Abstract
4-Hydroxyalkenal species are a class of peroxidative products of polyunsaturated fatty acids, which serve as “toxic second messengers” in cellular systems. Investigation of their cellular role is hindered due to the lack of sensitive, reliable, robust method for identification and quantification of these metastable metabolites. Herein, we explored the facile Michael adduct of carnosine with 4-hydroxyalkenal species and developed a sensitive, facile, shotgun lipidomics-based method for quantification of these compounds directly from organic solvent lipid extracts of biological samples. In the study, we extensively examined the factors that may affect the accurate quantification of 4-hydroxyalkenal species and found that this method possessed high reproducibility (<8%) and nearly 3 orders of linear dynamic range with a limit of quantification at lower than 0.56 fmol/μl. Mass levels of 4-hydroxyalkenal species in various biological samples, including mouse heart, kidney, liver, and skeletal muscle were determined by this developing method. In addition, the effects of sample collection methods and sample storage time on 4-hydroxyalkenal mass levels were also determined. We believe that development of this novel methodology should provide a powerful tool for us to better understand the role of 4-hydroxyalkenal species in biological processes.
4-Hydroxyalkenal species are a class of peroxidation products of polyunsaturated fatty acids (PUFAs) resulting from a variety of complex enzymatic and non-enzymatic reactions during diverse physiological and pathophysiological processes1, 2. The non-enzymatic peroxidation pathway of n-3 and n-6 PUFAs generates 4-hydroxy-2E-hexenal (4-HHE) and 4-hydroxy-2E-nonenal (4-HNE), respectively, and the enzymatically regulated peroxidation pathway of n-6 PUFA ultimately produces 4-HNE and 4-hydroxy-2E,6Z-dodecadienal3. Accordingly, these peroxidation pathways could be significantly enhanced under patho(physio)logical conditions in which large amounts of reactive oxygen species (ROS) are present4.
Most of these α,β-unsaturated aldehyde species are metastable, which reactively form covalent adducts with macromolecules (e.g., proteins and nucleic acids), thereby leading to inhibition of protein and DNA synthesis, dysregulation of enzyme activities, alteration in mitochondrial coupling, etc.1, 5–7. These species could also easily diffuse from its origin site to propagate the oxidative injury, thereby serving as “toxic second messenger”8. Several lines of evidence indicate that accumulation of the modified proteins with these reactive aldehyde species is manifest in cells during aging or under oxidative stress and that such modification and accumulation are linked to the pathogenesis of numerous diseases, such as atherosclerosis, diabetes, muscular dystrophy, rheumatoid arthritis, actinic elastosis, and neurodegenerative diseases (i.e., Alzheimer’s disease, Parkinson’s disease, and cerebral ischemia)1, 2, 9.
Varieties of methodologies have recently been developed and used to identify and quantitate 4-HNE alone or along with other aldehydes present in biological samples, including gas chromatography-mass spectrometry (GC-MS)10–12, high performance liquid chromatography (HPLC)13–15, HPLC combined with GC-MS16, electrospray ionization (ESI)-MS after direct infusion17, and LC-MS/MS18–22. In the methods using GC-MS, 4-HNE and other aldehydes were converted into their O-pentafluorobenzyloxime derivatives, and then detected by selected ion monitoring (SIM)10, 12 or stable isotope dilution MS11, 16. These approaches involved laborious multi-step chemical modification. 4-HNE could be derivatized with 2,4-dinitrophenylhydrazine (DNPH)13, 15 or fluorescently labeled with 4-(2-carbazoylpyrrolidin-1-yl)-7-nitro-2,1,3-benzoxadiazole14. The derivatives were then separated by HPLC and detected by electrochemical or laser induced fluorescence detection to increase the sensitivity of the methodologies. Those methods did not adequately provide structural details regarding the 4-hydroxyalkenals. ESI18, 20–22 or atmospheric chemical ionization19 has been coupled with LC to analyze 4-HNE and other biologically relevant aldehydes. Those aldehydes were derivatized with either cyclohexanedione18 or DNPH19, 20 and then detected by selected reaction monitoring. Alternatively, those aldehydes have been detected by LC-MS in tissue samples without derivatization22. In those methods, benzaldehyde18, heptanal and octanal20, or 3-non-2-enone22 was used as internal standard(s) for quantitation, respectively. 4-HNE was also directly detected by SIM on the [M+H]+ monoisotopic species for 4-HNE with d11-4-HNE as an internal standard by ESI-MS after direct infusion without any derivatization17. However, it only determined the 4-HNE content in biological samples with a relatively low sensitivity.
Based on the important roles of 4-hydroxyalkenals, especially 4-HNE, in the biological systems, it is critical to have a sensitive, simple, high throughput, and accurate method for identification and quantification of 4-hydroxyalkenal species in biological samples. In the current study, we explored the facile Michael adduct of carnosine with 4-hydroxyalkenal species (e.g., 4-HNE)23 and found that (1) ionization of the formed adducts by ESI-MS is substantially enhanced in comparison to native 4-hydroxyalkenal species in the positive-ion mode and (2) product-ion MS analysis of carnosine-adducted 4-hydroxyalkenal species displayed many abundant, informative, and characteristic fragment ions which could be exploited to identify and quantify these facile oxidation metabolites in the presence of their stable isotope-labeled counterparts. Based on these findings, we developed a sensitive, facile, shotgun lipidomics-based method for quantification of these compounds directly from chloroform extracts of biological samples after a simple step derivatization with carnosine. This method not only allows us to accurately quantify the levels of 4-HNE in the biological samples, but also enables us to discover the existence of other 4-hydroxyalkenal species (e.g., 4-hydroxy-nondienal (4-HNDE) and 4-hydroxy-dodecatrienal (4-HDTE)) and to assess the levels of these species. In the current study, the factors that may affect the accurate quantification of 4-hydroxyalkenal species including reproducibility, limit of quantification, linear dynamic range, extraction efficiency, the effects of ion suppression, etc. on quantification were examined extensively. Mass levels of 4-hydroxyalkenals in various biological samples, including mouse heart, kidney, liver, and skeletal muscle were determined by this developing method. Moreover, the effects of sample collection methods and sample storage time on 4-hydroxyalkenal mass levels were also determined.
Materials and Methods
Materials
Commercially available 4-hydroxyalkenals including 4-HHE (5 mg in 0.5 ml ethanol), 4-HNE (10 mg in 1 ml ethanol), and d3-4-HNE (500 μg in 1 ml methyl acetate) were purchased from Cayman Chemical Co. (Ann Arbor, MI) and used without further quantification. Stock solutions for these 4-hydroxyalkenal species were prepared in methanol, and all of the 4-hydroxyalkenals were stored at −80 °C. L-Carnosine (~ 99%) was purchased from Sigma-Aldrich (St. Louis, MO). All solvents used for lipid extraction and preparation and for MS analysis were obtained from Burdick and Jackson (Muskegon, MI), except formic acid obtained from Thermo Fisher Scientific, Inc. (Fair Lawn, NJ).
Preparation of lipid extracts from biological samples
Mice (male, C57BL/6, 3 months of age) were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were sacrificed by asphyxiation with carbon dioxide or cervical dislocation. Heart, kidney, liver, and skeletal muscle tissues were dissected, perfused with phosphate-buffered saline to remove blood, blotted with Kim-wipes to remove excess buffer, and then immediately freeze-clamped at the temperature of liquid nitrogen. All of biological tissue samples were stored at −80 °C. Wafers were pulverized into a fine powder with a stainless steel mortar and pestle. All animal procedures were performed in accordance with the “Guide for the Care and Use of Laboratory Animals” (National Academy of Science, 1996) and were approved by the “Animals Studies Committee” at Sanford-Burnham Medical Research Institute.
Tissue fine powders were weighed and homogenized in phosphate-buffered saline. Protein assay on the homogenates was performed by using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) with bovine serum albumin as a standard. All determined lipid levels were normalized to the protein content.
Fine powder of each individual biological tissue sample (~ 10 mg) was weighed into a disposable glass culture test tube, and d3-4-HNE was added as an internal standard prior to lipid extraction for quantitation of 4-hydroxyalkenals. Lipid extraction was performed by using a modified Bligh and Dyer procedure as described previously24. Each lipid extract was resuspended into a volume of 100 μl chloroform/methanol (1:1, v/v) per mg protein, and flushed with nitrogen, capped, and stored at −20 °C.
Derivatization of 4-hydroxyalkenals with carnosine
A certain amount of lipids (equivalent to 0.5 mg of tissue protein content) was transferred from the stock solution of lipid extracts (see above) to a disposable culture glass test tube with cap. To individual lipid solution was added 400 μl of water and 50 μl of carnosine aqueous solution (75 mM) or specified. After flushed with nitrogen and capped, the reaction mixtures were incubated at 37 °C for 24 hrs. The reaction was terminated by addition of 300 μl of water and 1.5 ml of chloroform/methanol (1:1, v/v) to each of sample tube and vortexing. The derivative products were extracted into the upper aqueous layer after centrifugation at 3,500 rpm for 10 min. This aqueous phase was washed three times by addition of chloroform (0.75 ml) each time. All the solvent phase extracts were discarded. The retained aqueous phase extracts were evaporated under a nitrogen stream with a water bath set at 40 °C. Each individual residue was reconstituted with 100 μl water/methanol (1:1, v/v), flushed with nitrogen, capped, and stored at −20 °C for ESI-MS analysis (typically analyzed on the same day).
Mass spectrometric analysis of carnosine adducts of 4-hydroxyalkenals
Individual carnosine-derived 4-hydroxyalkenal solution prepared above was further diluted with methanol/water/formic acid (80/20/0.1, v/v/v) to a final concentration of 100 to 1000 fmol of d3-4-HNE/μl for direct infusion. Mass spectrometric analysis was performed on a QqQ mass spectrometer (Thermo TSQ VANTAGE, San Jose, CA) equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY). An ionization voltage of 1.4 kV and a gas pressure of 0.25 psi on the NanoMate apparatus were employed for the MS analyses. The device was controlled by Chipsoft 8.3.1 software. All MS or tandem MS analyses were operated under Xcalibur software as described previously25. Typically, a 1-min period of signal averaging from 1 s per scan in the profile mode was used for each MS spectrum, and a 3-min period of signal averaging from 1 s per scan was employed for each tandem MS spectrum. For tandem MS analysis in neutral-loss, precursor-ion, or product-ion mode, collision gas (argon) pressure was set at 1.0 mTorr, and collision energy was specified in each experiment. A mass resolution setting of 0.7 Thomson was used for both MS and tandem MS analyses.
Results and Discussion
Derivatization of 4-hydroxyalkenals with carnosine
Enrichment of the low abundance 4-hydroxyalkenals by chromatography is not very practical due to their meta-stability. Moreover, although there exist both aldehyde and hydroxyl groups in each molecule of 4-hydroxyalkenals, their ionization efficiency by ESI-MS is also not good enough. These obstacles hamper the accurate analysis of these bioactive lipid peroxidation metabolites. A logical solution to resolve these obstacles is to develop a facile derivative method which could stabilize these reactive metabolites, enhance their ionization efficiency by ESI-MS, and provide characteristic fragments in tandem mass spectrometry.
To this end, we recognized that 4-hydroxyalkenal species could be readily derivatized through Michael adduct between C-3 of 4-hydroxyalkenals and the imidazole nitrogen of carnosine followed with a natural arrangement to form a hemi-acetal derivative as described previously 23. The primary amine in 4-hydroxyalkenal-carnosine adduct (Figure 1B) can be easily protonated under acidic conditions. Therefore, the carnosine acts as a polar group to enhance ionization in the positive-ion mode by ESI-MS. In addition, we found that the formed derivatives were stable at −20 °C for a few weeks as examined.
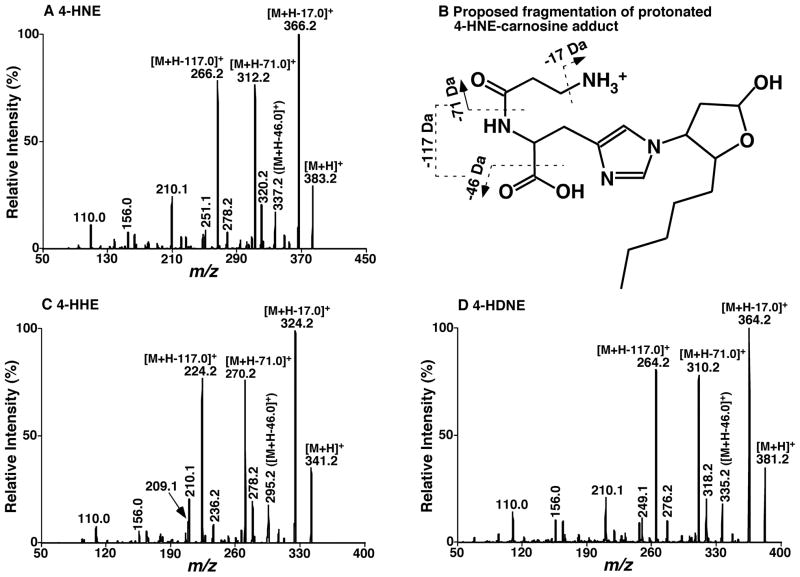
Figure 1.
Representative product-ion mass spectrometric analyses of carnosine-adducted 4-hydroxyalkenal species. Carnosine-4-hydroxyalkenal adducts were prepared by incubating individual 4-hydroxyalkenal species with carnosine at 37°C for 24 hrs as described under “Materials and Methods”. Product-ion ESI-MS analyses of carnosine adducts of 4-hydroxynonenal (4-HNE, Panel A), 4-hydroxyhexenal (4-HHE, Panel C), and 4-hydroxynondienal (4-HNDE, Panel D) were performed on a QqQ mass spectrometer as described under “Materials and Methods”. The fragmentation pattern of protonated 4-HNE-carnosine adduct was proposed (Panel B).
In the study, we optimized the ratio of carnosine vs. 4-hydroxyalkenals for derivatization. This ratio is critical for quantification of 4-hydroxyalkenal species since a high ratio favors complete derivatization whereas a lower ratio favors the reduction of ion suppression from the co-existence of carnosine with carnosine-adducted 4-hydroxyalkenals in the prepared samples. In addition, the levels of 4-hydroxyalkenals present in biological samples may vary in a board range. To optimize this ratio, we derivatized a lipid mixture containing mouse liver lipid extract and 4-hydroxyalkenals (i.e., 4-HNE and d3-4-HNE) at a molar ratio of 50:1 with varied amounts of carnosine (from 50 to 50,000 times of the 4-hydroxyalkenal levels). After working up the derivatization, we examined the ionization stability and efficiency with these samples. We found that the derivatization with the amount of carnosine in 500 times of the 4-hydroxyalkenal levels gave the best results (i.e., highest signals with low baseline levels). This amount of carnosine represents an addition of 3.75 μmol of carnosine to the lipid extracts of biological samples with 0.5 mg protein content.
Characterization of 4-hydroxyalkenal-carnosine adducts by ESI-MS/MS
Product-ion ESI-MS analysis of the protonated 4-HNE-carnosine adduct at m/z 383.2 after collision-induced dissociation displayed numerous abundant, informative, and characteristic product ions (Figure 1A). These ions include those very abundant ones at m/z 366.2, 312.2 and 266.2, corresponding to the neutral loss of 17.0 (ammonia), 71.0, and 117.0 Da, respectively (Figure 1B). Product-ion ESI-MS analyses of protonated other 4-hydroxyalkenal-carnosine adducts displayed essentially identical fragmentation patterns to that of 4-HNE (Figures 1C and 1D compared to Figure 1A). For example, very abundant product ions at m/z 324.2, 270.2 and 224.2 resulted from fragmentation of the ion at m/z 341.2 (i.e., protonated 4-HHE-carnosine adduct) (Figure 1C) and product ion mass spectrum of protonated 4-HNDE adduct at m/z 381.2 showed very abundant fragment ions at m/z 364.2, 310.2 and 264.2 (Figure 1D). These fragments correspond to the neutral losses of 17.0, 71.0, and 117.0 Da from their respective precursor ions and resulted from the carnosine-adducted polar group. Other abundant fragment ions include those corresponding to the neutral losses of 46.0 and 63.0 Da and the one at m/z 210.1. Therefore, a combined detection of a few of these fragment ions can be used to specifically identify the presence of carnosine adducts of 4-hydroxyalkenal species whereas the structures of 4-hydroxyalkenal species can be readily deduced from the detected molecular weight and the knowledge of naturally-existing PUFA structures.
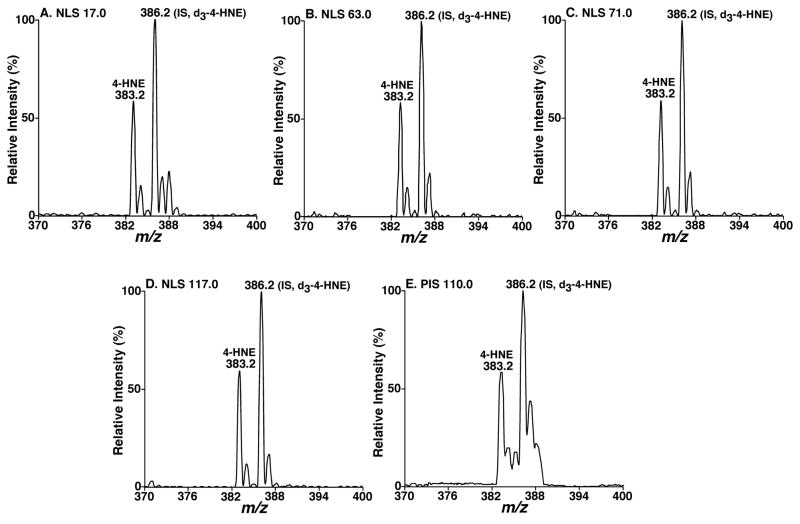
Because the fragment ions corresponding to the neutral losses of 17.0, 63.0, 71.0, and 117.0 Da are abundant and specific to carnosine adducts so that it is logical to perform neutral-loss scanning (NLS) of these neutral fragments to identify the presence of 4-hydroxyalkenal species (Figures 2A–2D). It is of interest that the tandem mass spectra of NLS 17.0, 63.0, 71.0, and 117.0 Da of a mixture of 4-HNE and d3-4-HNE at 0.56:1 molar ratio displayed an essentially identical intensity ratio of 4-HNE adduct vs. d3-4-HNE adduct in all the spectra under experimental conditions (Figures 2A–2D). This observation indicates that NLS of 17.0, 63.0, 71.0, and 117.0 Da can be used not only to identify 4-hydroxyalkenal compounds, but also possibly to quantify these species in comparison to their isotope labeled counterparts. A representative precursor-ion scan (PIS) of 110.0 Thomson was also acquired from the mixture for comparison (Figure 2E). This example indicates that an essentially identical intensity ratio of 4-HNE adduct vs. d3-4-HNE adduct could be obtained even from relatively low abundant fragments although the resolution in precursor ion mass spectrum was not as good as those in neutral-loss mass spectra as previously discussed26.
Figure 2.
Comparisons between different neutral loss mass spectra of different neutral fragments from a mixture of 4-HNE and d3-4-HNE after carnosine derivatization. Carnosine adducts of a mixture of 4-HNE and d3-4-HNE (a molar ratio of 0.56:1) were prepared by incubating the mixture with carnosine at 37°C for 24 hrs as described under “Materials and Methods”. Neutral loss scans of 17.0 Da (Panel A), 63.0 Da (Panel B), 71.0 Da (Panel C), and 117.0 Da (Panel D), and precursor-ion scan of 110.0 Thomson (Panel E) were acquired after direct infusion of the derivatized mixture solution at collision energy of 16, 27, 23, 28, and 26 eV, respectively, and collision gas pressure of 1 mTorr.
Quantitation of 4-hydroxyalkenals by ESI-MS/MS
To determine the possibility of neutral loss analysis of lipid extracts for quantification of 4-hydroxyalkenal species, we examined the limit of quantification and the linear dynamic range of the method with mixtures of 4-HNE and d3-4-HNE (0.56:1 molar ratio) at different concentrations by using NLS of 71.0 and 117.0 Da. These NLS mass spectra represent the best sensitivity and specificity for identification and quantitation of 4-hydroxyalkenal-carnosine adducts with optimized collision energy of 23 and 28 eV for NLS of 71.0, and 117.0 Da, respectively, and collision gas pressure (argon) at 1 mTorr. Under these conditions, the limit of quantification of 4-hydroxyalkenal species was found to be much lower than 560 amol/μL (Figure S1). We found that neutral loss tandem MS analysis of the mixtures at the different concentrations always gave rising to the virtually constant ratios of ion peaks of 4-HNE and d3-4-HNE which corresponds to the molar ratio of the mixture and yielded a broad linear dynamic range (Figure S1). Similar results from a mixture of 4-HHE and d3-4-HNE at a constant molar ratio were also obtained through NLS of 17.0, 63.0, 71.0, and 117.0 Da, indicating that the ionization and fragmentation of 4-hydroxyalkenal species largely depend on the carnosine adducts. Therefore, quantitation of other 4-hydroxyalkenal species than 4-HNE without their isotope labeled counterpart standards could also be conducted with d3-4-HNE as an internal standard after we determined the extraction recovery of other 4-hydroxyalkenal species relative to the d3-4-HNE standard (see below).
We further examined the correlation of different molar ratios between 4-HNE and d3-4-HNE or between 4-HHE and d3-4-HNE with the different ratios of these paired ion peak intensities which were determined by averaging the ratios obtained from neutral-loss MS analysis of 71.0 and 117.0 Da. Quantitative relationships of the ion peak intensity ratios vs. the molar ratios of 4-HNE and d3-4-HNE or 4-HHE and d3-4-HNE were obtained (Figure S2). It should be pointed out that correction for 13C isotopologue difference in the case of 4-HHE relative to d3-4-HNE should be considered for quantitation of 4-HHE by using d3-4-HNE as an internal standard as described previously26. Linear correlation coefficients (γ2) of 0.9995 or 0.9981, slope of 1.01 or 1.0287, and intercepts of -0.0311 or -0.0175 for the pair of 4-HNE and d3-4-HNE or of 4-HHE and d3-4-HNE, respectively, were obtained (Figure S2F). The high correlation between 4-HHE and d3-4-HNE further indicated that the presence of a few of methylene groups between 4-HHE and 4-HNE does not apparently affect the quantitation of 4-HHE through NLS of 71.0 and 117.0 Da in the positive ion mode. These results suggest that the levels of other 4-hydroxyalkenal species (such as 4-HNDE and 4-HDTE) present in biological samples could also be similarly assessed to that of 4-HHE by using d3-4-HNE as an internal standard. It should be emphasized that it would always be the best to use isotope labeled 4-hydroxyalkenal analogs if available as internal standards to quantify the levels of endogenous 4-hydroxyalkenal counterparts present in biological samples.
Examination of the factors that might affect the accurate quantitation of 4-hydroxyalkenals
The extraction recovery of 4-hydroxyalkenal species relative to d3-4-HNE was examined since there exist different numbers of methylene groups in 4-hydroxyalkenals from the internal standard, d3-4-HNE. The recovery of 4-HHE was examined by the described extraction methods as follows. The internal standard (d3-4-HNE, 1.0 nmol/mg protein) was added to a liver homogenate. The homogenized samples with and without addition of 4-HHE were extracted and the amounts of 4-HHE were determined by averaging the ratios obtained from neutral-loss MS analysis of 71.0 and 117.0 Da in comparison to the internal standard. The recovery was determined by comparison of the increased levels of 4-HHE from endogenous 4-HHE content with that of spiked 4-HHE. A 97% recovery of 4-HHE from the homogenized liver samples was obtained.
Majority of lipids were extracted to the chloroform layer after derivatization. The existence of these lipids in the aqueous phase is minimal. To determine the influence of other lipids potentially co-existing with 4-hydroxyalkenal-carnosine adducts in aqueous solution on quantitation of 4-hydroxyalkenals, we mixed various amounts of 4-HNE-carnosine adduct with a liver lipid extract in the presence of a certain amount of d3-4-HNE-carnosine adduct. Specifically, in a liver lipid extract (~ 250 pmol/μl), d3-4-HNE-carnosine adduct was spiked as a fixed concentration of 20 pmol/μl whereas 4-HNE-carnosine adduct was added by varying from 1.0 to 300 pmol/μl. The prepared solutions were diluted to a total lipid concentration of < 50 pmol/μl prior to direct infusion to the mass spectrometer with a nanomate device. NLS of 71.0 and 117.0 Da with these solutions was performed to determine the peak intensity ratios of 4-HNE and d3-4-HNE (Figure S3). A linear relationship between the molar ratio and the peak intensity ratio of the spiked 4-HNE to d3-4-HNE was obtained (Figure S3F). This result indicates that the effects of a residual amount of other lipids co-existing with 4-hydroxyalkenal-carnosine adducts on quantitation of these adducts can essentially be neglected and that shotgun lipidomics of these compounds can be practically performed. This is largely due to that the carnosine adduct plays a major role in ionization of these carnosine-adducted 4-hydroxyalkenal species and that there co-exists an isotope labeled internal standard during sample preparation.
We also compared the sensitivity of the derivatives with other reagents for quantification of 4-hydroxyalkenal levels in biological samples since previous studies demonstrated that N-acetyl-cysteine27, DNPH13, 15, 19, 20, and trichlorophenylhydrazine (TCPH)28 could also form adducts with 4-hydroxyalkenal species. We found that the adducts of 4-HNE with these reagents gave rise to much lower ion intensities than that of carnosine adduct in the survey scan mass spectra under similar experimental conditions. This result likely resulted from either the lower derivatization efficiency of 4-hydroxyalkenals with these reagents or the lower ionization efficiency of 4-hydroxyalkenal adducts with these reagents in comparison to that of carnosine or a combination of these potentials.
Quantitation of 4-hydroxyalkenals in lipid extracts of various biological samples
During our quantitation of 4-hydroxyalkenal levels in the lipid extracts of biological samples by using the developing approach, we determined the influence of methods for sacrificing animals on the mass levels of 4-hydroxyalkenals. Two groups of mice were sacrificed by either cervical dislocation or asphyxiation with carbon dioxide. These methods represent some of the most commonly-used ones in animal research. We found that the levels of 4-hydroxyalkenal species in the lipid extracts of biological samples collected after cervical dislocation was normally lower than those collected by asphyxiation with carbon dioxide (Table 1). This is likely due to the incomplete mitochondrial respiration during asphyxiation with carbon dioxide, leading to the increased electron leakage and ROS production which results in the higher levels of 4-hydroxyalkenals. The values of 4-HNE levels reported in literature are in a board range from 0.002 to 3.2 nmol/mg protein in the biological samples5, 17, 20. Our results well fell into these ranges.
Table 1.
Comparison of 4-hydroxyalkenal mass levels in lipid extracts of various mouse tissue samples collected by cervical dislocation or asphyxiation with CO2 under different storage timea.
| Method (Storage time) | Molecular Speciesb | #1 | #2 | #3 | Mean±SEM | #1 | #2 | #3 | Mean±SEM |
|---|---|---|---|---|---|---|---|---|---|
| Heart | Liver | ||||||||
| Cervical dislocation (1 day) | HHE | 0.53 | 0.67 | 0.40 | 0.53±0.06 | 0.14 | 0.08 | 0.19 | 0.14±0.02 |
| HNDE | 0.64 | 0.58 | 0.59 | 0.60±0.01 | 0.13 | 0.12 | 0.09 | 0.11±0.01 | |
| HNE | 0.32 | 0.28 | 0.31 | 0.30±0.01 | 0.42 | 0.46 | 0.32 | 0.40±0.03 | |
| HDTE | 0.32 | 0.14 | 0.31 | 0.26±0.04 | 0.07 | 0.15 | 0.05 | 0.09±0.02 | |
| CO2 (1 day) | HHE | 1.19 | 0.92 | 0.69 | 0.93±0.17 | 0.14 | 0.09 | 0.15 | 0.13±0.02 |
| HNDE | 0.84 | 0.96 | 0.85 | 0.88±0.04 | 0.19 | 0.10 | 0.26 | 0.18±0.06 | |
| HNE | 0.47 | 0.43 | 0.41 | 0.43±0.02 | 0.39 | 0.31 | 0.42 | 0.37±0.04 | |
| HDTE | 0.20 | 0.39 | 0.48 | 0.36±0.10 | 0.04 | 0.04 | 0.11 | 0.06±0.03 | |
| CO2 (30 day) | HHE | 0.69 | 0.78 | 0.81 | 0.76±0.03 | 0.03 | 0.06 | 0.07 | 0.05±0.01 |
| HNDE | 0.69 | 0.61 | 0.67 | 0.66±0.02 | 0.03 | 0.07 | 0.07 | 0.06±0.01 | |
| HNE | 0.35 | 0.28 | 0.31 | 0.31±0.01 | 0.12 | 0.15 | 0.19 | 0.15±0.02 | |
| HDTE | 0.24 | 0.20 | 0.19 | 0.21±0.01 | 0.01 | 0.03 | 0.03 | 0.03±0.01 | |
| Kidney | Muscle | ||||||||
| Cervical dislocation (1 day) | HHE | 0.17 | 0.11 | 0.22 | 0.16±0.02 | 0.09 | 0.13 | 0.09 | 0.10±0.01 |
| HNDE | 0.31 | 0.23 | 0.31 | 0.29±0.02 | 0.15 | 0.23 | 0.14 | 0.17±0.02 | |
| HNE | 0.46 | 0.40 | 0.41 | 0.42±0.01 | 0.15 | 0.12 | 0.17 | 0.15±0.01 | |
| HDTE | 0.22 | 0.27 | 0.16 | 0.22±0.03 | 0.10 | 0.07 | 0.07 | 0.08±0.01 | |
| CO2 (1 day) | HHE | 0.31 | 0.39 | 0.26 | 0.32±0.05 | 0.34 | 0.55 | 0.31 | 0.40±0.09 |
| HNDE | 0.27 | 0.35 | 0.30 | 0.31±0.03 | 0.31 | 0.52 | 0.41 | 0.42±0.07 | |
| HNE | 0.32 | 0.40 | 0.36 | 0.36±0.03 | 0.35 | 0.39 | 0.43 | 0.39±0.03 | |
| HDTE | 0.09 | 0.13 | 0.17 | 0.13±0.03 | 0.20 | 0.28 | 0.20 | 0.22±0.03 | |
The mass levels of 4-hydroxyalkenal species in nmol/mg protein were determined by averaging the levels determined from both NLS71 and NLS117 of individual lipid extract as described in the text.
The abbreviations of 4-hydroxyalkenal molecular species (i.e., 4-HHE, 4-HNDE, 4-HNE, and 4-HDTE) denote 4-hydroxyhexenal, 4-hydroxynondinenal, 4-hydroxynoneal, and 4-hydroxydodecatrienal, respectively.
We further examined the reproducibility of the developed method with two sets of experiments. First, we determined the 4-HNE levels of mouse heart, liver, kidney, and muscle from one preparation of a tissue sample, but analyzed each sample for three separate times, each of which 4-HNE content was the average of the levels determined from NLS of 71 and 117 Da. We found the variation of 4-HNE levels of these tissue samples was within 8% of experimental error. In the second set of experiments, we separately prepared and derivatized 4-hydroxyalkenals in lipid extracts of mouse heart, liver, kidney, and muscle from three different animals with carnosine. Again, the hydroxyalkenal mass content of each individual sample was the average of the levels determined from NLS of 71 and 117 Da. We found the variation of these mass levels of each tissue type was within 15% of experimental error (Table 1). These results suggest that the experimental errors of the method were largely from sample preparation. We believe that the variation introduced from the protein assay contributes the major part of the demonstrated reproducibility of the method.
We tested the effects of tissue storage time on the mass levels of endogenous 4-hydroxyalkenal species. Mouse heart and liver samples were extracted on the same day of the dissection or after being stored at −80 °C for a month. After quantification of the levels of 4-hydroxyalknenal species, we found that the 4-hydroxyalkenal levels decreased as the storage time in both heart and liver tissues (Table 1). The results suggest that the high chemical reactivity of those metastable metabolites could still slowly react with other macromolecules or degraded in those tissues even at −80 °C.
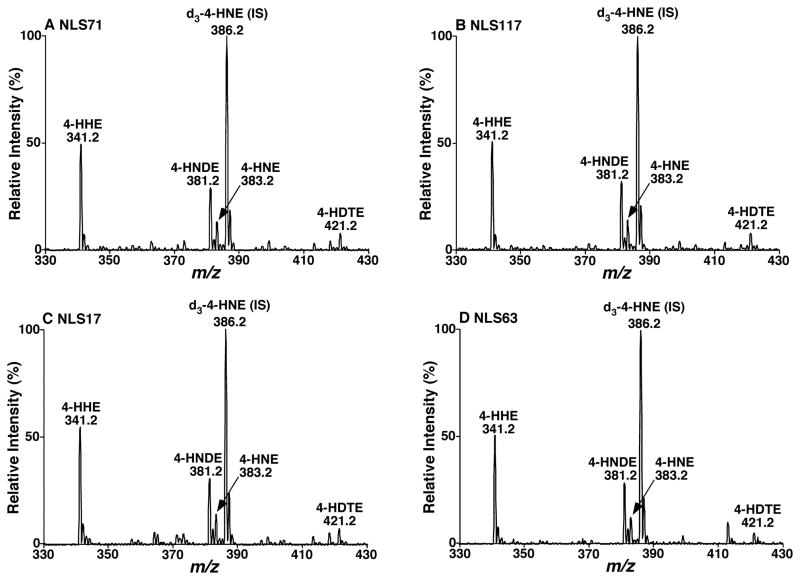
In the analyses of 4-hydroxyalkenal-carnosine adducts from biological sample preparations by tandem MS, NLS of 71.0 and 117.0 Da displayed other ion peaks corresponding to 4-HHE, 4-HNDE, and 4-HDTE in the mass range in addition to 4-HNE (Figures 3A and 3B). The presence of these 4-hydroxyalkenal species was confirmed not only with the identical ion peak intensity ratios of these ions relative to the selected internal standard (i.e., d3-4-HNE), but also with other tandem MS analyses (e.g., NLS of 17.0 and 63.0 Da) (Figures 3C and 3D). Based on the determined extraction recovery and the unaffected ionization efficiency of carnosine-adducted 4-HHE, 4-HNDE, and 4-HDTE relative to that of 4-HNE, we assessed the levels of these 4-hydroxyalkenal species in mouse heart, liver, kidney, and skeletal muscle (Table 1). To the best of our knowledge, the mass levels of most of these species have not been reported previously.
Figure 3.
Representative tandem mass spectrometric analysis of carnosine-derivatized 4-hydroxyalkenal species from mouse myocardial lipid extracts. Lipid extracts of mouse myocardium were performed by a modified Bligh-Dyer method and derivatized with carnosine as described under “Materials and Methods”. Neutral loss scans of 71.0 (Panel A), 117.0 (Panel B), 17.0 (Panel C), and 63.0 Da (Panel D) were acquired on a QqQ mass spectrometer after direct infusion of the derivatized lipid extract at collision energy of 23, 28, 16 and 27 eV, respectively. The abbreviations of 4-HHE, 4-HNDE, 4-HNE, and 4-HDTE denote 4-hydroxyhexenal, 4-hydroxynondinenal, 4-hydroxynoneal, and 4-hydroxydodecatrienal, respectively. “IS” stands for internal standard.
In summary, our multi-dimensional MS-based shotgun lipidomics technology platform26, 29, 30 was extended to characterization and quantitation of 4-hydroxyalkenal species in crude lipid extracts of biological samples. Facile and specific derivatization of 4-hydroxyalkenal species with carnosine was exploited in the developing method. This derivatization procedure leads to stabilizing 4-hydroxyalkenal species for prevention of their losses during analysis, enhancing their ionization by ESI-MS, and increasing the specificity with a unique set of fragment ions. We believe that with the development of this novel methodology, the role of 4-hydroxyalkenal species in biological processes could be further investigated.
Supplementary Material
Acknowledgments
This work was supported by National Institute on Aging Grants R01 AG31675 and Intramural institutional research funds.
References
- 1.Poli G, Schaur RJ. IUBMB Life. 2000;50:315–321. doi: 10.1080/713803726. [DOI] [PubMed] [Google Scholar]
- 2.Uchida K. Free Radic Biol Med. 2000;28:1685–1696. doi: 10.1016/s0891-5849(00)00226-4. [DOI] [PubMed] [Google Scholar]
- 3.Riahi Y, Cohen G, Shamni O, Sasson S. Am J Physiol Endocrinol Metab. 2010;299:E879–886. doi: 10.1152/ajpendo.00508.2010. [DOI] [PubMed] [Google Scholar]
- 4.Yun MR, Park HM, Seo KW, Lee SJ, Im DS, Kim CD. Free Radic Res. 2010;44:742–750. doi: 10.3109/10715761003758122. [DOI] [PubMed] [Google Scholar]
- 5.Esterbauer H, Schaur RJ, Zollner H. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 6.Parola M, Bellomo G, Robino G, Barrera G, Dianzani MU. Antioxid Redox Signal. 1999;1:255–284. doi: 10.1089/ars.1999.1.3-255. [DOI] [PubMed] [Google Scholar]
- 7.Echtay KS. Free Radic Biol Med. 2007;43:1351–1371. doi: 10.1016/j.freeradbiomed.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Uchida K, Shiraishi M, Naito Y, Torii Y, Nakamura Y, Osawa T. J Biol Chem. 1999;274:2234–2242. doi: 10.1074/jbc.274.4.2234. [DOI] [PubMed] [Google Scholar]
- 9.Stadtman ER. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 10.Luo XP, Yazdanpanah M, Bhooi N, Lehotay DC. Anal Biochem. 1995;228:294–298. doi: 10.1006/abio.1995.1353. [DOI] [PubMed] [Google Scholar]
- 11.Bruenner BA, Jones AD, German JB. Anal Biochem. 1996;241:212–219. doi: 10.1006/abio.1996.0402. [DOI] [PubMed] [Google Scholar]
- 12.Kawai Y, Takeda S, Terao J. Chem Res Toxicol. 2007;20:99–107. doi: 10.1021/tx060199e. [DOI] [PubMed] [Google Scholar]
- 13.Goldring C, Casini AF, Maellaro E, Del Bello B, Comporti M. Lipids. 1993;28:141–145. doi: 10.1007/BF02535778. [DOI] [PubMed] [Google Scholar]
- 14.Liu YM, Jinno H, Kurihara M, Miyata N, Toyo’oka T. Biomed Chromatogr. 1999;13:75–80. doi: 10.1002/(SICI)1099-0801(199902)13:1<75::AID-BMC817>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Uchida T, Gotoh N, Wada S. Lipids. 2002;37:621–626. doi: 10.1007/s11745-002-0941-z. [DOI] [PubMed] [Google Scholar]
- 16.Selley ML, Bartlett MR, McGuiness JA, Hapel AJ, Ardlie NG. J Chromatogr B Biomed Sci Appl. 1989;488:329–340. doi: 10.1016/s0378-4347(00)82957-6. [DOI] [PubMed] [Google Scholar]
- 17.Gioacchini AM, Calonghi N, Boga C, Cappadone C, Masotti L, Roda A, Traldi P. Rapid Commun Mass Spectrom. 1999;13:1573–1579. doi: 10.1002/(SICI)1097-0231(19990815)13:15<1573::AID-RCM675>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien-Coker IC, Perkins G, Mallet AI. Rapid Commun Mass Spectrom. 2001;15:920–928. doi: 10.1002/rcm.324. [DOI] [PubMed] [Google Scholar]
- 19.Andreoli R, Manini P, Corradi M, Mutti A, Niessen WM. Rapid Commun Mass Spectrom. 2003;17:637–645. doi: 10.1002/rcm.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams TI, Lovell MA, Lynn BC. Anal Chem. 2005;77:3383–3389. doi: 10.1021/ac048265+. [DOI] [PubMed] [Google Scholar]
- 21.Honzatko A, Brichac J, Picklo MJ. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;857:115–122. doi: 10.1016/j.jchromb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Warnke MM, Wanigasekara E, Singhal SS, Singhal J, Awasthi S, Armstrong DW. Anal Bioanal Chem. 2008;392:1325–1333. doi: 10.1007/s00216-008-2383-3. [DOI] [PubMed] [Google Scholar]
- 23.Aldini G, Carini M, Beretta G, Bradamante S, Facino RM. Biochem Biophys Res Commun. 2002;298:699–706. doi: 10.1016/s0006-291x(02)02545-7. [DOI] [PubMed] [Google Scholar]
- 24.Christie WW, Han X. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. 4. The Oily Press; Bridgwater, England: 2010. [Google Scholar]
- 25.Han X, Yang K, Gross RW. Rapid Commun Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han X, Gross RW. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 27.Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, Ojika M, Yodoi J, Uchida K. J Biol Chem. 2009;284:28810–28822. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangalli L, Chiesa LM, Passero E, Manzocchi A, Maffeo G, Biondi PA. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;796:201–207. doi: 10.1016/j.jchromb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Yang K, Cheng H, Gross RW, Han X. Anal Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han X, Yang K, Gross RW. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.