Abstract
The pathways by which Merkel cell polyomavirus (MCV) infection contributes to the formation of Merkel cell carcinomas are important for understanding the pathogenesis of these cancers. We hypothesized that MCV T antigen suppresses normal responses to ultraviolet radiation (UVR)-induced DNA damage. An MCV-infected cell line (MKL-1) exhibited UVR hypersensitivity, impaired repair of DNA lesions and cell cycle arrest following UVR, as well as reduced levels of the DNA damage recognition protein, XPC. When ectopically expressed in uninfected UISO cells, mutant but not wild-type T antigen resulted in loss of repair of UVR-induced cyclobutane pyrimidine dimers and reductions in XPC, p53 and p21 levels, whereas both wild-type and mutant T antigen inhibited cell cycle arrest following UVR. Similarly, only mutant T antigen in normal fibroblasts inhibited DNA repair and XPC expression, while both mutant and wild-type T antigens produced cell cycle dysregulation. Wild-type T antigen expression produced large T, 57kT and small T antigens while mutant T antigen was only detectable as a truncated large T antigen protein. Expression of wild-type large T antigen but not small T antigen inhibited the G1 checkpoint in UISO cells, but neither wild-type large T nor small T antigens affected DNA repair, suggesting that large T antigen generates cell cycle defects, and when mutated may also impair DNA repair. These results indicate that T antigen expression by MCV can inhibit key responses to UVR-induced DNA damage and suggest that progressive MCV-mediated abrogation of genomic stability may be involved in Merkel cell carcinogenesis.
Keywords: Merkel cell carcinoma, polyomavirus, DNA repair, cell cycle, ultraviolet radiation
INTRODUCTION
Merkel cell carcinoma (MCC) is an aggressive and unusually lethal type of non-melanoma skin cancer 1. Although rare, MCC has increased in incidence by four-fold over a 20 year period since 1986, particularly in immunosuppressed populations2–7. Unfortunately, MCC is also difficult to treat, and patient survival is disappointing 2,3. Recently, an important advance came with the discovery that the majority of MCCs are associated with a novel polyomavirus, designated Merkel cell polyomavirus (MCV) 8,9. Viral DNA is clonally integrated into the MCC genome, consistent with MCV infection occurring early in if not prior to tumorigenesis, and thus it may play an important if not causative role in MCC pathogenesis8,10,11.
The wild-type MCV T antigen locus produces a transcript that can be alternatively spliced to encode large T (LT), 57 kDa (57kT), and small T (sT) antigen proteins 9. In all MCV-positive MCCs examined thus far, the MCV T antigen gene is mutated due to pyrimidine substitutions that are consistent with ultraviolet radiation (UVR)-induced mutagenesis and that ultimately result in a prematurely truncated LT protein 9. The truncations remove the LT helicase domain necessary for viral replication, as well as sequences that, at least in the homologous LT from simian virus 40 (SV40), correspond to domains that directly bind p53 12. However, the remaining N-terminal region preserves a DnaJ domain and a retinoblastoma protein (Rb) binding domain 9. Interestingly, although incapable of viral replication, MCC cell lines that harbor MCV uniformly appear to require truncated LT expression for their own continued proliferation and survival, indicating that LT affects cell cycle regulation and apoptosis9,13. However, the full scope of biological activities of MCV LT as well as other T antigen isoforms is still relatively unexplored, particularly with regard to mechanisms that may be important to the carcinogenic process, including those that contribute to genomic instability following UVR.
MCC has been associated with numerous genetic changes, but the pathways by which these mutations occur are unclear 14–17. However, because MCC typically arises on sun-exposed areas of fair-skinned individuals and is associated with pyrimidine-type transition mutations, a role for solar UVR-induced DNA damage in MCC pathogenesis has long been speculated 1,2,14,16. The major UVR-induced DNA photoproducts are cyclobutane pyrimidine dimers (CPDs) and pyrimidine(6-4)pyrimidone photoproducts (6-4 PPs). These DNA lesions are repaired by nucleotide excision repair (NER). One NER pathway—global genomic repair (GGR)—removes lesions throughout the entire genome 18. GGR is dependent on multiple proteins, one of which—the XPC protein—functions as a key component of the DNA damage recognition step 19. In addition to genetic loss of normal XPC expression occurring in xeroderma pigmentosum, selective loss of GGR also occurs in cells that are infected with well-known viruses that target two key tumor suppressor proteins, p53 and Rb, that are important for both normal GGR as well as cell cycle arrest following DNA damage. For example, expression of human papillomavirus type 16 E6 oncoprotein in fibroblasts results in p53 degradation and loss of GGR and DNA damage-induced cell cycle checkpoints 20–22. Expression of human papillomavirus E7 oncoprotein, which inhibits Rb, has also been reported to impair GGR, although Rb itself has variably been reported to regulate GGR in both a positive and negative manner 23–26. Importantly, SV40-transformed cell lines also exhibit significantly impaired GGR of CPDs 27.
The relationship between MCV and DNA damage in the infected cells remains unknown. However, LT from SV40 is known to impair many facets of the DNA damage response in large part by disrupting the functions of Rb and p53 28. Thus, critical pathways such as DNA repair and cell cycle arrest in response to DNA damage may also be targeted by MCV T antigens. We hypothesized that, based on its homology to SV40, MCV acting through T antigen could impair normal cellular responses to UVR-induced DNA damage. We report that the presence of MCV is associated with loss of GGR as well as absence of normal cell cycle arrest following UVR, and that expression of a tumor-derived truncated T antigen is sufficient to produce defects in GGR and replication arrest following DNA damage, but that wild-type T antigen activity appears to be restricted to disruption of cell cycle control. These results suggest specific routes by which MCV infection and mutations in the T antigen may lead to acquisition of additional genetic changes associated with MCC 14,16.
MATERIALS AND METHODS
Cells and UVR irradiation
MKL-1 cells (gift from P. Moore and Y. Chang, University of Pittsburgh) were grown as floating multicellular aggregates in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) with a doubling time of 5 days 29. UISO-MCC-1 cells (henceforth referred to as UISO cells, and gift from T.K. Das Gupta, University of Illinois) were grown as adherent monolayers in minimal essential medium supplemented with 15% FBS and non-essential amino acids with a doubling time of 3 days 30. Normal human dermal fibroblasts (NHFs) were isolated from neonatal foreskins and grown in DMEM supplemented with 10% FBS. Cells were irradiated by placing them in a thin layer of phosphate-buffered saline (PBS) and exposing them to a germicidal lamp (predominantly 254 nm) as previously described 21. UVR dosage was monitored by an IL1400A photometer equipped with a SEL240 detector (International Light, Inc., Newburyport, MA).
Viability assay
Following irradiation with 0–10 J/m2 UVR in PBS, cells treated in triplicate were returned to their normal media and allowed to incubate for 5 days before treatment in media containing 50 μg/mL thiazolyl blue for 4 hours, as previously described 21. Cells were washed in PBS, and the formazan salt precipitates were dissolved in dimethyl sulfoxide and quantified by absorbance at 540 nm.
Transfection
The expression vectors, TAg206, TAg350, LT206, and sTco (a codon-optimized vector henceforth designated as sT) were gifts from P. Moore and Y. Chang, University of Pittsburgh, and are based on the pcDNA6 vector and respectively encode the full-length wild-type MCV T antigen locus, a mutated T antigen locus from the MCC350 tumor, a wild-type LT, and wild-type sT 9,31,32. For experiments with TAg206 or TAg350 or the corresponding control empty vector, pcDNA6 (Invitrogen, Carlsbad, CA), 2 μg of the vector were suspended with 106 cells and electroporated using an Amaxa Nucleofector system (Lonza, Basel, Switzerland). UISO cells were electroporated in Nucleofector Solution L using Program X-005. NHFs were treated according to directions for the NHDF Nucleofector kit. Following electroporation, cells were transferred to 35 mm dishes and allowed to attach overnight in their respective media. Media was refreshed the following day, and cells were used for experiments 48 hours following electroporation. For experiments involving pcDNA6, LT206 and sT vectors, cells were transfected in the presence of 1.8 μg/mL of vector and Lipofectamine 2000 (Invitrogen, 2.1μL/mL) for 4 hours according to the manufacturer’s instructions, after which cells were returned to their normal medium and allowed to incubate for an additional 40 hours before irradiation.
GGR assay
As described previously, cells were pre-labeled with methyl-[3H]thymidine (Perkin-Elmer, Boston, MA), irradiated with 10 J/m2 UVR, and allowed to repair for up to 24 hours before DNA preparation 21,33. For experiments in which cells were transfected with an expression vector, transfection occurred following pre-label with methyl-[3H]thymidine and 48 hours before irradiation. An immunoblot assay was used to probe damaged DNA using monoclonal antibodies (MBL International, Woburn, MA) against CPD (TDM-2, 1:5000) and 6-4PP (6-4M2, 1:5000), followed by anti-mouse secondary antibodies conjugated to horseradish peroxidase, and then detection by enhanced chemiluminescence and imaging (Fuji LAS 3000, Tokyo, Japan). Individual samples were excised from the blot for scintillation counting to correct for replication. Results are the average of two or three experiments analyzed in triplicate.
Western immunoblotting
As previously described, protein concentrations in whole cell lysates were determined by the bicinchoninic acid assay (Pierce, Rockford, IL) 21,33. Equal quantities of total protein were separated on sodium dodecyl sulfate-polyacrylamide 8% gels, and blotted onto polyvinyldifluoride membranes (Bio-Rad, Hercules, CA) 21,34. Membranes were probed with monoclonal antibodies to MCV LT (CM2B4 31, 1:200, Santa Cruz Biotechnology, Santa Cruz, CA), MCV sT (1:250, gift of Y. Chang and P. Moore)35, XPC (1:1000, Genetex, TX), the V5 epitope (1:1000, Bethyl Laboratories, Montgomery, TX), p53 (DO-1, 1:500, Santa Cruz Biotechnology), p21 (1:200, BD Biosciences, San Diego, CA) and actin (1:3000, Sigma, St. Louis, MO) followed by secondary antibodies conjugated to horseradish peroxidase and detected by enhanced chemiluminescence.
RT-PCR
RNA from cells was prepared and analyzed by RT-PCR as described previously 21,33,36. The following primers were used: LT forward: 5′ CCTCTGGGTATGGGTCCTTCTCA 3′. LT reverse: 5′ ATGGTGTTCGGGAGGTATATC 3′. sT forward: 5′ CCCAAGTAGGAGGAAATCCA 3′. sT reverse: 5′ TTGTCTCGCCAGCATTGTAG 3′.
Flow cytometry
UISO cells and transfected UISO cells and NHFs were first trypsinized. Cells were mechanically disaggregated by pipetting, washed in PBS, and then fixed by the addition of ethanol to a final concentration of 70% (v/v). Following centrifugation, samples were resuspended and incubated for 1 hour at 37°C in PBS containing 100 μg/mL RNase A and 20 μg/mL propidium iodide. Cells were filtered through a 100 μm Nylon mesh and a minimum of 104 cells were analyzed on a Becton-Dickinson FACScan.
Statistical analysis
The unpaired t-test was used to compare treatment groups, and was performed using Kaleidagraph 4.1 (Synergy Software, Reading, PA).
RESULTS
Hypersensitivity to UVR in MCV-positive cells
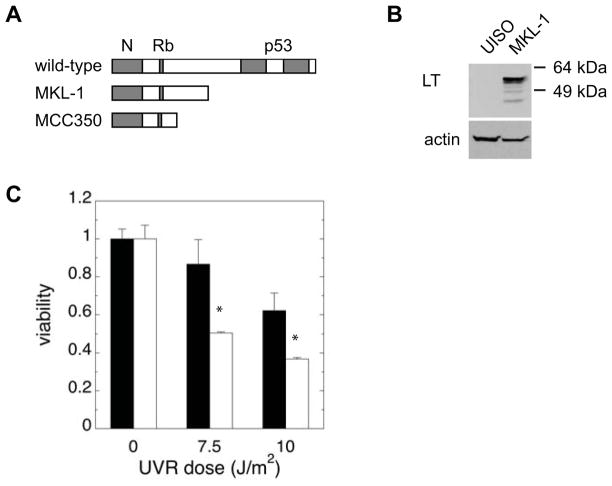
We first compared the post-UVR survival of an MCC cell line, MKL-1 29, that harbors MCV to that of another MCC cell line, UISO 30, that is uninfected. As previously described, MKL-1 cells but not UISO cells are infected with MCV, reflected by expression of LT that was detected using an antibody directed against an epitope within the MCV LT that is preserved in truncated LT proteins (Figure 1A,B) 9,13. The LT expressed by MKL-1 cells is known to be truncated relative to the full-length wild-type protein9,13 (Figure 1A and data not shown). When exposed to increasing doses of UVR, MKL-1 cells exhibited moderate hypersensitivity relative to UISO, with 60% of the viability of UISO cells at 10 J/m2 (Figure 1C). These results suggested that MKL-1 cells may have a defective response to DNA damage that affects survival following UVR.
Figure 1.
MKL-1 cells are sensitive to UVR. A) Schematic depiction of MCV LT gene structure, indicating relative positions of conserved N-terminal sequences, Rb-binding and p53-binding motifs, and truncating mutations in MKL-1 and MCC350. B) Western blot of LT expression in UISO and MKL-1. C) UVR survival. UISO (black bars) and MKL-1 (white bars) cells were irradiated with increasing doses of UVR and allowed to incubate for 5 days before viability was assessed with thiazolyl blue. The asterisk (*) denotes p<0.02.
Poor global genomic repair in MCV-positive cells
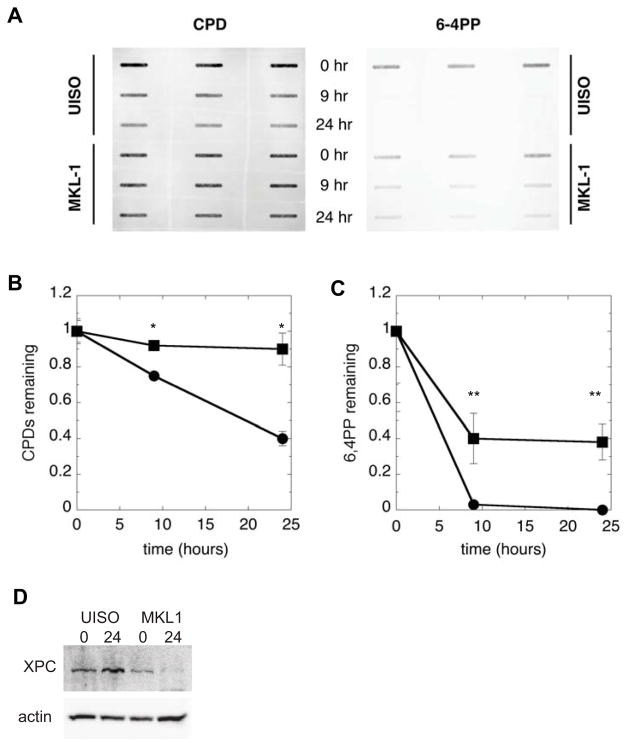
To determine whether differences in NER exist between MKL-1 and UISO cells, we measured the GGR kinetics for the common UVR-induced DNA photoproducts (Figure 2A–C). Following exposure to 10 J/m2 UVR, approximately 40% of the initially formed CPDs remained in UISO cells by 24 hours, corresponding to 60% repair. Nearly all 6-4PPs were repaired by 9 hours in UISO cells. These DNA repair kinetics are typical of those our lab has observed in other repair-proficient cell types resident in skin, such as fibroblasts and keratinocytes 21. In contrast, MKL-1 cells repaired only 10% of CPDs and ~60% of 6-4PPs by 24 hours following UVR.
Figure 2.
MKL-1 cells are impaired in GGR. UISO and MKL-1 cells, pre-labeled with tritiated thymidine, were irradiated with 10 J/m2 UVR and allowed to repair for up to 24 hours before harvesting and analysis. A) Typical slot blot immunoassay result using monoclonal antibodies against CPDs and 6-4PPs. Quantification of results for B) CPDs and C) 6-4PPs in UISO (●) and MKL-1 (■) cells, corrected for replication by normalizing to 3H signal. The asterisk (*) denotes p<0.001 and the double asterisk (**) denotes p<0.01. D) Western blot of XPC levels in UISO and MKL-1 at 0 and 24 hours following 10 J/m2 UVR.
When XPC was examined, levels of XPC were moderately reduced in MKL-1 cells at baseline, relative those in UISO cells, (Figure 2D). As has been described in other cell types such as fibroblasts and keratinocytes, XPC levels were induced by 24 hours following UVR in UISO cells21. Interestingly, following UVR, XPC levels in MKL-1 cells actually decreased to nearly undetectable levels, and were significantly reduced from induced XPC levels in UISO cells (Figure 2D). Overall, these results indicate that MKL-1 cells have a significant defect in GGR, both relative to UISO cells and to other DNA repair-proficient cells.
Loss of G1 arrest in MKL-1 cells following UVR
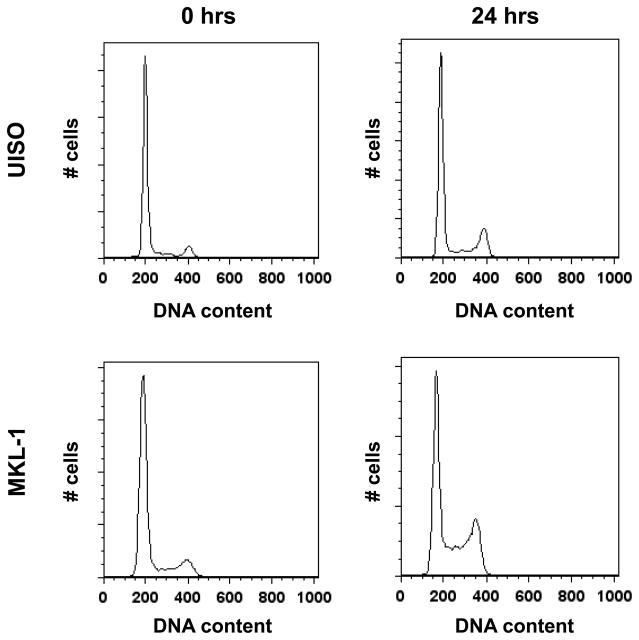
Because the MCV LT possesses homology to the SV40 LT whose actions also include disruption of cell cycle arrest, we also examined cells for loss of cell cycle checkpoints following UVR (Figure 3, Table 1). By 24 hours following UVR, UISO cells exhibited a modest decrease in the G1 population, a slight increase in the S-phase population, and a two-fold increase in the G2/M population over baseline. In contrast, while MKL-1 cells also had an almost two-fold increase in the G2/M population by 24 hours post-UVR, they were notable for a much larger reduction of cells in G1 and a nearly three-fold increase in the S-phase population. These results indicate a notable absence of G1 arrest in MKL-1 cells following UVR and the relative preservation of cell cycle arrest in UISO cells, suggesting that MCV infection may be associated with disruption of the G1/S cell cycle checkpoint.
Figure 3.
Loss of UVR-induced cell cycle arrest in MKL-1. UISO and MKL-1 cells were irradiated with 10 J/m2 UVR, and harvested immediately or allowed to incubate for 24 hours before flow cytometric analysis. The percentage of cells in each cell cycle phase is indicated in Table 1.
Table 1.
Cell cycle distributions 24 hours following UVR (%)
| Cell | Phase | 0 hrs | 24 hours | p |
|---|---|---|---|---|
| UISO | G0/G1 | 83.3 | 71.5 | |
| S | 8.0 | 9.2 | ||
| G2/M | 9.2 | 19.2 | ||
| MKL-1 | G0/G1 | 79.4 | 53.9 | |
| S | 6.0 | 17.9 | ||
| G2/M | 14.3 | 27.8 | ||
|
| ||||
| UISO + pcDNA6 | G0/G1 | 76.3 ± 1.8 | 70.2 ± 2.8 | |
| S | 11.7 ± 2.8 | 13.4 ± 5.0 | ||
| G2/M | 12.0 ± 0.9 | 16.6 ± 2.0 | ||
| UISO + TAg206 | G0/G1 | 74.6 ± 4.9 | 66.6 ± 0.2 | |
| S | 7.2 ± 1.2 | 9.0 ± 4.0 | ||
| G2/M | 18.1 ± 3.5 | 24.4 ± 3.9 | ||
| UISO + TAg350 | G0/G1 | 72.0 ± 0.1 | 49.2 ± 0.8 | 0.001 |
| S | 11.2 ± 2.0 | 19.7 ± 7.4 | ||
| G2/M | 16.8 ± 3.0 | 31.2 ± 8.2 | 0.02 | |
|
| ||||
| NHF + pcDNA6 | G0/G1 | 77.7 ± 3.1 | 70.4 ± 4.4 | |
| S | 9.2 ± 1.8 | 7.0 ± 2.6 | ||
| G2/M | 13.4 ± 1.5 | 22.3 ± 3.3 | ||
| NHF + TAg206 | G0/G1 | 56.8 ± 0.5 | 44.1 ± 0.4 | 0.001 |
| S | 18.9 ± 1.3 | 25.8 ± 3.9 | ||
| G2/M | 25.3 ± 0.4 | 30.0 ± 3.4 | ||
| NHF + TAg350 | G0/G1 | 68.9 ± 2.9 | 51.3 ± 5.7 | 0.05 |
| S | 12.8 ± 1.3 | 20.0 ± 1.6 | 0.009 | |
| G2/M | 19.3 ± 3.4 | 28.6 ± 2.2 | 0.04 | |
In some cases, the sum of percentages do not exactly equal 100 due to rounding errors and averaging of independent experiments. Where indicated, errors are given as standard deviations of 2 or 3 independent experiments.
MCV T antigen inhibits DNA repair and cell cycle arrest following UVR
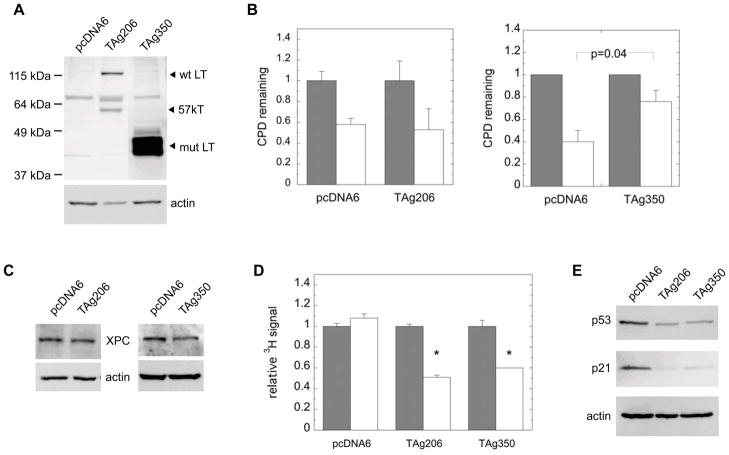
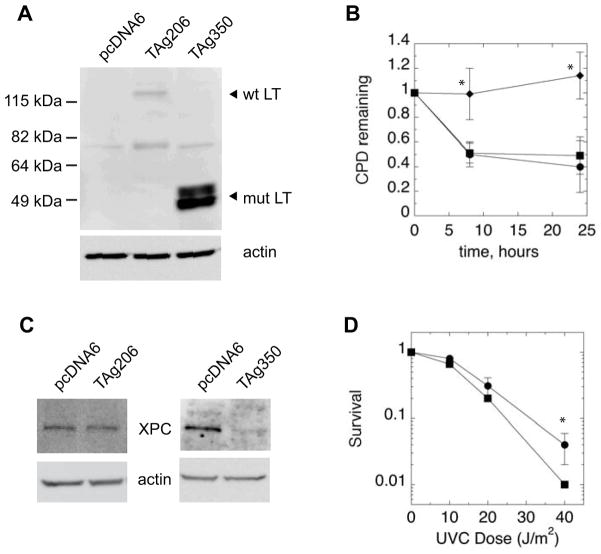
To further test the specific role of T antigen in inhibiting the DNA damage response, we expressed both wild-type and mutant T antigen from another MCC tumor in UISO cells. UISO cells were transfected with expression vectors encoding the wild-type T antigen (TAg206) or T antigen derived from the MCC350 tumor (TAg350), whose LT is more severely truncated than that of MKL-1’s, but similarly preserves the Rb binding domain (Figure 1A) 9. UISO cells transfected with the T antigen vectors express LT whereas cells transfected with a control vector do not (Figure 4A) 31. Cells transfected with TAg206 but not TAg350 resulted in two bands, with the upper and lower bands previously assigned as the LT and 57kT isoforms, respectively 9,31. Expression of the truncated LT from TAg350 was noticeably greater than from TAg206, as has been described previously 9. The vectors express the LT as a fusion protein with the V5 epitope at the C-terminus, and Western blots probed with anti-V5 antibodies also produced nearly identical results (data not shown). Following irradiation with UVR, UISO cells expressing the empty vector control repaired approximately 50% of CPD by 24 hours, similar to the parental UISO cells (Figures 2B and 4B). Cells expressing wild-type T antigen also repaired similarly to the control empty vector. In contrast, UISO cells transfected with TAg350 repaired less than 25% of CPDs by 24 hours, indicating loss of GGR. Since the number of cells that could be transfected was limiting, we did not examine repair of 6-4PP which requires larger quantities of DNA for immunodetection. We also examined expression of the XPC protein and found that UISO cells treated with TAg350 exhibited 70±15% (p=0.04) of the XPC levels of control cells transfected with an empty vector (Figure 4C). Cells expressing the wild-type T antigen exhibited no differences in XPC levels relative to control samples, correlating with their normal repair kinetics. These results suggested that mutant T antigen from TAg350 which is expressed as a truncated LT, but not the full-length wild-type protein, is sufficient to reduce repair of CPDs in association with reductions in XPC levels.
Figure 4.
Ectopic T antigen expression in UISO cells. Cells were transfected with the expression vectors, TAg206 or TAg350, which encode the wild-type or MCC350 tumor T antigen, respectively, or with an empty vector, pcDNA6, which served as a control. A) Western blot of LT and 57kT expression in UISO cells, 2 days following transfection, showing wild-type (wt) and truncated mutant (mut) protein. The ~75 kDa band present in all samples is a non-specific signal. B) For GGR, experiments, cells were pre-labeled with tritiated thymidine prior to transfection with either pcDNA6, TAg206 or TAg350. 2 days following transfection, cells were irradiated with UVR and harvested immediately (black bars) or allowed to repair for 24 hours (white bars) before assay of CPDs. C) Western blot of XPC and actin expression in UISO cells 2 days following transfection with pcDNA6, TAg206, or TAg350. D) Relative 3H specific activity of DNA from UISO cells transfected for 2 days with pcDNA6 or TAg350 harvested either immediately (black bars) or 24 hours (white bars) following UVR. Asterisks (*) denote differences from control with p<0.005. E) Western blot of p53 and p21 in UISO cells 2 days following transfection with pcDNA6, TAg206, or TAg350, with actin used as a loading control.
During the GGR assay, cells were labeled with tritiated thymidine prior to but not following UVR exposure in order to correct for post-UVR replication. Because equivalent amounts of DNA were assayed on the GGR immunoblot, reductions in the specific activity of harvested DNA reflect DNA replication in control and T antigen-positive UISO cells following UVR (Figure 4D). The relative 3H specific activity in DNA from control and LTAg-expressing UISO cells was the same prior to UVR, and did not change in control cells following UVR. In contrast, after UVR the relative specific activity of DNA from both wild-type and truncated mutant T antigen-expressing UISO cells was diluted to 50–60% that of the controls by 24 hours. These results indicate that following UVR-induced DNA damage control UISO cells transfected with empty vector ceased DNA replication while cells expressing T antigen continued to replicate. To further confirm these findings, we examined identically treated cells by flow cytometry (Table 1). Similar to the parental UISO cells, by 24 hours following UVR, cells transfected with the empty vector exhibited some reduction in the G0/G1 population, little change in the S-phase population, and a modest increase in the G2/M population. In contrast, UISO cells expressing TAg206 and particularly TAg350 vectors, had larger reductions in the G0/G1 population and increases in the G2/M population, while TAg350 resulted in increased numbers of S-phase cells after UVR exposure.
We also examined levels of p53 protein, as well as its downstream target, p21, which is important in cell cycle arrest following DNA damage (Figure 4E). Interestingly, both wild-type and mutant T antigen expression reduced expression of p53 as well as p21 in UISO cells. Taken together, these results suggest that both wild-type and mutant MCV T antigens abrogate cell cycle arrest at the G1 checkpoint in response to UVR, allowing cells to continue DNA replication despite persistent DNA damage.
Mutant MCV T antigen impairs repair, cell cycle control, and UVR resistance in fibroblasts
UISO cells, while not infected with MCV, are nonetheless derived from a MCC tumor, and thus may not necessarily be representative of normal, non-transformed cells’ responses to T antigen expression. Although Merkel cells are presumed to be the originating cells for MCC, normal human Merkel cells have proven difficult to isolate and culture in vitro 37,38. Thus, full-length wild-type and mutant T antigen encoded by TAg206 and TAg350, respectively, were instead expressed in normal human dermal fibroblasts (Figure 5A). The 57kT isoform was poorly expressed in fibroblasts. TAg350 again was more strongly expressed than TAg206, as has been observed in other cell types 9. Following UVR, fibroblasts expressing the control vector repaired nearly 50% of CPDs, as did cells expressing the wild-type T antigen (Figure 5B). However, cells expressing TAg350 exhibited no significant repair, recapitulating the results observed in UISO cells. Additionally, cells expressing mutant but not wild-type T antigen exhibited markedly reduced levels of the XPC protein, representing a >80% reduction relative to the control levels (Figure 5C). These results indicate that inhibition of GGR is likely a general feature of mutant T antigen from MCC350 regardless of cell type and degree of cell transformation.
Figure 5.
Ectopic T antigen expression in normal human fibroblasts. Cells were transfected with vectors TAg206 or TAg350 or with empty control vector, pcDNA6. A) Western blot of LT expression in fibroblasts, 2 days following transfection, with indicated positions of wild-type (wt) and truncated mutant (mut) LT from TAg206 and TAg350, respectively. The band at ~75 kDa present in all three samples represents non-specific staining in NHFs. B) For GGR experiments, cells were transfected with control pcDNA6 (●), or TAg206 (■) or TAg350 (□). 2 days following transfection, cells were irradiated with UVR and either harvested immediately or allowed to repair for 24 hours before assay of CPDs. Asterisks (*) indicate differences with the control with p ≤ 0.02. C) Western blot of XPC and actin expression in fibroblasts 2 days following transfection with pcDNA6, TAg206, or TAg350. D) UVR survival. NHFs expressing either pcDNA6 (●) or TAg350 (■) were irradiated with increasing doses of UVR and allowed to incubate for 5 days before viability was assessed with thiazolyl blue. Asterisks (*) indicate differences with the control with p=0.04.
When NHFs expressing wild-type and mutant T antigen were examined by flow cytometry, significant cell cycle distribution aberrations were apparent (Table 1). Relative to the control cells, both TAg206 and TAg350 treatment resulted in much higher baseline proportions of cells in S and G2/M phases. These cell cycle distributions were further skewed 24 hours following UVR, with further depletion of G1 and increases in S and G2/M relative to control cells (p=0.02), indicating a lack of baseline cell cycle regulation as well as checkpoint activation following DNA damage.
Given the large degrees of impairment in DNA repair and cell cycle arrest observed in NHFs treated with mutant T antigen, we ascertained whether this was associated with UVR sensitivity (Figure 5D). NHFs treated with TAg350 exhibited ~70% and 25% of the viability of controls at 20 and 40 J/m2, respectively. These results suggest that the mutant T antigen sensitizes cells to DNA damage.
Role of LT and sT in UISO cells
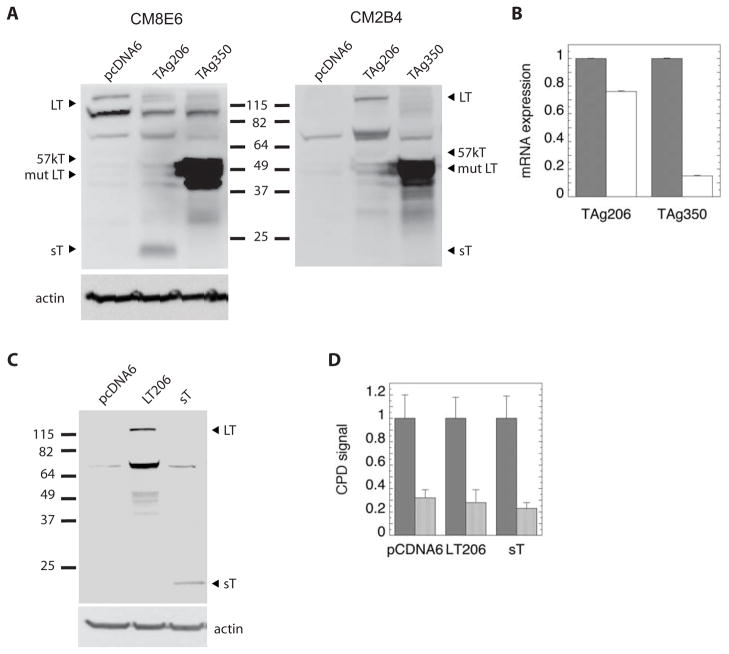
TAg206 and TAg350 encode genomic T antigen sequences. Thus, TAg206 is capable of generating full-length LT, 57kT and sT, while the TAg350 mutation is expected to generate a truncated LT, normal sT, and no 57kT. An antibody (CM8E6) that detects an epitope common to LT, 57kT and sT confirmed that TAg206 does indeed express all three T antigen isoforms (Fig. 6A). The identity of the band assigned as sT was confirmed by stripping the blot originally probed with CM8E6 antibody, re-probing it with the CM2B4 antibody that recognizes only LT and 57kT, and observing the disappearance of the sT band. In contrast, TAg350 appears to only express mutant LT protein. While the absence of 57kT was expected in TAg350-treated cells, interestingly no sT was detectable by Western blotting, and levels of sT transcript detected by RT-PCR were seven-fold lower than those observed with TAg206 (Fig. 6B). These results suggest that the aberrant cell cycle and DNA repair phenotypes associated with TAg350 are likely due to mutant LT, although the data leave open the possibility that undetectable levels of sT protein may be exert an effect.
Figure 6.
LT and sT expression in UISO cells. A) UISO cells were transfected with vectors TAg206 or TAg350 or with empty control vector, pcDNA6 and analyzed 2 days later for expression of T antigen isoforms by Western blot. The blot was probed first with the pan-T antigen antibody, CM8E6, then stripped and re-probed with the CM2B4 antibody that recognizes only LT and 57kT. Actin served as a loading control. The bands at ~100 and ~75 kDa represent non-specific bands. B) Cells transfected with TAg206 or TAg350 were also analyzed by RT-PCR for LT (gray bars) and sT (white bars). C) UISO cells were transfected with pcDNA6, LT206, and sT vectors and probed with the CM8E6 antibody by Western blotting. D) Cells were treated with pcDNA6, LT206 and sT vectors for 48 hours, then irradiated with UVR and harvested immediately (gray bars) or allowed to repair for 24 hours (white bars) before assay of CPDs.
In an effort to further specify the activities of LT and sT, we separately expressed wild-type LT and sT in UISO cells using expression vectors encoding the cDNAs for each (Fig. 6C). In principle, the wild-type LT transcript is available to be alternatively spliced to the 57kT sequence, but under the conditions employed, 57kT protein was undetectable in transfected UISO cells. Cells expressing LT or sT did not exhibit any impairment in their ability to repair CPDs (Fig. 6D). However, relative to pcDNA6-treated controls, cells expressing LT did have an increase in their S phase population following UVR, suggesting a G1 checkpoint defect (Table 2). Cells expressing sT alone had a higher proportion of cells in S phase relative to controls, but did not exhibit cell cycle arrest failure following UVR
Table 2.
Cell cycle distribution 24 hours following UVR (%)
| Cell | Phase | 0 hrs | 24 hours |
|---|---|---|---|
| UISO + pcDNA6 | G0/G1 | 89.9 | 91.0 |
| S | 5.4 | 5.6 | |
| G2/M | 4.6 | 3.4 | |
| UISO + LT206 | G0/G1 | 88.3 | 83.6 |
| S | 6.4 | 10.7 | |
| G2/M | 5.3 | 5.8 | |
| UISO + sT | G0/G1 | 85.3 | 83.2 |
| S | 8.8 | 6.2 | |
| G2/M | 6.0 | 10.6 |
DISCUSSION
The direct study of MCV’s role in MCC pathogenesis offers the unique opportunity to evaluate the properties and role of a polyomavirus which is directly relevant to human disease. Our results demonstrate that an MCC cell line infected with MCV is significantly impaired in its ability to respond to UVR-induced DNA damage relative to a separate uninfected MCC cell line. For example, while uninfected UISO cells possessed GGR kinetics for both CPDs and 6-4PPs that are typical for other repair-proficient cells, the magnitudes of repair deficiency observed in infected MKL-1 cells are comparable to those observed in cells derived from patients with the hereditary disease, xeroderma pigmentosum, which is characterized by a loss of NER 39,40. As far as can be ascertained from the original report describing MKL-1 cells, the donor patient was not clinically affected by xeroderma pigmentosum 29. Nonetheless, while the UISO and MKL-1 cell lines appeared to exhibit very different repair and cell cycle arrest responses, they are derived from separate donors and are quite different in their morphologic and culture characteristics, and thus it is not possible to conclude from these data alone that MCV specifically abrogates the DNA damage response.
Our experiments expressing T antigen in UISO cells as well as in normal fibroblasts were able to more directly assess the ability of MCV to abrogate the normal cellular response to UVR in a common genetic and culture media background. The results indicate that a mutant T antigen derived from MCV is alone sufficient to cause repair and cell cycle arrest defects in MCV-negative MCC cells, recapitulating a similar degree of impairment observed in MCV-infected MKL-1 cells. The defects in repair are associated with reductions in levels of the XPC protein which is involved in the initial DNA damage recognition step of GGR 19. XPC levels were approximately correlated with GGR activity, as levels of CPD repair were significantly affected in mutant T antigen-expressing fibroblasts in which XPC levels were barely detectable while both repair and XPC levels in UISO cells were less dramatically affected by mutant T antigen.
It is also notable that mutant T antigen generating a truncated LT significantly impaired GGR of CPDs, whereas expression of the wild-type T antigen from either genomic sequences or of wild-type LT did not. It is possible that truncating mutations of LT acquire an activity not present in the full-length wild-type protein. The more severely truncated LT from MCC350 may in fact be a special case, as it has been demonstrated to lack a nuclear localization signal, and thus may have a different set of targets than those of longer LTs 41. It is also possible that the reduced expression level of the wild-type relative to the mutant LT in our experimental system may explain their differing repair phenotypes, though this seems unlikely for two reasons. First, cells transfected with up to 6-fold higher quantities of TAg206 did express larger quantities of LT protein, but still did not result in detectable changes in repair kinetics or XPC expression (S. K. Demetriou and D. H. Oh, unpublished results). Second, and most importantly, under conditions in which repair was unaffected, cells expressing the wild-type T antigen exhibited prominent defects in p53 and p21 along with cell cycle dysfunction that are comparable to those found in cells expressing the T antigen mutant. Thus, our results suggest that differential activities of wild-type and mutant LT in the response to DNA damage are a result of their intrinsic properties.
While evidence that sT is an oncoprotein has recently been reported 32, we did not observe that sT expression had a detectable effect on GGR of CPDs or in cell cycle arrest following UVR. Indeed, sT was also not significantly expressed from TAg350, although the reasons are unclear. TAg350 possesses polymorphisms in the sequences encoding sT 9, but these are synonymous with wild-type and not predicted to affect the ability to correctly splice and generate sT; however, it is possible that they affect stability of the sT transcript. In any case, our results taken together suggest that LT is capable of disrupting cell cycle arrest following UVR-induced DNA damage, and that when mutated may also impair GGR.
Although XPC appears to be one downstream target affected by mutant LT, it is also possible that other NER proteins are affected. In any case, the results suggest that mutant LT expression is capable of disrupting GGR even in normal, untransformed cells, and thus could be involved in creating a state of genomic instability as an early event in MCC genesis. This scenario is also consistent with the existence of UVR-type signature mutations in MCC 16.
The related SV40 LT is well-known to disrupt the DNA damage response, in large part by binding to the Rb and p53 tumor suppressors, and these may also be targets for the MCV LT that lead to functional impairment of GGR and cell cycle arrest following UVR. MCV LT, including the truncated mutant encoded by TAg350, has been reported to bind to Rb, and loss of Rb function is associated with loss of the G1 checkpoint in response to DNA damage 9,42. Although there are conflicting reports on the precise effect of Rb on GGR activity, there is evidence that Rb can positively regulate GGR 23–25,43. The similar actions of both wild-type and mutant LT in disrupting cell cycle arrest in cells are consistent with Rb binding as the mediator of the cell cycle-related actions of LT. Thus, it is possible that LT-mediated disruption of Rb alone could account for the observed effects in the DNA damage response.
On the other hand, both the MKL-1 and UISO cells possess wild-type p53 whose function may also be targeted by LT 16, as observed in Figure 4D. Loss of GGR and cell cycle arrest in response to UVR-induced DNA damage occurs with loss of p53 in fibroblasts 21,22,27,28 as well as in UISO cells (S.K. Demetriou and D.H. Oh, unpublished results). It is notable that the MCV LT mutants that have been identified in MCC, including those from MKL-1 and MCC350, typically lose their C-terminal domains that are homologous to those in SV40 which directly bind p53 9. However, even without direct p53 binding, SV40 LT is still able to antagonize p53 function due to an activity that has been localized to the N-terminus of that LT, and such an activity may also exist for MCV LT 44,45. Nevertheless, the ability of wild-type LT to diminish p53 levels comparable to the mutant MCV LT without having a discernible effect on CPD repair renders p53 a less likely sole mediator of the repair deficits created by the mutant LT9.
While MCV appears to be relatively widespread in humans, MCCs are rare, suggesting that MCV infection alone is insufficient to transform cells 46,47. Other genetic changes are likely required for Merkel cell carcinogenesis 1,2,14–17. Our data suggest possible pathways by which MCV genomic integration and subsequent T antigen mutation progressively interfere with key mechanisms of genomic instability. Wild-type T antigen expression upon early infection may lead to abrogation of cell cycle arrest following DNA damage. Once mutated to become a truncated protein, however, LT in particular may acquire additional functionality allowing it to inhibit both GGR and cell cycle checkpoint activation. These LT activities that impair the UVR DNA damage response could provide an explanation for the overall distribution of MCC on chronically sun-exposed skin. These results suggest impairment in at least two processes—DNA repair and cell cycle arrest—by which MCV infection may create a state of genomic instability in normal cells following solar UVR exposure, inappropriately allowing cells to replicate unrepaired DNA and predisposing them to accumulate additional key carcinogenic mutations. Although Shuda et al. have reported an apparently low mutation rate in MCC cDNAs that seems inconsistent with a mutator phenotype 9, this result was based on analysis of a single tumor distinct from MKL-1 or MCC350. Further, a mutator phenotype would be expected to produce random mutations that, unless critical for transformation, would likely be averaged out in any analysis of a cDNA library from a genetically heterogeneous tumor 48.
Whether all MCC T antigens and LT generally abrogate the DNA damage response to UVR and other forms of DNA damage, and the precise mechanism by which they do so, remain to be systematically tested. However, impairment of DNA repair and cell cycle checkpoints in MCC tumors that are infected with MCV may be potentially targeted by therapies that induce DNA damage, preferentially sensitizing MCC cells relative to normal tissue. The presence of MCV infection has been associated with a better prognosis for patients with MCC and it remains to be seen whether positive treatment outcomes are associated with DNA damaging therapeutic modalities that exploit defective DNA damage responses in MCV-infected tumor cells 49,50.
Acknowledgments
We thank Drs. Yuan Chang and Patrick Moore for providing MKL-1 cells, the TAg206, TAg350, LT206 and sT vectors, and the CM8E6 antibody; Dr. T.K. Das Gupta for UISO cells; and R. Griby of the Northern California Institute for Research and Education for assistance with flow cytometry. Work was supported by an HIV-associated malignancies pilot project award from NCI (P30 CA82103), a University of California Cancer Research Coordinating Committee grant, and a VA Merit Award (all to DHO).
References
- 1.The Rockville Merkel Cell Carcinoma Group. Merkel cell carcinoma: Recent progress and current priorities on etiology, pathogenesis, and clinical management. J Clin Oncol. 2009;27:4021–6. doi: 10.1200/JCO.2009.22.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agelli M, Clegg LX, Becker JC, Rollison DE. The etiology and epidemiology of Merkel cell carcinoma. Curr Probl Cancer. 2010;34:14–37. doi: 10.1016/j.currproblcancer.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Albores-Saavedra J, Batich K, Chable-Motero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases; a population based study. J Cutan Pathol. 2009;37:20–7. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 4.Hodgson NC. Merkel cell carcinoma: Changing incidence trends. J Surg Oncol. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 5.Koljonen V, Kukko H, Tukiainen E, Böhling T, Sankila R, Pukkala E, Sihto H, Joensuu H, Kyllönen L, Mäkisalo H. Incidence of Merkel cell carcinoma in renal transplant recipients. Nephrol DIal Transplant. 2009;24:3231–5. doi: 10.1093/ndt/gfp334. [DOI] [PubMed] [Google Scholar]
- 6.Penn I, First MR. Merkel’s cell carcinoma in organ recipients: Report of 41 cases. Transplantation. 1999;68:1717–21. doi: 10.1097/00007890-199912150-00015. [DOI] [PubMed] [Google Scholar]
- 7.Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. Merkel cell carcinoma and HIV infection. Lancet. 2002;359:497–8. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- 8.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci. 2008;105:16272–7. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laude HC, Jonchère B, Maubec E, Carlotti A, Marinho E, Couturaud B, Peter M, Sastre-Garau X, Avril MF, Dupin N, Rozenberg F. Distinct Merkel cell polyomavirus molecular features in tumour and non-tumour specimens from patients with Merkel cell carcinoma. PLoS Pathog. 2010;6:e1001076. doi: 10.1371/journal.ppat.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sastre-Garau X, Peter M, Avril MF, Laude h, Couturier J, Rozenberg F, Almeida A, Boitier F, Carlotti A, Couturaud B, Dupin N. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J Pathol. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- 12.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–82. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–72. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Gele M, Kaghad M, Leonard JH, Van Roy N, Naeyaert JM, Geerts ML, Van Belle S, Cocquy V, Bridge J, Sciot R, De Wolf-Peeters C, De Paepe A, et al. Mutation analysis of p73 and TP53 in Merkel cell carcinoma. Br J Cancer. 2000;82:823–6. doi: 10.1054/bjoc.1999.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Gele M, Leonard JH, Van Roy N, Van Limbergen H, Van Belle S, Cocquyt V, Salwen HR, De Paepe A, Speleman F. Combined karyotyping, CGH and M-FISH analysis allows detailed characterization of unidentified chromosomal rearrangements in Merkel cell carcinoma. Int J Cancer. 2002;101:137–45. doi: 10.1002/ijc.10591. [DOI] [PubMed] [Google Scholar]
- 16.Popp S, Waltering S, Herbst C, Moll I, Boukamp P. UV-B-type mutations and chromosmal imbalances indicate common pathways for the the development of Merkel and skin squamous cell carcinomas. Int J Cancer. 2002;99:352–60. doi: 10.1002/ijc.10321. [DOI] [PubMed] [Google Scholar]
- 17.Paulson KG, Lemos BD, Feng B, Jaimes N, Peñas PF, Bi X, Maher E, Cohen L, Leonard JH, Granter SR, Chin L, Nghiem P. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-myc. J Invest Dermatol. 2009;129:1547–55. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeijmakers JHJ. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 19.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker PJ, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–32. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 20.El-Mahdy MA, Hamada FM, Wani MA, Zhu Q, Wani AA. p53-degradation by HPV-16 E6 preferentially affects the removal of cyclobutane pyrimidine dimers from non-transcribed strand and sensitizes mammary epithelial cells to UV-radiation. Mutat Res. 2000;459:135–45. doi: 10.1016/s0921-8777(99)00066-x. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson BE, Oh DH. Proficient global nucleotide excision repair in human keratinocytes but not fibroblasts deficient in p53. Cancer Res. 2005;65:8723–9. doi: 10.1158/0008-5472.CAN-05-1457. [DOI] [PubMed] [Google Scholar]
- 22.Ford JM, Baron EL, Hanawalt PC. Human fibroblasts expressing the human papillomavirus E6 gene are deficient in global genomic nucleotide excision repair and sensitive to ultraviolet irradiation. Cancer Res. 1998;58:599–603. [PubMed] [Google Scholar]
- 23.Therrien J-P, Drouin R, Baril C, Drobetsky EA. Human cells compromised for p53 function exhibit defective global and transcription-coupled nucleotide excision repair, whereas cells compromised for pRb function are defective only in global repair. Proc Natl Acad Sci. 1999;96:15038–43. doi: 10.1073/pnas.96.26.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin PS, McPherson LA, Chen AY, Sage J, Ford JM. The role of the retinoblastoma/E2F1 tumor suppressor pathway in the lesion recognition step of nucleotide excision repair. DNA Repair. 2009;8:795–802. doi: 10.1016/j.dnarep.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy TM, Kumar MS, Smith ML. Rb stabilizes XPC and promotes cellular NER. Anticancer Res. 2010;30:2438–88. [PubMed] [Google Scholar]
- 26.Prost S, Lu P, Caldwell H, Harrison D. E2F regulates DDB2: Consequences for DNA repair in Rb-deficient cells. Oncogene. 2007;26:3572–81. doi: 10.1038/sj.onc.1210151. [DOI] [PubMed] [Google Scholar]
- 27.Bowman KK, Sicard D, Ford JM, Hanawalt PC. Reduced global genomic repair of ultraviolet light-induced cyclobutane pyrimidine dimers in simian virus 40-transformed human cells. Mol Carcinog. 2000;29:17–24. [PubMed] [Google Scholar]
- 28.Pipas JM, Levine AJ. Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol. 2001;11:23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- 29.Rosen ST, Gould VE, Salwen HR, Herst CV, Le Beau MM, Lee I, Bauer K, Marder RJ, Anderson R, Kies MS, Moll R, Franke WW, et al. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Lab Invest. 1987;56:302–12. [PubMed] [Google Scholar]
- 30.Ronan SG, Green AD, Shilkaitis A, Huang T-SW, Das Gupta TK. Merkel cell carcinoma: In vitro and in vivo characteristics of a new cell line. J Am Acad Dermatol. 1993;29:715–22. doi: 10.1016/0190-9622(93)70236-m. [DOI] [PubMed] [Google Scholar]
- 31.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras M-T, Tolstov YL, Gjoerup O, Mansukani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–9. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–34. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh DH, Yeh K. Differentiating human keratinocytes are deficient in p53 but retain global nucleotide excision repair following ultraviolet radiation. DNA Repair. 2005;4:1149–59. doi: 10.1016/j.dnarep.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Ferguson BE, Li H, Dong TK, Hsiao JL, Oh DH. Impaired repair of cyclobutane pyrimidine dimers in human keratinocytes deficient in p53 and p63. Carcinogenesis. 2008;29:70–5. doi: 10.1093/carcin/bgm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83:12118–28. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mogha A, Fautrel A, Mouchet N, Guo N, Corre S, Adamski H, Watier E, Misery L, Galibert MD. Merkel cell polyomavirus small T antigen mRNA level is increased following in vivo UV-radiation. PLoS One. 2010;5:e11423. doi: 10.1371/journal.pone.0011423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boulais N, Pereira U, Lebonvalley N, Gobin E, Dorange G, Rougier N, Chesne C, Misery L. Merkel cells as putative regulator cells in skin disorders: An in vitro study. PLoS One. 2009;4:e6528. doi: 10.1371/journal.pone.0006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimohira-Yamasaki M, Toda S, Narisawa Y, Sugihara H. Merkel cell-nerve interactino undergoes formation of a synapse-like structure in a primary culture. Cell Strutc Funct. 2006;31:39–45. doi: 10.1247/csf.31.39. [DOI] [PubMed] [Google Scholar]
- 39.Emmert S, Kobayashi N, Khan SG, Kraemer KH. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc Natl Acad Sci USA. 2000;97:2151–6. doi: 10.1073/pnas.040559697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci. 1999;96:424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Hein J, RIchardson SCW, Basse PH, Toptan T, Moore PS, Gjoerup OV, Chang Y. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. J Biol Chem. 2011;286:17079–90. doi: 10.1074/jbc.M110.192856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukas J, Muller H, Bartkova J, Spitkovsky D, Kjerulff AA, Jansen-Durr P, Strauss M, Bartek J. DNA tumor virus oncoprotyeins and retinoblastoma gene mutations share the ability to relieve the cell’s requirement for cyclin D1 function in G1. J Cell Biol. 1994;125:625–38. doi: 10.1083/jcb.125.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prost S, Lu P, Caldwell H, Harrison D. E2F regulates DDB2: Consequences for DNA repair in Rb-deficient cells. Oncogene. 2006 doi: 10.1038/sj.onc.1210151. Advance online publication, 18 December 2006. [DOI] [PubMed] [Google Scholar]
- 44.Quartin RS, Cole CN, Pipas JM, Levine AJ. The amino-terminal functions of the simian virus 40 large T antigen and required to overcome wild-type p53-mediated growth arrest of cells. J Virol. 1994;68:1334–41. doi: 10.1128/jvi.68.3.1334-1341.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rushton JJ, Jiang D, Srinivasan A, Pipas JM, Robbins PD. Simian virus 40 T antigen can regulate p53-mediated transcription independent of binding p53. J Virol. 1997;71:5620–3. doi: 10.1128/jvi.71.7.5620-5623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer. 2009;125:1250–6. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer J, Nghiem P, Galloway DA. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst. 2009;101:15101522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: Clinical implications. Cancer Res. 2008;68:3551–7. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- 49.Sihto H, Kukko H, Koljonen V, Sankila R, Böhling T, Joensuu H. Clinical factors associated with Merkel cell polyomavirus infection in Merkel cell carcinoma. J Natl Cancer Inst. 2009;101:938–45. doi: 10.1093/jnci/djp139. [DOI] [PubMed] [Google Scholar]
- 50.Bhatia K, Goedert JJ, Modali R, Preiss L, Ayers LW. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. Int J Cancer. 2010;126:2240–6. doi: 10.1002/ijc.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]