Abstract
Purpose
There is a large interindividual variability in dexmedetomidine dose requirements for sedation of patients in intensive care units (ICU). Cytochrome P450 2A6 (CYP2A6) mediates an important route of dexmedetomidine metabolism, and genetic variation in CYP2A6 affects the clearance of other substrate drugs. We examined whether CYP2A6 genotypes affect dexmedetomidine disposition.
Methods
In 43 critically ill ICU patients receiving dexmedetomidine infusions adjusted to achieve the desired level of sedation, we determined a median of 5 plasma dexmedetomidine concentrations each. Forty subjects were genotyped for five common CYP2A6 alleles and grouped into normal (n=33), intermediate (n=5), and slow metabolizers (n=2).
Results
Using a Bayesian hierarchical nonlinear mixture model, estimated dexmedetomidine clearance was 49.1 L/hr (posterior mean; 95% credible interval, 41.4 to 57.6 L/hr). There were no significant differences in dexmedetomidine clearance among normal, intermediate, and slow CYP2A6 metabolizer groups.
Conclusion
Genetic variation in CYP2A6 is not an important determinant of dexmedetomidine clearance in ICU patients.
Keywords: CYP2A6, Dexmedetomidine, Pharmacogenetics, Bayesian Modeling
Introduction
Dexmedetomidine (Precedex®, Abbott Laboratories, Abbott Park, IL, USA), a centrally acting selective α2-adrenoceptor agonist, is used for sedation in intensive care units and as an adjunct to anaesthesia during operative procedures [1]. Like other sedatives used in current practice, there is significant interindividual variation in plasma dexmedetomidine clearance [2, 3] and such interindividual variability in the pharmacokinetics of drugs may be due to genetic or environmental factors. Variants of genes encoding drug metabolizing enzymes often contribute to the observed interindividual variability in drug disposition and responses [4–6].
There is limited information about the metabolism of dexmedetomidine. One of its important pathways of metabolism is aliphatic hydroxylation, mediated primarily by the enzyme cytochrome P450 2A6 (CYP2A6), to generate 3-hydroxy dexmedetomidine and other metabolites [7]. There is significant genetic variation in CYP2A6 which contributes to interindividual differences in the pharmacokinetics of substrate drugs such as nicotine [8–12]; total and CYP2A6-mediated nicotine clearance is reduced by 40% and 50%, respectively, in subjects carrying one or two loss-of-function alleles or two decreased-function alleles [11]. The effect of genetic variants in CYP2A6 on dexmedetomidine disposition has not been studied, and there are currently no recommendations for dose adjustment in carriers of loss-of-function alleles [7]. The prolonged infusion of dexmedetomidine is an increasingly popular sedative strategy in critically ill patients [13, 14], a patient group with high inter- and intraindividual variability in drug disposition. We therefore examined whether CYP2A6 genotype is a clinically meaningful contributor to the interindividual variability in dexmedetomidine clearance in critically ill patients receiving therapeutic dexmedetomidine infusions in the ICU.
Materials and Methods
Subjects
We studied 43 patients in medical and surgical ICUs who received dexmedetomidine as part of a study examining the effects of different sedatives on acute brain dysfunction in mechanically ventilated patients [13]. The study was approved by the Institutional Review Boards of Vanderbilt University, Nashville, TN, and University of Toronto, Toronto, Ontario. Subjects or surrogates of critically ill patients gave written informed consent.
Study procedures
All medical and surgical ICU patients at the Vanderbilt Medical Center requiring mechanical ventilation for more than 24 hours were eligible to participate in a study which compared the effects of sedation with dexmedetomidine or lorazepam on acute brain dysfunction [13]. Patients were excluded if any of the following conditions were present: neurological disease (for example, previous stroke or cerebral palsy), active seizures, Childs-Pugh class B or C liver disease, moribund state with planned withdrawal of life support, family or physician refusal, alcohol abuse, active myocardial ischemia, 2nd/3rd degree heart block, severe dementia, benzodiazepine dependency, pregnancy or lactation, severe hearing disabilities, and inability to understand English. Patients in the dexmedetomidine arm of this study were started on a dose rate of 0.15 μg kg−1 hr−1, and ICU nurses were allowed to titrate the infusion to a maximum of 1.5 μg kg−1 hr−1 to achieve the sedation goal set by the patient’s medical team using the Richmond Agitation-Sedation Scale (RASS) [15]. Dexmedetomidine was infused as needed until extubation or for the maximum duration approved by the Food and Drug Administration for this specific study (120 hours). Concomitant medications were recorded from the patient files. Blood for dexmedetomidine concentrations was drawn twice daily, once in the morning and once in the evening, though some variability occurred for logistic reasons. Details of the study protocol are described elsewhere [13].
Genotyping
Genomic DNA samples from 42 patients (DNA from 1 patient was not available for genotyping) were genotyped for CYP2A6 alleles *2, *4, *9, *12 and *17. These variants have allele frequencies > 4.0 % in Caucasians and African-Americans and are known to affect drug metabolism [12]. Genotyping was based on previously published 2-step polymerase chain reaction amplification assays, where the first step is CYP2A6 gene-specific and the second step is allele-specific [16–19]. Forty (96 %) patients were successfully genotyped for each CYP2A6 allele; DNA from two patients did not amplify in the genotyping assays and DNA was not available from one patient. The frequency of genotypes categorized by ethnicity is shown in Tables 1 and 2.
Table 1.
Demographic characteristics of ICU patients
| Parameter | Mean ± SD/n |
|---|---|
| Age (yrs) | 57.1 ± 15.0 |
| Range | 20 to 85 |
| Sex (Males/Females) | 25/18 |
| Height (m) | 1.70 ± .12 |
| Range | 1.52 to 1.90 |
| Weight (kg) | 84.0 ± 21.9 |
| Range | 53 to 159 |
| CYP2A6 genotype | NM/IM/SM |
| Total (n=40)* | 33/5/2 |
| Caucasians (n=38) | 32/5/1 |
| African-Americans (n=2) | 1/0/1 |
| Smokers/Non-smokers | 12/31 |
| Dexmedetomidine clearance (L/hr) ** | 48.5 ± 4.4 (40.3 to 57.6) |
| Dexmedetomidine volume of distribution (L)** | 76.6 ± 31.0 (24.4 to 141.4) |
DNA from one subject was not available, and genotyping failed in two subjects.
Data represent posterior mean ± standard deviation and the 95 % credible interval in parenthesis. The values were estimated from 43 subjects.
NM = Normal metabolizer; IM = Intermediate metabolizer; SM = Slow metabolizer.
Table 2.
CYP2A6 genotypes
| Genotype | Functional genotype group | ICU Patients* (n=40) | Total (%) | |
|---|---|---|---|---|
| Whites (n=38) | Blacks (n=2) | |||
| *1/*1 | NM | 32 | 1 | 33 (82.5%) |
| *1/*9 | IM | 5 | 0 | 5 (12.5%) |
| *1/*4 | SM | 1 | 0 | 1 (2.5%) |
| *9/*9 | SM | 0 | 1 | 1 (2.5%) |
DNA from one subject was not available, and genotyping failed in two subjects
NM = Normal metabolizer; IM = Intermediate metabolizer; SM = Slow metabolizer.
We grouped CYP2A6 genotypes into normal, intermediate, and slow metabolizers according to previous studies that were based on the CYP2A6-mediated metabolism of nicotine [11, 20]. Those without any detected variant alleles, the CYP2A6 *1/*1 genotype, were defined as normal metabolizers and assumed to have 100 % enzyme activity. Those with one allele associated with decreased functional activity, represented by CYP2A6*1/*9 and CYP2A6*1/*12 genotypes, were defined as intermediate metabolizers (≈80 % enzyme activity), while slow metabolizers (<50 % enzyme activity) included any subject with one or two loss-of-activity alleles (*2 and *4) [21, 22], or two alleles with decreased function (*9 and *17) [23, 24], e.g. CYP2A6 *1/*4, *1/*2, *1/*17, *2/*2, *9/*9, *9/*17 and *17/*17 [11, 20].
Plasma dexmedetomidine determination
Plasma dexmedetomidine concentrations were measured by reversed-phase high-performance liquid chromatography with tandem mass spectrometric detection (LC-MS/MS; SCIEX API 365 instrument, Foster City, CA, USA). The method, a modification from a published procedure [25], is described in detail elsewhere [26]. The lower limit of quantitation of the assay was 0.02 ng/mL. The within- and between-run precision of the assay (coefficient of variation) was within 8 % in the relevant concentration range.
Statistical analysis
Initial population PK modeling with conventional residual error models implemented in NONMEM [27] revealed a very poor fit due to outliers. Thus, to accommodate outliers using a more flexible residual error model, we fitted the dexmedetomidine PK data using a Bayesian hierarchical mixture PK model, details of which can be found in Choi et al [28]. The benefit of this model is that outliers do not need to be arbitrarily excluded, while their inclusion is not expected to yield significant bias. As the structural part of model, a one-compartmental multiple infusions model [29] was chosen since a two-compartmental model did not improve the fit. Clearance and volume of distribution were allowed to be correlated and assumed to follow a multivariate log-normal distribution. All priors were assumed to be flat. We fitted the model using Markov chain Monte Carlo methods implemented with PKBugs 1.1 [30] and WinBUGS 1.4.3 [31].
The parameters of interest were the differences in logarithm of dexmedetomidine clearance between intermediate and normal metabolizers, and between slow and normal metabolizers. Demographic covariates such as age, sex, weight, and smoking status were considered as potential confounding factors and included in the model. Several diagnostics for goodness-of-fit showed that our model fit the data well. Data are presented as posterior means, standard deviations, medians, and 95% credible intervals, which are analogues of the means, medians and the 95% confidence intervals in the traditional (frequentist) statistical approach.
Results
Subject Characteristics and genotypes
Tables 1 and 2 show demographic data and genotype data for the study cohort (n=43), categorized according to ethnicity. The mean (±SD) APACHE II and SOFA scores for these 43 subjects were 28 ± 8 and 10 ± 3, respectively. None of the patients received potent inducers (e.g., dexamethasone, rifampin, barbiturates) or potent (tranylcypromine, miconazole) or moderately strong inhibitors (e.g., ketoconazole, isoniazid, desipramine)32 concomitantly with dexmedetomidine.
CYP2A6 genetic variants and dexmedetomidine clearance
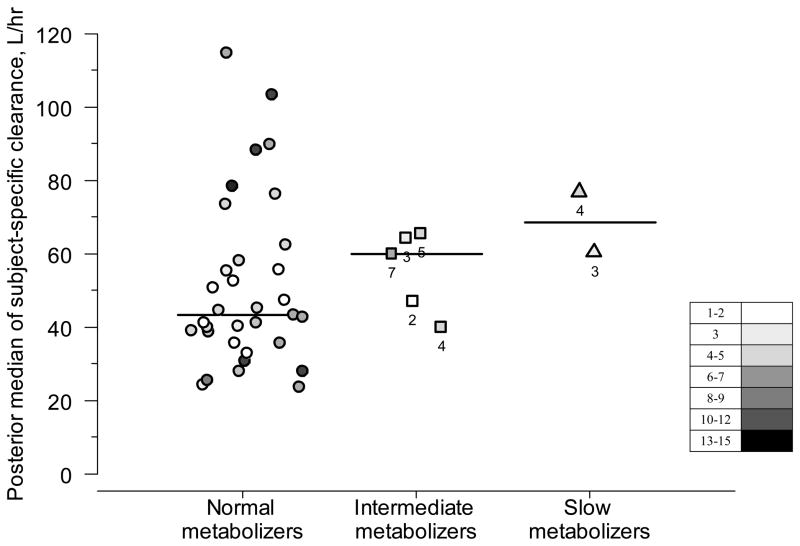
The estimated population mean for dexmedetomidine clearance was 49.1 L/hr (posterior mean; 95% credible interval, 41.4 to 57.6 L/hr). Figure 1 shows the posterior medians for subject-specific clearance of dexmedetomidine according to genotype. The posterior medians for population clearance of dexmedetomidine did not differ significantly among normal metabolizers (48.5 L/hr; 95 % credible interval 39.8 to 58.7 L/hr; n=33), intermediate metabolizers (56.9 L/hr; 31.6 to 94.4 L/hr; n=5) and slow metabolizers (86.2 L/hr; 26.9 to 218.7 L/hr; n=2). To better demonstrate the clinical significance of the differences among metabolizer groups, we calculated the credibility intervals (converted from the log-transformed back into the original scale) for the ratios of the clearance between intermediate and normal metabolizers (95% CI, 0.66 to 2.26) and between slow and normal metabolizers (0.44 to 3.92). Furthermore, using these posterior probabilities, we calculated that the probabilities of there being a ≥30% reduction of clearance in intermediate metabolizers and of ≥50% reduction in slow metabolizers (compared to normal metabolizers) were 3.9% and 8.6%, respectively.
Figure 1. Posterior medians of subject-specific clearance of dexmedetomidine according to genotype in intensive care patients.
Each dot represents one patient. The darker the dots, the larger the number of samples per patient: the points with white color represent 1–2 samples/patient and black ones 15 samples/patient (see inbox legend). For intermediate and slow metabolizers, the number of samples is also presented. For normal metabolizers, the median number of samples was 5 (range, 1 to 15) per patient. Horizontal bars represent medians for subject-specific clearances in each metabolizer group.
Other factors affecting dexmedetomidine concentrations
Clearance of dexmedetomidine was not associated with age, sex, or smoking, but there was evidence for a positive association of borderline statistical significance between clearance and body weight (Table 3).
Table 3. Determinants of dexmedetomidine clearance in ICU patients.
The table presents posterior means and 95 % credible intervals for the parameters from the Bayesian hierarchical nonlinear mixture model
| Parameter | Posterior mean | 95 % credible interval |
|---|---|---|
| Smoking | 0.147 | −0.246 to 0.545 |
| Weight | 0.008 | −0.0003 to 0.016 |
| Age | −0.002 | −0.017 to 0.012 |
| Genotype (SM vs NM) | 0.321 | −0.690 to 1.348 |
| Genotype (IM vs NM) | 0.139 | −0.473 to 0.779 |
The following equation takes into account the posterior means and was used to determine the subject-specific clearance.
Cli = 0.147×Smoking + 0.008×Weight – 0.002×Age + 0.321×SM + 0.139×IM, where Cli is subject specific clearance, Smoking is an indicator for smoking status (Smoking = 1 if a subject is smoker, and Smoking = 0 for nonsmoker), Weight is body weight in kg, Age is age in years, SM is an indicator for slow metabolizer (SM=1 if a subject is slow metabolizer, and SM = 0 otherwise) and IM is an indicator for intermediate metabolizer (IM = 1 if a subject is intermediate metabolizer, and IM = 0 otherwise).
Discussion
Little is known about the variability in dexmedetomidine disposition in critically ill patients. CYP2A6 contributes to dexmedetomidine metabolism via aliphatic hydroxylation, and functional genetic variants in CYP2A6 could therefore affect dexmedetomidine clearance. The major new finding of this study is that in the complex setting of the ICU, CYP2A6 genotype is not an important determinant of the highly variable plasma dexmedetomidine disposition.
Dexmedetomidine is the D-enantiomer of medetomidine. Following intravenous administration, dexmedetomidine has a rapid distribution phase and undergoes extensive biotransformation involving direct glucuronidation and CYP2A6-mediated aliphatic hydroxylation in the liver. Its reported clearance varies from 39.0 L/hr in healthy subjects [7] to 48.3 L/hr in ICU patients [33], with a mean terminal elimination half-life of 2.0 hours in healthy subjects [7] and 3.1 hours in ICU patients [33]. In our study in ICU patients, the estimated posterior means of dexmedetomidine clearance was 49.1 L/hr (95 % credible interval, 41.4 to 57.6 L/hr), within the expected range.
CYP2A6 is a highly polymorphic gene, and duplications, deletions, and variants resulting in decreased enzymatic activity have been characterized [10, 11, 19]. CYP2A6 variants can have functional effects on CYP2A6 metabolic activity and are known to affect the disposition of another important CYP2A6 substrate, nicotine [10–12], explaining some of the interindividual and interethnic variability in nicotine clearance. In contrast, the role of CYP2A6 variants in dexmedetomidine disposition has not been investigated. We therefore examined whether common functional CYP2A6 variants [12] affect dexmedetomidine clearance in ICU patients and found no significant association between CYP2A6 intermediate and slow metabolizer groups and dexmedetomidine clearance in these patients. Several reasons for this should be considered.
Our cohort provided the opportunity to study patients receiving therapeutic doses of dexmedetomidine, but this occurred in a complex clinical setting. Thus, it is possible that any effect of CYP2A6 genotype was obscured by the many clinical variables, including concomitant medications, which could affect dexmedetomidine disposition in these patients. Therefore, our study does not exclude the possibility that a small effect of CYP2A6 genetic variation might be detected in a more homogenous subject population investigated under controlled experimental circumstances. Nevertheless, our results suggest that in the complex clinical setting of the ICU, CYP2A6 genotype has no clinically important effect on dexmedetomidine disposition and does not need to be accounted for.
The individual clearance estimates illustrated in Fig. 1 indicated a trend in the opposite direction from that originally hypothesized, but these differences were not statistically significant. Our ability to examine differences in dexmedetomidine clearance among CYP2A6 metabolizer groups was limited by the small sample size, with only five and two subjects having intermediate and slow metabolizer genotypes, respectively. However, based on our data we could confidently rule out a ≥30% reduction of clearance in intermediate metabolizers and of ≥50% reduction in slow metabolizers (compared to normal metabolizers), an effect that is comparable in magnitude to that of the CYP2C9*3 variant on warfarin maintenance dose. We cannot confidently rule out smaller effects of genotypes on dexmedetomidine clearance; however, considering the wide intra- and inter-patient variability in factors potentially affecting the pharmacokinetics and pharmacodynamics of dexmedetomidine in the ICU population, such effects – if they exist - would likely be of only limited clinical significance.
Our data was collected in a busy real-life ICU setting and included extreme outliers that were likely artifactual. Conventional population PK analysis yielded biased estimates. Thus, to minimize this bias, we used a new statistical model which robustly accommodated the artifactual outliers. Nevertheless, bias in our estimates of PK parameters cannot be ruled out given the limitations of data inherent in the study design.
There are no precise estimates of the contribution of the CYP2A6 metabolic pathway to the total clearance of dexmedetomidine in vivo, although in vitro studies suggest that it may be smaller than that of N-glucuronidation [7]. Nevertheless, since in vitro putative metabolic pathways may not relate well to in vivo drug disposition, in vivo studies in patients most likely to receive the drug provide valuable information. In our study, we did not determine dexmedetomidine metabolite concentrations, and therefore our study does not provide information on the relative contribution of aliphatic hydroxylation and glucuronidation pathways to dexmedetomidine metabolism.
In conclusion, in the complex clinical setting of the ICU, common genetic variants in CYP2A6, the enzyme mediating one major route of the metabolism of dexmedetomidine, did not have a clinically important contribution to the wide interindividual variability of dexmedetomidine clearance. Thus, in critically ill patients, CYP2A6 genotyping appears to be of little value for the individualization of the dexmedetomidine dosing regimen in a particular patient.
Acknowledgments
We would like to thank Dr. Päivi Poijärvi-Virta, Ph.D. (Researcher, Clinical Research Services Turku, University of Turku, Turku, Finland) for the technical performance of the dexmedetomidine assay, and Ewa Hoffmann, M.Sc. (Research Associate, Centre for Addiction and Mental Health and University of Toronto, Toronto, Ontario, Canada) and Qian Zhou, Ph.D. (Research Associate, Centre for Addiction and Mental Health and University of Toronto, Toronto, Ontario, Canada) for their assistance with the genotyping.
Funding
This work was supported by Vanderbilt Clinical and Translational Science Award from the National Center for Research Resources [1 UL 1 RR024975]; National Institutes of Health grants [P01 HL56693, R21 AG034412, GM31304, and DA 020830]; and Center for Addiction and Mental Health and the Canadian Institutes for Health Research grant [MOP86471]. Dr. Pandharipande is supported via the Veterans Affairs Career Development Award, the American Society of Critical Care Anaesthesiologists-Foundation for Anaesthesia Education and Research Mentored Research Grant and the Vanderbilt Physician Scientists Development Award. Dr. Ely is supported by a Veterans Affairs Merit Award and National Institutes of Health grant [R01 AG027472-01A1]. Dr. Tyndale is a recipient of a Canada Research Chair in Pharmacogenetics. Dr. Choi is supported by National Institutes of Health grant [R21 AG034412]. Dr. Stein is the recipient of the Dan May Chair in Medicine.
Footnotes
Competing interests
Dr. Tyndale is a shareholder and chief scientific officer of Nicogen Inc., a company focused on the development of novel smoking cessation therapies; no funds were received from Nicogen for these studies, nor was this manuscript reviewed by other people associated with Nicogen. Dr. Tyndale is a paid advisor to pharmaceutical companies that market or are developing smoking cessation medications.
The laboratory of Dr. Scheinin has contract research relationships with Orion Corporation (Espoo, Finland) and Hospira (Lake Forest, IL, USA). Hospira has a license agreement with Orion Corporation concerning dexmedetomidine (Precedex®). Dr. Scheinin has received speaker fees and consulting fees from Orion Corporation, Dr. Pandharipande has received research funding and honoraria from Hospira Inc., and Dr. Ely has received honoraria and grants from Hospira, Lilly, Pfizer, Glaxo Smith Kline, and Aspect Medical. None of the other authors has a conflict of interest relevant to the work presented.
References
- 1.Gerlach AT, Dasta JF. Dexmedetomidine: an updated review. Ann Pharmacother. 2007;41(2):245–252. doi: 10.1345/aph.1H314. [DOI] [PubMed] [Google Scholar]
- 2.Talke P, Richardson CA, Scheinin M, Fisher DM. Postoperative pharmacokinetics and sympatholytic effects of dexmedetomidine. Anesth Analg. 1997;85(5):1136–1142. doi: 10.1097/00000539-199711000-00033. [DOI] [PubMed] [Google Scholar]
- 3.Vilo S, Rautiainen P, Kaisti K, Aantaa R, Scheinin M, Manner T, Olkkola KT. Pharmacokinetics of intravenous dexmedetomidine in children under 11 yr of age. Br J Anaesth. 2008;100(5):697–700. doi: 10.1093/bja/aen070. [DOI] [PubMed] [Google Scholar]
- 4.Klotz U. The role of pharmacogenetics in the metabolism of antiepileptic drugs: pharmacokinetic and therapeutic implications. Clin Pharmacokinet. 2007;46(4):271–279. doi: 10.2165/00003088-200746040-00001. [DOI] [PubMed] [Google Scholar]
- 5.Hawwa AF, Collier PS, Millership JS, McCarthy A, Dempsey S, Cairns C, McElnay JC. Population pharmacokinetic and pharmacogenetic analysis of 6-mercaptopurine in paediatric patients with acute lymphoblastic leukaemia. Br J Clin Pharmacol. 2008;66(6):826–837. doi: 10.1111/j.1365-2125.2008.03281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, Kim RB, Roden DM, Stein CM. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358(10):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Precedex® product label. Abbott Park, IL, USA: Abbott Laboratories; 2008. [Google Scholar]
- 8.Perez-Stable EJ, Herrera B, Jacob P, III, Benowitz NL. Nicotine metabolism and intake in black and white smokers. JAMA. 1998;280(2):152–156. doi: 10.1001/jama.280.2.152. [DOI] [PubMed] [Google Scholar]
- 9.Caraballo RS, Giovino GA, Pechacek TF, Mowery PD, Richter PA, Strauss WJ, Sharp DJ, Eriksen MP, Pirkle JL, Maurer KR. Racial and ethnic differences in serum cotinine levels of cigarette smokers: Third National Health and Nutrition Examination Survey, 1988–1991. JAMA. 1998;280(2):135–139. doi: 10.1001/jama.280.2.135. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, Kwon JT, McLeod HL, Yokoi T. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80(3):282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Benowitz NL, Swan GE, Jacob P, III, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80(5):457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther. 2005;77(3):145–158. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Pandharipande PP, Pun BT, Herr DL, Maze M, Girard TD, Miller RR, Shintani AK, Thompson JL, Jackson JC, Deppen SA, Stiles RA, Dittus RS, Bernard GR, Ely EW. Effect of sedation with dexmedetomidine vs lorazepam on acute brain dysfunction in mechanically ventilated patients: the MENDS randomized controlled trial. JAMA. 2007;298(22):2644–2653. doi: 10.1001/jama.298.22.2644. [DOI] [PubMed] [Google Scholar]
- 14.Riker RR, Shehabi Y, Bokesch PM, Ceraso D, Wisemandle W, Koura F, Whitten P, Margolis BD, Byrne DW, Ely EW, Rocha MG. Dexmedetomidine vs midazolam for sedation of critically ill patients: a randomized trial. JAMA. 2009;301(5):489–499. doi: 10.1001/jama.2009.56. [DOI] [PubMed] [Google Scholar]
- 15.Sessler CN, Gosnell MS, Grap MJ, Brophy GM, O’Neal PV, Keane KA, Tesoro EP, Elswick RK. The Richmond Agitation-Sedation Scale: validity and reliability in adult intensive care unit patients. Am J Respir Crit Care Med. 2002;166(10):1338–1344. doi: 10.1164/rccm.2107138. [DOI] [PubMed] [Google Scholar]
- 16.Mwenifumbo JC, Myers MG, Wall TL, Lin SK, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6*7, CYP2A6*8 and CYP2A6*10 as assessed with a novel haplotyping method. Pharmacogenet Genomics. 2005;15(3):189–192. doi: 10.1097/01213011-200503000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14(9):615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Dempsey D, Tutka P, Jacob P, III, Allen F, Schoedel K, Tyndale RF, Benowitz NL. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Mwenifumbo JC, Al KN, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, Tyndale RF. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008;29(5):679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 20.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11(4):400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 21.Yamano S, Tatsuno J, Gonzalez FJ. The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes. Biochemistry. 1990;29(5):1322–1329. doi: 10.1021/bi00457a031. [DOI] [PubMed] [Google Scholar]
- 22.Nunoya K, Yokoi T, Takahashi Y, Kimura K, Kinoshita M, Kamataki T. Homologous unequal cross-over within the human CYP2A gene cluster as a mechanism for the deletion of the entire CYP2A6 gene associated with the poor metabolizer phenotype. J Biochem. 1999;126(2):402–407. doi: 10.1093/oxfordjournals.jbchem.a022464. [DOI] [PubMed] [Google Scholar]
- 23.Kiyotani K, Yamazaki H, Fujieda M, Iwano S, Matsumura K, Satarug S, Ujjin P, Shimada T, Guengerich FP, Parkinson A, Honda G, Nakagawa K, Ishizaki T, Kamataki T. Decreased coumarin 7-hydroxylase activities and CYP2A6 expression levels in humans caused by genetic polymorphism in CYP2A6 promoter region (CYP2A6*9) Pharmacogenetics. 2003;13(11):689–695. doi: 10.1097/00008571-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Fukami T, Nakajima M, Yoshida R, Tsuchiya Y, Fujiki Y, Katoh M, McLeod HL, Yokoi T. A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clin Pharmacol Ther. 2004;76(6):519–527. doi: 10.1016/j.clpt.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Ji QC, Zhou JY, Gonzales RJ, Gage EM, El Shourbagy TA. Simultaneous quantitation of dexmedetomidine and glucuronide metabolites (G-Dex-1 and G-Dex-2) in human plasma utilizing liquid chromatography with tandem mass spectrometric detection. Rapid Commun Mass Spectrom. 2004;18(15):1753–1760. doi: 10.1002/rcm.1548. [DOI] [PubMed] [Google Scholar]
- 26.Snapir A, Posti J, Kentala E, Koskenvuo J, Sundell J, Tuunanen H, Hakala K, Scheinin H, Knuuti J, Scheinin M. Effects of low and high plasma concentrations of dexmedetomidine on myocardial perfusion and cardiac function in healthy male subjects. Anesthesiology. 2006;105(5):902–910. doi: 10.1097/00000542-200611000-00010. [DOI] [PubMed] [Google Scholar]
- 27.Beal S, Sheiner L, Boeckmann A, editors. NONMEM Users Guides. Ellicott City, MD: ICON Development Solutions; 1989. [Google Scholar]
- 28.Choi L, Caffo BS, Kohli U, Pandharipande P, Kurnik D, Ely EW, Stein CM. A Bayesian hierarchical nonlinear mixture model in the presence of artifactual outliers in a population pharmacokinetic study. J Pharmacokinet Pharmacodyn. 2011;38(5):613–636. doi: 10.1007/s10928-011-9211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibaldi M, Perrier D. Pharmacokinetics 1975 [Google Scholar]
- 30.Lunn DJ, Wakefield J, Thomas A, Best N, Spiegelhalter D. PKBugs User Guide. Dept. Epidemiology & Public Health. Imperial College School of Medicine; London: 1999. [Google Scholar]
- 31.Spiegelhalter D, Thomas A, Best N, Lunn D. WinBUGS User Manual, version 1.4. MRC Biostatistics Unit, Institute of Public Health and Department of Epidemiology and Public Health, Imperial College School of Medicine; London: 2003. [Google Scholar]
- 32.Lacy CF, Armstrong LL, Goldman MP, Lance LL, editors. Drug Information Handbook. 15. LexiComp Inc; Hudson, OH: 2007. Cytochrome P450 Enzymes: Substrates, Inhibitors, and Inducers; pp. 1899–1912. [Google Scholar]
- 33.Venn RM, Karol MD, Grounds RM. Pharmacokinetics of dexmedetomidine infusions for sedation of postoperative patients requiring intensive care. Br J Anaesth. 2002;88(5):669–675. doi: 10.1093/bja/88.5.669. [DOI] [PubMed] [Google Scholar]