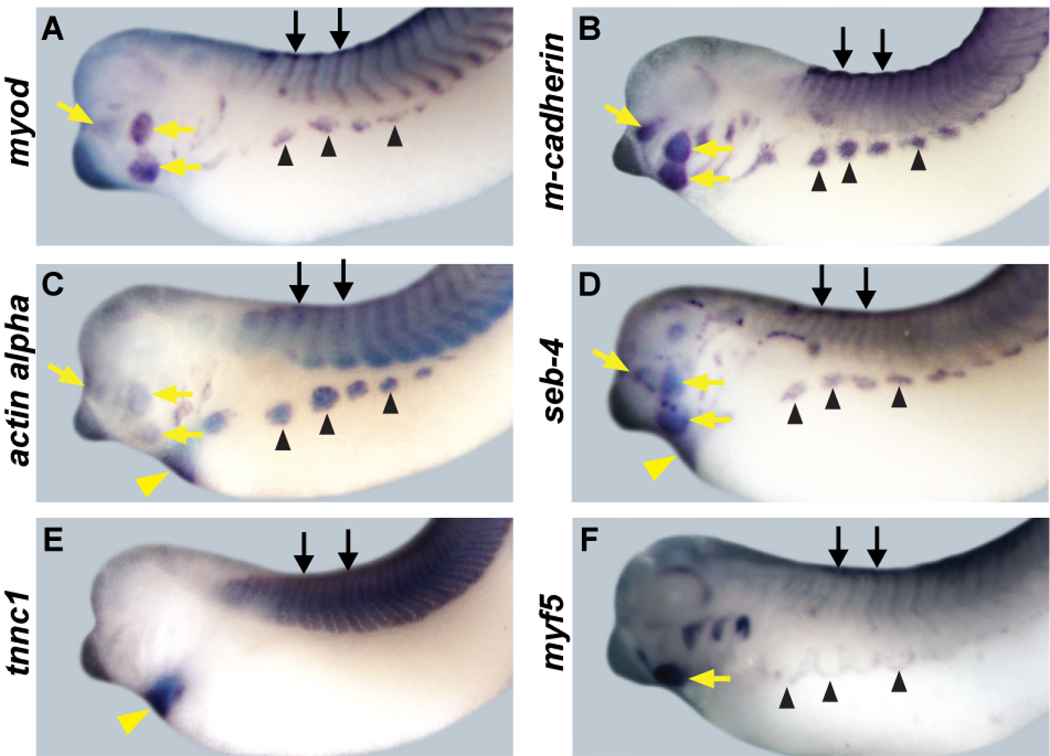
We next compared the expression domains of these candidate target genes (Figure 4) with those of ebf2 and ebf3 (Figure 2) by WM-ISH at stage 35~37. We chose this stage because most structures that will give rise to skeletal muscles, including somites (black arrows), migrating hypaxial muscle anlagen (black arrowheads) and developing jaw muscle (yellow arrows), are clearly detectable at this stage. All six genes are expressed in the tissues that will give rise to skeletal muscle ((Dosch et al., 1997; Fetka et al., 2000; Hopwood et al., 1989; Hopwood et al., 1991; Martin and Harland, 2001) and Figure 4). myod is expressed in a strong central band in the somites, with especially strong expression at the dorsal and ventral lips. It is also expressed in jaw muscle and migrating hypaxial muscle anlagen (Dosch et al., 1997; Martin and Harland, 2001), and Figure 4A). m-cadherin is expressed in a weaker central band in the somites, and with diffuse expression throughout the somites. It is also expressed in jaw muscle and migrating hypaxial muscle anlagen (Figure 4B). The expression patterns of actin alpha and seb-4 in muscle tissue are very similar to that of myod. They are expressed in the somites, jaw muscle and the migrating hypaxial muscle anlagen ((Fetka et al., 2000; Sturgess et al., 1980) and Figure 4, C and D). tnnc1 is expressed in the somites at this stage (Figure 4E). myf5 is expressed in the somites, jaw muscle and migrating hypaxial muscle anlagen, but the expression pattern of myf5 is different from other candidate targets in the migrating hypaxial muscle anlagen, in that it appears to be at the leading edge, rather than within the bulk of the anlagen. Expression of myf5 is also weaker than that of the other candidate targets at this stage ((Dosch et al., 1997; Martin and Harland, 2001) and Figure 4). actin alpha, seb-4 and tnnc1 are expressed in heart, which is composed of cardiac muscle (Figure 4 C-E, yellow arrowheads). These skeletal muscle expression patterns of myod, m-cadherin, actin alpha, seb-4, tnnc1 and myf5 are very similar to expression of ebf2 and ebf3, including expression in somites and migrating hypaxial muscle anlagen. This strong correlation suggests that the candidate targets we have identified by microarray could be in vivo targets of EBF activity during Xenopus muscle development.
Figure 4. Expression patterns of muscle target genes.
In stage 35–37 embryos myod, m-cadherin, actin alpha, seb-4, tnnc1 and myf5 are all expressed in skeletal muscle including somites (black arrows), migrating hypaxial muscle anlagen (black arrowheads) and jaw muscle (yellow arrows). myod, m-cadherin, actin alpha, seb-4, and myf5 (A-D and F) are expressed in the somites, migrating hypaxial muscle anlagen and jaw muscle, and these expression patterns overlap with those of ebf2 and ebf3 (Figure 2). m-cadherin (B) is weakly expressed in a central band in somites, with expression throughout the somite. myf5 (F) expression in somites is weaker than other genes at this stage, and is expressed at the leading edge of migrating hypaxial muscle. tnnc1 (E) is expressed in the somites. actin alpha, seb-4, and tnnc1 are expressed in the heart (yellow arrowheads). All embryos show lateral views.
EBF2 and EBF3 are sufficient and required for the expression of muscle candidate targets in vivo
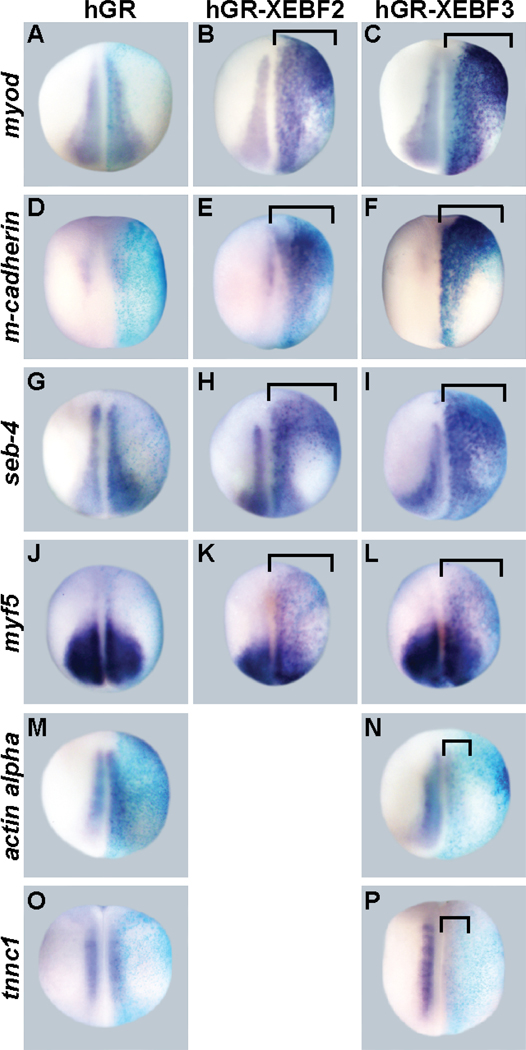
To determine if EBF2 and EBF3 are sufficient for driving the expression of the candidate target genes in vivo, we examined the expression level of candidate targets after overexpression of hGR-XEBF2 or hGR-XEBF3 in Xenopus embryos. Overexpression was achieved by injection of mRNA into one cell of two-cell stage embryos, followed by treatment of the embryos with DEX from the gastrula stage (stage 11/11.5) to the neurula stage (stage 14/15) (about 6~8 hours at room temperature). We found that overexpression of EBF2 or EBF3 caused upregulation of myod (16/16 embryos for hGR-XEBF2 and 16/16 embryos for hGR-XEBF3), m-cadherin (28/33 and 15/24), seb-4 (18/18 and 24/24), and myf5 (33/50 and 30/48) (Figure 5, A-L, brackets), which supports the microarray data, and suggests that EBF activity is sufficient to drive expression of these candidate genes in vivo. However, the expression of actin alpha (17/24) and tnnc1 (36/48) were downregulated by EBF3 (Figure 5, M-P, brackets) suggesting that actin alpha and tnnc1 are not positively regulated by EBF activity in vivo at these early developmental stages. To more closely mimic the stages used for the animal caps for the microarray screen and to test for stage-dependent effects, we repeated the experiment but incubated with DEX from the blastula stage (stage 9), for 4.5 hours (to the gastrula stage (stage 10.5~11)), exactly as was done for the microarray. Consistent with the previous conditions, myod (27/27), seb-4 (24/26) and myf5 (27/30) were upregulated by EBF3 (data not shown). Actin alpha (28/28) and tnnc1 (25/25) were not upregulated by EBF3, consistent with the conclusion that these genes are not regulated by EBF activity in vivo at these stages. However, in contrast with the previous experiment the expression level of m-cadherin was not upregulated (26/26), suggesting that a longer induction period or a later developmental stage may be required for this gene to be induced by EBF activity in vivo.
Figure 5. EBF2 and EBF3 are sufficient for muscle target gene expression.
hGR-XEBF2 or hGR-XEBF3 mRNA were injected into one cell of two-cell stage embryos, followed by DEX treatment from the late gastrula stage (stage 11/11.5) to the neurula stage (stage 14/15). hGR mRNA was injected in control embryos. β-gal mRNA was coinjected as a marker of the injected side. In all panels the right side is the injected side, showing the blue color of X-gal staining. The (purple) expression levels of myod (B and C), m-cadherin (E and F), seb-4 (H and I), and myf5 (K and L) are strongly upregulated by EBF2 and EBF3 (brackets), while expression of hGR alone does not change the expression level of the target genes (A, D, G, and J). The expression of actin alpha (N) and tnnc1 (P) is downregulated by EBF3. All panels show dorsal views.
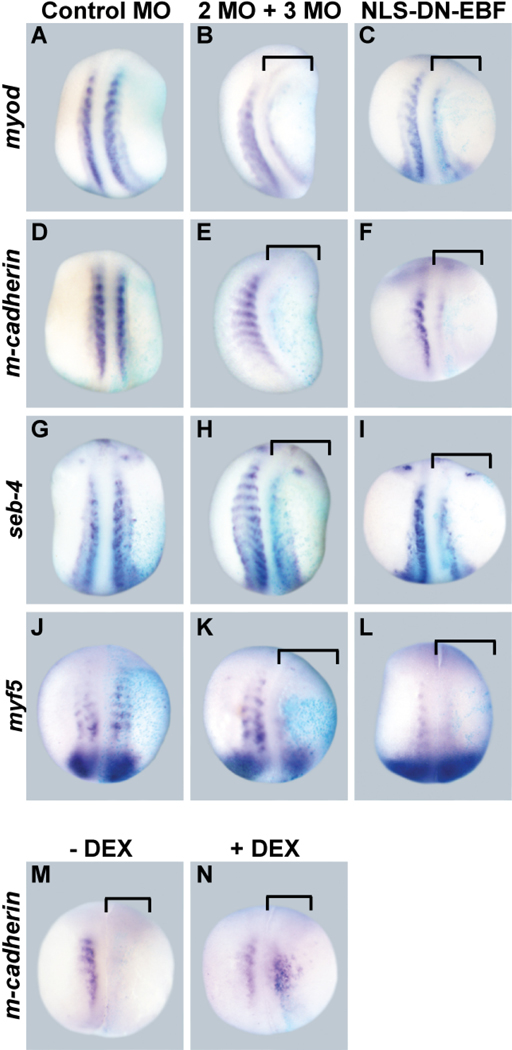
To determine if EBF2 and EBF3 activity is required for the expression of the four upregulated candidate target genes in vivo, we examined their expression level after knockdown of EBF2 and EBF3, targeting both factors together since they often act redundantly. To block EBF2 and EBF3 function, EBF2 MO and EBF3 MO were coinjected into two vegetal cells of eight-cell stage embryos. The endogenous expression level of candidate target genes was examined at the early tail bud stage (stage 20/21), a stage in which the anterior somites are clearly formed, and the expression of each target is apparent (Figure 6). After knockdown of both EBF2 and EBF3, the expression of myod (16/18 embryos), m-cadherin (7/8), seb-4 (6/10), and myf5 (14/17) were downregulated (Figure 6, B, E, H, and K, brackets). Control MO did not change the expression levels of these genes (Figure 6, A, D, G, and J). These four genes were also downregulated by expression of NLS-DN-EBF; MyoD (16/17), m-cadherin (15/15), seb-4 (16/18), myf5 (12/17) (Figure 6, C, F, I, and L, and brackets). These knockdown experiments suggest that EBF factors are required for the expression of each of our candidate targets in vivo.
Figure 6. EBF2 and EBF3 are necessary for muscle target gene expression.
(A-L) Two vegetal cells of eight-cell stage embryos were injected with either XEBF2 MO and XEBF3 MO together, or control MO. β-gal mRNA was coinjected as a marker of the injected side. The expression level of target genes was examined at stage 20/21. In all panels the right side is the injected side, showing the blue color of X-gal staining. The (purple) expression levels of myod (B and C), m-cadherin (E and F), seb-4 (H and I), and myf5 (K and L) are downregulated by XEBF2 MO and XEBF3 MO together or by NLS-DN-EBF (brackets), while control MO does not change their expression levels (A, D, G, and J). (M, N) Two vegetal cells of eight-cell stage embryos were coinjected with XEBF2 MO, XEBF3 MO and hGR-XEBF2 mRNA, followed by DEX treatment from the late gastrula stage (stage 11/11.5) to the early tailbud stage (stage 20). The expression of m-cadherin is downregulated without EBF activity (M), but expression was rescued in the presence of EBF activity (N). All panels show dorsal views.
Previously we have shown that EBF2 MO and EBF3 MO specifically block EBF activity during Xenopus neuronal development (Green and Vetter, 2011). To demonstrate the specificity of the MOs during muscle development, we performed an mRNA rescue experiment. EBF2 MO and EBF3 MO were coinjected with hGR-XEBF2 mRNA, which does not have overlapping sequence with the MOs, and then half of the embryos were treated with DEX from the gastrula stage (stage 11.5) to the early tailbud stage (stage 20) and the other half were used as controls. Compared to the strong downregulation of m-cadherin seen with morpholino alone (19/25), the expression level of m-cadherin was rescued in the presence of active hGR-XEBF2 (20/24) (Figure 6, M and N). This data demonstrates that the EBF2 MO and EBF3 MO specifically block EBF activity during Xenopus muscle development. The disrupted somite segmentation caused by the morpholinos was not rescued, but this is not surprising since we have found that somite segmentation can be disrupted by both gain and loss of EBF activity.
EBF3 can induce the expression of myod and myf5 in the absence of protein synthesis
We next determined if EBF proteins can drive the expression of the four candidate targets directly, in the absence of protein synthesis. Briefly, we prepared animal caps expressing hGR-XEBF3 then added cycloheximide (CHX), an inhibitor of protein synthesis, prior to DEX treatment (see also (Green and Vetter, 2011)). Animal caps were collected at stage 9 and divided into four groups: untreated controls (−C−D), DEX alone (−C+D), CHX alone (+C−D), and both CHX and DEX (+C+D). The expression levels of the four candidate targets were then examined by RT-QPCR (Figure 7).
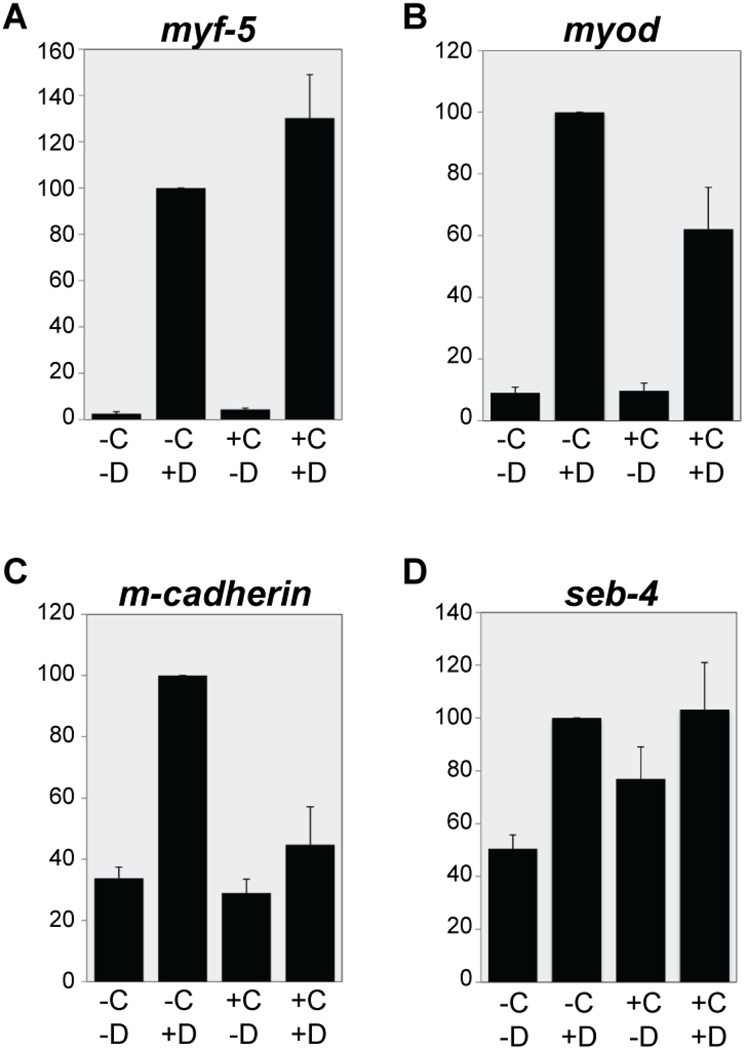
Figure 7. The identification of direct and indirect candidate targets of EBF3.
hGR-XEBF3 mRNA was injected into one-cell stage embryos, and animal caps were collected at the blastula stage (stage 9). The animal caps were divided into four groups, based on CHX and DEX treatment: −C−D, −C+D, +C−D, and +C+D. After a 3.5-hour incubation with CHX and/or a 3-hour incubation with DEX, total RNA was isolated from the animal caps and RT-QPCR was conducted with the isolated RNA. The expression level was normalized with the expression level of histone h4 and then normalized to the expression level of −C+D, for each gene, at 100 arbitrary units. The expression level of myf5 in the condition of +C+D is comparable to the condition of −C+D (A) and the expression level of myod in the condition of +C+D is only partially reduced compared to the condition of −C+D (B). The expression level of m-cadherin in the condition of +C+D is similar to the levels of the two control conditions (C). The expression level of seb-4 in +C+D is similar to the expression level in −C+D but also similar to the expression level in one control condition, +C−D so it is not conclusively a direct or indirect target of EBF activity. Error bars represent standard error of the mean. N = 5 replicates, 20 to 30 animal caps per condition.
The expression level of myf5 in the condition of +C+D was comparable to the condition of -C+D (Figure 7A), which suggests that myf5 is a direct target. The expression level of myod in the condition of +C+D was partially reduced compared to the levels in the condition of -C+D (Figure 7B), which suggests that EBF3 primarily drives the expression of myod directly, but that there may also be an indirect component. The expression level of m-cadherin in the condition of +C+D was similar to the two control conditions, −C−D and +C−D (Figure 7C), so we conclude that m-cadherin is likely to be an indirect target of EBF3. Finally the results for seb-4 were inconclusive since the expression level of seb-4 in the condition of +C+D is similar to the expression level in the condition of −C+D but also similar to the expression level in one control condition, +C−D (Figure 7D).
MYOD can upregulate the expression of ebf2 and ebf3 in vivo in a positive feedback loop
In Drosophila, the MYOD ortholog Nautilus can drive expression of the ebf ortholog collier (Dubois et al., 2007). We therefore asked whether MYOD can also regulate ebf gene transcription in Xenopus. MYOD-hGR (Kolm and Sive, 1995) mRNA was injected into one cell of two-cell stage embryos, followed by treatment of embryos with DEX from the gastrula stage (stage 11/11.5) to the neurula stage (stage 14/15). The expression levels of ebf2 and ebf3 were then examined by WM-ISH. The expression levels of both ebf2 and ebf3 were upregulated by activated MYOD (31/31 for ebf2 and 19/19 for ebf3, Figure 8, brackets). Combined with our result that EBF activity directly drives myod expression, this suggests that MYOD and EBF may have a reciprocal transcriptional interaction in vertebrates.
Figure 8. MYOD drives expression of ebf2 and ebf3.
(A-D) MYOD-hGR mRNA or control, hGR mRNA was injected into one cell of two-cell stage embryos, followed by DEX treatment from the late gastrula stage (stage 11/11.5) to the neurula stage (stage 14/15). β-gal mRNA was coinjected as a marker of the injected side. In all panels the right side is the injected side, showing the light blue color of X-gal staining. The expression of ebf2 (B) and ebf3 (D) is strongly upregulated by activated MYOD-hGR (brackets), while control hGR injection does not change the expression level of ebf2 (A) or ebf3 (C). All panels show dorsal views. (E, F) Two vegetal cells of eight-cell stage embryos were co-injected with XEBF2 MO, XEBF3 MO and MYOD-hGR mRNA, followed by DEX treatment from the late gastrula stage (stage 11/11.5) to the early tailbud stage (stage 20). The expression level of m-cadherin is downregulated without MYOD activity (E), but the expression level was rescued in the presence of MYOD activity (F).
Since we found above that m-cadherin is an indirect target of EBF activity, and since MYOD and EBF appear to have a reciprocal transcriptional relationship in Xenopus embryos, we next examined whether MYOD could restore m-cadherin expression, which is normally lost when EBF activity is blocked. We coinjected EBF2 MO and EBF3 MO with MYOD-hGR mRNA, and treated with DEX to activate MYOD. While the expression level of m-cadherin was strongly downregulated by the morpholinos in the absence of active MYOD (8/12), the expression level of m-cadherin was rescued in the presence of active MYOD (14/18), (Figure 8, E and F). This data is consistent with results showing that m-cadherin is a direct target of MYOD in Xenopus embryos (Hsiao and Chen, 2010). Overall, these results suggest that m-cadherin expression is dependent on both EBF activity and MYOD activity, and that EBF acts through MYOD to promote m-cadherin expression.
Discussion
While EBF proteins have well documented roles in neural and B cell development, nothing has been reported about their role in muscle development in vertebrates. Our findings that the ebf2 and ebf3 genes are extensively expressed in developing muscle tissue in Xenopus, and that EBF activity is required for the normal development of muscle tissue reveal an unexpected role for EBF transcription factors in vertebrate muscle development. We have identified multiple genes, with known function in muscle tissue, to be downstream of EBF transcriptional activity. These genes represent potential routes whereby EBF activity can help regulate commitment, differentiation, and migration of muscle cells. The candidate target genes that were found in the microarray screen (see Green and Vetter, 2011, and GEO database GSE25734; http://www.ncbi.nlm.nih.gov/geo/) but that we did not test further in vivo also open additional potential routes for exploration of EBF activity during muscle development.
EBF proteins function in muscle cell determination to drive myod and myf5 expression directly
Our discovery that MYOD and MYF5 appear to be direct candidate transcriptional targets of EBF activity demonstrates a potentially important role for EBF proteins in muscle cell determination. MRFs including MYOD, MYF5, Myogenin and MRF4 are bHLH transcription factors that form heterodimers with other bHLH proteins, such as the ubiquitously expressed E proteins. These MRFs are critical for driving transcription of muscle-related genes (Biressi et al., 2007; Buckingham, 2001; Chanoine and Hardy, 2003; Pownall et al., 2002; Shih et al., 2008). In particular, MyoD, Myf5 double knockout mice display a complete absence of muscle cells (Rudnicki et al., 1993).
In this study we show that in Xenopus myod and myf5 are candidate targets of EBF proteins, and that ebf2 and ebf3 can in turn be regulated by MYOD. MYOD is expressed in early presomitic mesoderm. ebf2 and ebf3 are detected by WM-ISH in pre-somitic mesoderm as well, but not at the early stages when myod is present. This suggests that EBF proteins are likely involved in maintaining and reinforcing the expression of myod rather than initiation of myod expression. Maintenance of myod expression by EBF proteins appears to be important in Xenopus, since we find that knockdown of EBF activity strongly reduces the expression of myod, and disrupts normal skeletal muscle development, including that of somites, hypaxial muscle and jaw muscle. Our study therefore suggests that EBF proteins are involved in Xenopus myogenic determination by maintaining and reinforcing the expression of myod and myf5. It is known that MYOD also can drive its own expression (Thayer et al., 1989; Weintraub et al., 1989), but in Xenopus this auto-regulation of MYOD may need to be reinforced by EBF regulation to give proper myogenic specification.
Our finding, conversely, that MYOD drives expression of ebf genes is analogous to the finding in Drosophila that the MYOD ortholog Nautilus drives collier expression (Dubois et al., 2007). In mouse, no muscle phenotype has been reported for knockouts of Ebf genes. However, there is redundancy of function for the Ebf genes, and since no Ebf1/Ebf2/Ebf3 triple knockout mouse has been described, the full contribution of EBF activity to mouse muscle development remains unknown. However, in two types of microarray screens for MYOD targets in mouse cultured cell lines, Ebf expression was either unchanged or even downregulated (Bergstrom et al., 2002). Also, Ebf genes were not found to be a MYOD target by ChIP analysis in a mouse cultured cell line (Cao et al., 2006). The fact that our findings are analogous to Drosophila but somewhat at odds with these microarray and ChIP experiments in mouse cultured cell lines could be due to species differences or to differences in experimental design. Thus, it remains to be determined whether EBF proteins play a role in muscle development in other vertebrate species.
EBF proteins function in muscle cell differentiation to drive m-cadherin and seb-4 expression
We show that knockdown of EBF activity leads to delayed migration of hypaxial muscle anlagen, defective somite organization, and reduced differentiation of skeletal muscle. The known functions of M-Cadherin and SEB-4 suggest that EBF proteins may control aspects of myoblast migration and differentiation through regulation of these and possibly additional target genes. However, MYOD and other MRFs can drive both m-cadherin expression and seb-4 expression (Hsiao and Chen, 2010; Li et al., 2010), and we found that EBF regulation of m-cadherin was likely indirect. Thus it seems likely that some aspects of muscle gene regulation by EBFs occur indirectly through MYOD and possibly through MYF5.
There is evidence that both M-Cadherin and SEB-4 could function as important downstream effectors of EBF activity in muscle cell development. M-Cadherin is an adhesion protein that is present in developing and adult skeletal muscle, and at the adult neuromuscular junction. During development, it is known to be involved in the differentiation of skeletal muscle, with special importance for myoblast fusion (Charrasse et al., 2007; Cifuentes-Diaz et al., 1996; Donalies et al., 1991; Moore and Walsh, 1993; Pouliot et al., 1994; Zeschnigk et al., 1995). There are also reports of its involvement in muscle cell migration in zebrafish somites (Cortes et al., 2003), and of its association with microtubules in a myoblast cell line (Kaufmann et al., 1999). Since we find that m-cadherin is expressed in Xenopus from as early as the presomitic mesoderm period and continuing through the events of muscle differentiation, it is possible that it is involved in steps including somite formation, hypaxial muscle migration, and maintenance of proper cell relationships during late myoblast differentiation.
It has been shown that the RNA binding protein SEB-4 is necessary for myogenesis (Li et al., 2010; Miyamoto et al., 2009). SEB-4 is likely involved in regulation of cytoskeletal events in muscle development, since it is a Xenopus homolog of the C. elegans protein SUP-12, which regulates splicing of unc-60 mRNA (Anyanful et al., 2004). UNC-60 is the ortholog of actin depolymerizing factor (ADF)/cofilin which controls actin filament dynamics (Bamburg, 1999; Bamburg et al., 1999; Maciver and Hussey, 2002). The seb-4 gene is expressed in the presomitic mesoderm from gastrulation, and its expression is restricted to somites, jaw muscle and myocardium at the tailbud stage ((Fetka et al., 2000) and Figure 4). Since it is expressed at the somite stage, and since somite rotation in Xenopus involves actin rearrangement (Kragtorp and Miller, 2006), seb-4 may be necessary for proper somite rotation.
The transcriptional relationship between EBF proteins and bHLH proteins
Our systematic study of transcriptional targets of EBF proteins, together with evidence from other reports and other species, is expanding the scope of evidence for reciprocal transcriptional relationships between EBF proteins and bHLH proteins involved in cell commitment and differentiation in multiple cell lineages. First, during neuronal development EBF proteins have been shown to act upstream of bHLH genes in multiple contexts. For example, EBF2 can drive expression of the proneural bHLH genes ngnr-1 and neurod in Xenopus (Dubois et al., 1998; Pozzoli et al., 2001). Additionally, misexpressed mouse Ebf1 drives expression of ngn1 and ngn2 in chick spinal cord (Garcia-Dominguez et al., 2003). We also recently found that the bHLH gene nscl-1 is transcriptionally regulated by EBF activity (Green and Vetter, 2011). These findings show striking similarity to what we report here, namely that the bHLH genes myod and myf5 are regulated by EBF2 and EBF3.
Conversely, there is also evidence that bHLH proteins can drive expression of ebf genes in multiple contexts. For example, in Xenopus, the bHLH transcription factors NGNR-1, NeuroD and ATH5 can upregulate ebf2 and ebf3 (Dubois et al., 1998; Logan et al., 2005; Pozzoli et al., 2001; Seo et al., 2007), and misexpressed ngn2 drives ebf1 and ebf3 expression in chick spinal cord (Garcia-Dominguez et al., 2003). In Drosophila, Nautilus drives collier expression (Dubois et al., 2007). Analogously, our current study shows that MYOD can drive expression of ebf2 and ebf3 in Xenopus embryos.
These studies support the idea that EBF proteins and bHLH proteins have reciprocal transcriptional relationships in multiple lineages. Because EBF proteins and bHLH proteins appear to control the expression of each other in positive feedback loops in both neuron and muscle tissues, and possibly in multiple species, we believe that there may be an ancient transcriptional relationship between these two gene families. Evidence also exists of reciprocal relationships between EBF and bHLH proteins in B cell development (Greenbaum and Zhuang, 2002; Kee and Murre, 1998; Kwon et al., 2008; Seet et al., 2004; Smith et al., 2002; Zhuang et al., 2004). All of these relationships appear to be primarily centered on stabilizing commitment of cells to a particular lineage. Interestingly, the potential spectrum of activities in muscle tissue suggested by our experiments, including stabilizing commitment, directing migration, and directing cytoskeletal organization, is very analogous to the range of activities driven by EBF proteins in neural development.
Research highlights.
-
>
Ebf genes are expressed in developing Xenopus muscle tissues.
-
>
EBF protein activity is required for normal Xenopus skeletal muscle development.
-
>
By microarray screen we found targets of EBF transcriptional activity that are muscle related.
-
>
EBF is sufficient and required for expression of some muscle related genes in vivo.
-
>
The EBF target MyoD upregulates the expression of ebf genes, demonstrating a feedback loop.
Supplementary Material
Supplemental Figure 1. EBF proteins, but not noggin alone, can activate myod in animal caps without inducing mesoderm
Noggin mRNA and/or hGR-Xebf3 mRNA were injected into embryos at the single-cell stage. At the blastula stage, the animal caps were dissected, then divided into two groups and either treated with DEX or left as untreated controls. Following a 4.5 hour incubation, total RNA was isolated and RT-PCR performed. The column labeled –RT is a negative control in which reverse transcriptase was omitted at the cDNA synthesis step. The column labeled UN is a control in which animal caps were obtained from uninjected control embryos. The column labeled Nog is a control demonstrating that Noggin alone does not induce expression of myod or m-actin, which is a marker of mesoderm. The columns labeled hGR-Xebf3 and Noggin+hGR-Xebf3 show that myod expression is induced only in the presence of active Xebf3 due to the addition of DEX (+D). The column labeled TE (Total embryo) is a cDNA mixture from stage 12 and stage 27, used as a positive control for myoD and m-actin expression. As a positive control for all samples, RT-PCR was performed for histone h4.
Acknowledgments
We thank Kathryn Moore and Eric Green for critical comments on the manuscript. We thank Brett Milash for assistance with the microarray data analysis. We thank Ombretta Pozzoli for the pCS2+hGR-MT-XEBF3 construct. This work was funded by NIH grant EY012274, and by University of Utah development funds to MLV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, Ono S. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J Cell Biol. 2004;167:639–647. doi: 10.1083/jcb.200407085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally-Cuif L, Dubois L, Vincent A. Molecular cloning of Zcoe2, the zebrafish homolog of Xenopus Xcoe2 and mouse EBF-2, and its expression during primary neurogenesis. Mech Dev. 1998;77:85–90. doi: 10.1016/s0925-4773(98)00144-0. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E, Ferrari S, Cusella-De Angelis MG, Tajbakhsh S, Cossu G. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev Biol. 2007;304:633–651. doi: 10.1016/j.ydbio.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Boudjelida H, Muntz L. Multinucleation during myogenesis of the myotome of Xenopus laevis: a qualitative study. Development. 1987;101:583–590. doi: 10.1242/dev.101.3.583. [DOI] [PubMed] [Google Scholar]
- Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. Embo J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. Embo J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoine C, Hardy S. Xenopus muscle development: from primary to secondary myogenesis. Dev Dyn. 2003;226:12–23. doi: 10.1002/dvdy.10206. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Comunale F, Fortier M, Portales-Casamar E, Debant A, Gauthier-Rouviere C. M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol Biol Cell. 2007;18:1734–1743. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Diaz C, Goudou D, Padilla F, Facchinetti P, Nicolet M, Mege RM, Rieger F. M-cadherin distribution in the mouse adult neuromuscular system suggests a role in muscle innervation. Eur J Neurosci. 1996;8:1666–1676. doi: 10.1111/j.1460-9568.1996.tb01310.x. [DOI] [PubMed] [Google Scholar]
- Cortes F, Daggett D, Bryson-Richardson RJ, Neyt C, Maule J, Gautier P, Hollway GE, Keenan D, Currie PD. Cadherin-mediated differential cell adhesion controls slow muscle cell migration in the developing zebrafish myotome. Dev Cell. 2003;5:865–876. doi: 10.1016/s1534-5807(03)00362-9. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–718. doi: 10.1016/s0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Vincent A. Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: transcriptional response to notch signalling. Development. 1999;126:1495–1504. doi: 10.1242/dev.126.7.1495. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Donalies M, Cramer M, Ringwald M, Starzinski-Powitz A. Expression of M-cadherin, a member of the cadherin multigene family, correlates with differentiation of skeletal muscle cells. Proc Natl Acad Sci U S A. 1991;88:8024–8028. doi: 10.1073/pnas.88.18.8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C. Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development. 1997;124:2325–2334. doi: 10.1242/dev.124.12.2325. [DOI] [PubMed] [Google Scholar]
- Dowell P, Cooke DW. Olf-1/early B cell factor is a regulator of glut4 gene expression in 3T3-L1 adipocytes. J Biol Chem. 2002;277:1712–1718. doi: 10.1074/jbc.M108589200. [DOI] [PubMed] [Google Scholar]
- Dubois L, Bally-Cuif L, Crozatier M, Moreau J, Paquereau L, Vincent A. XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus. Curr Biol. 1998;8:199–209. doi: 10.1016/s0960-9822(98)70084-3. [DOI] [PubMed] [Google Scholar]
- Dubois L, Enriquez J, Daburon V, Crozet F, Lebreton G, Crozatier M, Vincent A. Collier transcription in a single Drosophila muscle lineage: the combinatorial control of muscle identity. Development. 2007;134:4347–4355. doi: 10.1242/dev.008409. [DOI] [PubMed] [Google Scholar]
- Dubois L, Vincent A. The COE--Collier/Olf1/EBF--transcription factors: structural conservation and diversity of developmental functions. Mech Dev. 2001;108:3–12. doi: 10.1016/s0925-4773(01)00486-5. [DOI] [PubMed] [Google Scholar]
- Elinson RP. Muscle development in a biphasic animal: the frog. Dev Dyn. 2007;236:2444–2453. doi: 10.1002/dvdy.21220. [DOI] [PubMed] [Google Scholar]
- Fetka I, Radeghieri A, Bouwmeester T. Expression of the RNA recognition motif-containing protein SEB-4 during Xenopus embryonic development. Mech Dev. 2000;94:283–286. doi: 10.1016/s0925-4773(00)00284-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Poquet C, Garel S, Charnay P. Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Development. 2003;130:6013–6025. doi: 10.1242/dev.00840. [DOI] [PubMed] [Google Scholar]
- Garel S, Marin F, Mattei MG, Vesque C, Vincent A, Charnay P. Family of Ebf/Olf-1-related genes potentially involved in neuronal differentiation and regional specification in the central nervous system. Dev Dyn. 1997;210:191–205. doi: 10.1002/(SICI)1097-0177(199711)210:3<191::AID-AJA1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev. 1998;77:95–141. doi: 10.1016/s0925-4773(98)00115-4. [DOI] [PubMed] [Google Scholar]
- Green YS, Vetter ML. EBF factors drive expression of multiple classes of target genes governing neuronal development. Neural Dev. 2011;6:19. doi: 10.1186/1749-8104-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S, Zhuang Y. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc Natl Acad Sci U S A. 2002;99:15030–15035. doi: 10.1073/pnas.232299999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- Hagman J, Gutch MJ, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. Embo J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. MyoD expression in the forming somites is an early response to mesoderm induction in Xenopus embryos. Embo J. 1989;8:3409–3417. doi: 10.1002/j.1460-2075.1989.tb08505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. Xenopus Myf-5 marks early muscle cells and can activate muscle genes ectopically in early embryos. Development. 1991;111:551–560. doi: 10.1242/dev.111.2.551. [DOI] [PubMed] [Google Scholar]
- Hsiao SP, Chen SL. Myogenic regulatory factors regulate M-cadherin expression by targeting its proximal promoter elements. Biochem J. 2010;428:223–233. doi: 10.1042/BJ20100250. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev Biol. 2001;232:327–338. doi: 10.1006/dbio.2001.0178. [DOI] [PubMed] [Google Scholar]
- Kahn BB. Type 2 diabetes: when insulin secretion fails to compensate for insulin resistance. Cell. 1998;92:593–596. doi: 10.1016/s0092-8674(00)81125-3. [DOI] [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Jan YN, Vetter ML. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- Kaufmann U, Kirsch J, Irintchev A, Wernig A, Starzinski-Powitz A. The M-cadherin catenin complex interacts with microtubules in skeletal muscle cells: implications for the fusion of myoblasts. J Cell Sci. 1999;112(Pt 1):55–68. doi: 10.1242/jcs.112.1.55. [DOI] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren A, Bengal E, Frank D. p38 MAP kinase regulates the expression of XMyf5 and affects distinct myogenic programs during Xenopus development. Dev Biol. 2005;288:73–86. doi: 10.1016/j.ydbio.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Kielbowna L. Cytological and cytophotometrical studies on myogenesis in Xenopus laevis. Zool Pol. 1966;17:247–255. [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies identify blastemal cells derived from dedifferentiating limb regeneration. Nature. 1984;308:67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Kragtorp KA, Miller JR. Regulation of somitogenesis by Ena/VASP proteins and FAK during Xenopus development. Development. 2006;133:685–695. doi: 10.1242/dev.02230. [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- Li HY, Bourdelas A, Carron C, Shi DL. The RNA-binding protein Seb4/RBM24 is a direct target of MyoD and is required for myogenesis during Xenopus early development. Mech Dev. 2010;127:281–291. doi: 10.1016/j.mod.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Liberg D, Sigvardsson M, Akerblad P. The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Mol Cell Biol. 2002;22:8389–8397. doi: 10.1128/MCB.22.24.8389-8397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan MA, Steele MR, Van Raay TJ, Vetter ML. Identification of shared transcriptional targets for the proneural bHLH factors Xath5 and XNeuroD. Dev Biol. 2005;285:570–583. doi: 10.1016/j.ydbio.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Lukin K, Fields S, Hartley J, Hagman J. Early B cell factor: Regulator of B lineage specification and commitment. Semin Immunol. 2008;20:221–227. doi: 10.1016/j.smim.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Hussey PJ. The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews3007. reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgaretti N, Pozzoli O, Bosetti A, Corradi A, Ciarmatori S, Panigada M, Bianchi ME, Martinez S, Consalez GG. Mmot1, a new helix-loop-helix transcription factor gene displaying a sharp expression boundary in the embryonic mouse brain. J Biol Chem. 1997;272:17632–17639. doi: 10.1074/jbc.272.28.17632. [DOI] [PubMed] [Google Scholar]
- Mariani FV, Choi GB, Harland RM. The neural plate specifies somite size in the Xenopus laevis gastrula. Dev Cell. 2001;1:115–126. doi: 10.1016/s1534-5807(01)00018-1. [DOI] [PubMed] [Google Scholar]
- Martin BL, Harland RM. Hypaxial muscle migration during primary myogenesis in Xenopus laevis. Dev Biol. 2001;239:270–280. doi: 10.1006/dbio.2001.0434. [DOI] [PubMed] [Google Scholar]
- Martin BL, Harland RM. A novel role for lbx1 in Xenopus hypaxial myogenesis. Development. 2006;133:195–208. doi: 10.1242/dev.02183. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Hidaka K, Jin D, Morisaki T. RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells. 2009;14:1241–1252. doi: 10.1111/j.1365-2443.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- Moody SA. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Moore R, Walsh FS. The cell adhesion molecule M-cadherin is specifically expressed in developing and regenerating, but not denervated skeletal muscle. Development. 1993;117:1409–1420. doi: 10.1242/dev.117.4.1409. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Pub; 1994. [Google Scholar]
- Pouliot Y, Gravel M, Holland PC. Developmental regulation of M-cadherin in the terminal differentiation of skeletal myoblasts. Dev Dyn. 1994;200:305–312. doi: 10.1002/aja.1002000405. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Pozzoli O, Bosetti A, Croci L, Consalez GG, Vetter ML. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev Biol. 2001;233:495–512. doi: 10.1006/dbio.2001.0230. [DOI] [PubMed] [Google Scholar]
- Prasad BC, Ye B, Zackhary R, Schrader K, Seydoux G, Reed RR. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development. 1998;125:1561–1568. doi: 10.1242/dev.125.8.1561. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. Embo J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Muscle development: forming the head and trunk muscles. Acta Histochem. 2008;110:97–108. doi: 10.1016/j.acthis.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J Immunol. 2002;169:261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- Sturgess EA, Ballantine JE, Woodland HR, Mohun PR, Lane CD, Dimitriadis GJ. Actin synthesis during the early development of Xenopus laevis. J Embryol Exp Morphol. 1980;58:303–320. [PubMed] [Google Scholar]
- Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Wang SS, Tsai RY, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa HJ, Cox JA, Takagi T. Diversity of the troponin C genes during chordate evolution. J Biochem. 1998;123:1180–1190. doi: 10.1093/oxfordjournals.jbchem.a022059. [DOI] [PubMed] [Google Scholar]
- Zeschnigk M, Kozian D, Kuch C, Schmoll M, Starzinski-Powitz A. Involvement of M-cadherin in terminal differentiation of skeletal muscle cells. J Cell Sci. 1995;108(Pt 9):2973–2981. doi: 10.1242/jcs.108.9.2973. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Jackson A, Pan L, Shen K, Dai M. Regulation of E2A gene expression in B-lymphocyte development. Mol Immunol. 2004;40:1165–1177. doi: 10.1016/j.molimm.2003.11.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. EBF proteins, but not noggin alone, can activate myod in animal caps without inducing mesoderm
Noggin mRNA and/or hGR-Xebf3 mRNA were injected into embryos at the single-cell stage. At the blastula stage, the animal caps were dissected, then divided into two groups and either treated with DEX or left as untreated controls. Following a 4.5 hour incubation, total RNA was isolated and RT-PCR performed. The column labeled –RT is a negative control in which reverse transcriptase was omitted at the cDNA synthesis step. The column labeled UN is a control in which animal caps were obtained from uninjected control embryos. The column labeled Nog is a control demonstrating that Noggin alone does not induce expression of myod or m-actin, which is a marker of mesoderm. The columns labeled hGR-Xebf3 and Noggin+hGR-Xebf3 show that myod expression is induced only in the presence of active Xebf3 due to the addition of DEX (+D). The column labeled TE (Total embryo) is a cDNA mixture from stage 12 and stage 27, used as a positive control for myoD and m-actin expression. As a positive control for all samples, RT-PCR was performed for histone h4.