Abstract
Bone homeostasis can be compromised by an increase in osteoclast-mediated resorption and/or a decrease in osteoblast-mediated bone deposition. While many efforts have focused on treating osteoclast resorption, there has been less emphasis on identifying strategies for promoting osteoblast function. Herein, we describe a high-throughput screening assay to select for small molecules that augment bone morphogenetic protein-2 (BMP-2)-mediated osteoblast lineage commitment. After an initial screen of 5405 compounds; consisting of FDA-approved drugs, known bioactives, and compounds with novel chemical makeup, we identified 45 small molecules that promoted osteoblast commitment. Of the 45 candidates, there was a broad array of classes that included nine retinoid analogs/derivatives and four immunosuppressants, notably rapamycin and FK-506, which were chosen for further study. Treatment of osteoblast precursor cells with rapamycin or FK-506, either alone, or synergistically with BMP-2, increased levels of phospho-Smad1/5/8 protein and transcription of Runx-2, Osx and Smad-7, consistent with a role in promoting osteoblast differentiation. Only FK-506 was able to enhance osteocalcin transcripts and alizarin red staining, both late markers for differentiation. When osteoblast differentiation was suppressed with exogenous TGF-β1 treatment, rapamycin (but not FK-506) was able to rescue expression of differentiation markers, indicating distinct but overlapping activity of these compounds. Collectively, these data add to an understanding of pathways engaged in osteoblastogenesis, support a role for non-redundant immunosuppressant signaling, and provide a novel approach for the discovery of potentially therapeutic compounds that affect bone remodeling.
INTRODUCTION
Loss of bone mass is an increasingly common morbidity in our aging society and is associated with an increased risk of fracture and frailty and, alarmingly, increased prevalence in chronic but treated disorders such as long-term HIV-1 infection and diabetes mellitus [1] [2]. Understanding the mechanism through which bone mass is regulated and the risk factors for bone dysregulation are critical challenges for developing new and effective therapeutics. There are substantial data supporting crosstalk between bone forming osteoblasts and bone resorbing osteoclasts, which allow for dynamic bone remodeling necessary for bone maintenance, strength and structural integrity ([3–5]. Net bone loss can occur if there is a loss in osteoblast activity or if there is an increase in osteoclast activity [6, 7]. The therapeutic focus has more recently been on blocking osteoclast activity (e.g., antibody to RANKL, Denosumab,[8]) and while these advances are promising, efforts to promote osteoblast activity may also be useful in establishing an arsenal of therapeutic options in countering bone loss.
High-throughput screens are becoming increasingly more common in their use to identify novel compounds as therapeutic. A clear advantage of this approach is that small molecule libraries have the capacity to probe cellular pathways to identify novel modulatory nodes with desirable effects. For example, a screen was recently done to identify anti-inflammatory compounds for use in cystic fibrosis and several compounds were discovered [9]. We adopted a similar strategy to identify osteoblast promoting compounds using a complex set of chemical libraries that range from FDA approved compounds to small molecules with unknown function.
Bone homeostasis is maintained by the proliferation and differentiation of osteoprogenitor cells, which will eventually guide the production of mineralized bone [10, 11]. Osteoblast differentiation can be induced by bone morphogenetic protein (BMP)-2, initiating a signal cascade that promotes osteoblast specific genes; including the transcription factor Runx2, a critical regulator, as well as the downstream transcription factor osterix (Osx) [12–15]. Mature osteoblasts then produce alkaline phosphatase (ALP) that can be measured via cell staining [16] and was used in this study to identify compounds that promote osteoblastogenesis. Late differentiation is induced by another transcription factor, osteocalcin (Ocn) and this allows the cells to eventually secrete proteins that form the mineralized extracellular matrix [12–15].
To find novel drugs that promote osteoblast differentiation, we began with the C2C12 cell line that is biased to become muscle but can be induced to differentiate along the osteoblast lineage in the presence of BMP-2 [17], with essentially no background ALP staining, allowing us to find drugs that would enhance this efficiency and therefore potentially enhance osteoblast differentiation of pre-osteoblasts. Once the libraries were screened, 45 compounds were identified as “positive hits” that enhanced ALP expression. We chose two hits; FK506 and rapamcyin, for further study because their effect on the BMP-2 pathway was unclear. Both of these immunosuppressive drugs have previously been implicated in osteoblastogenesis, however the results have been contradictory and the mechanisms of action remain unclear [18–23]. Additionally, transforming growth factor (TGF)-β1 is an anti-inflammatory cytokine that can function as both an antagonist [24–27] and agonist [28–30] on bone differentiation, depending on context. We therefore utilized TGFβ1 under antagonistic conditions to further explore rapamycin and FK506 activity under conditions that attenuate bone.
MATERIALS AND METHODS
Cell Culture
C2C12
The C2C12 myoblast cell line (ATCC) was maintained in growth medium (GM) that consisted of High Glucose DMEM (Gibco), 10% Fetal Bovine Serum (Gibco), and 1% pen/strep (Invitrogen). To induce osteoblast formation, cells were allowed to reach 50% confluency, washed with PBS (1x) and switched to differentiation medium (DM) that consisted of Low Glucose DMEM, 2% Horse Serum (Gibco), 1% pen/strep and lyophilized Bone Morphogenetic Protein-2 (BMP-2) (Genscript #Z00327), reconstituted in 20mM acetic acid, at a concentration range of 20–200ng/mL [17]. Negative controls were switched to DM but only received 20mM acetic acid. Cells were passaged approximately every two days and were kept below 70% confluency, as per manufacturer instructions.
MC3T3-E1
MC3T3-E1 Subclone 4 pre-osteoblast cells (ATCC #CRL-2593) were maintained in Minimum Essential Medium, Alpha modification (Invitrogen #A10490-01) containing 10% FBS and 1% penicillin/streptomycin. Osteoblast induction was performed by supplementing the medium with 100ng/mL BMP-2, as previously described [31]. All cell lines used were maintained in a 37°C incubator at 5% CO2.
Chemical Libraries
Chemical libraries used in this study were prepared by the Center for Molecular Discovery at Boston University Medical Center. The libraries consisted of 640 FDA approved compounds (Enzo Life Sciences), 480 ICCB known bioactive compounds (Enzo Life Sciences), 446 compounds from the NIH Clinical Collection (BioFocus DPI), ~2,000 Natural-like compounds from the Center for Chemical Methodology and Library Development at Boston University, and a diversity subset of 2,000 compounds (Chembridge).
BMP-2 and Compound Stimulation
384-square well plates (BD) were used for primary screening. PBS was added to the outermost wells to reduce edge effects. In each plate, 40 wells were used as positive controls, 28 wells were used as negative controls, and the center 240 wells were used for compound testing. All plates were screened in duplicate. The inner 308 wells received 750 cells per well and were allowed to adhere in GM for 24 hours. GM was then removed, wells were washed once with PBS, and DM was added to the inner 308 wells. Negative control wells received DM. Positive control wells received BMP-2 in DM. Test wells received BMP-2 and compounds at a target concentration of 1uM. DMSO was used as the vehicle for compound addition in this study. DMSO was also added to positive and negative control wells in equimolar amounts.
ALP and DAPI Staining
Cells were initially fixed with a fixative solution that consisted of a 3:10:26 ratio mixture of 37% formaldehyde:citrate:acetone. Colorimetric detection of osteoblasts was achieved using the Alkaline Phosphatase (ALP) kit obtained from Sigma-Aldrich (Catalog #86C). Cell number was determined based on nuclei staining with DAPI nucleic acid stain as per manufacturers instructions (Invitrogen).
Image Acquisition and Analysis
Well images identifying ALP+ cells were acquired using a MIAS-2 plate reader, as per manufacturer instructions (Digilab, Holliston, MA).
Search Strategies (ImageJ, Digilab, Visual inspection)
Acquired images were assessed using three different search strategies (ImageJ software, Digilab software, and systematic scanning by eye); each having different sensitivities and specificities with regard to this assay.
ImageJ Analysis
Well images were opened with the image software platform ImageJ. Images were normalized on a per plate basis and, since ALP positive cells become darker in a gray scale image, the image darkness was assessed. Images were analyzed using an Area Fraction method whereby a minimum pixel darkness threshold was applied for all image wells. All pixels that were as dark or darker than the threshold applied were converted to black. All pixels that did not meet the threshold were converted to white. The binary image was then assessed to see percentage of pixels of the image that were black (i.e. Area Fraction). Average Area Fraction from duplicate compound wells was divided by average duplicate nuclei count (as measured by DAPI count in ImageJ) to get Area Fraction on a per-cell basis. This ratio (Area Fraction: Nuclei count) was then compared to positive control wells. Ratios for the positive control wells, per plate, were determined and any compound wells that were greater than three standard deviations above the average ratio of positive control wells were considered potential augmenters of bone formation and were compared with the two other search strategies.
Digilab Analysis
Well images were analyzed using eaZYX Image Analyzer software (Digilab, Holliston, MA). Cell number was determined by counting the number of DAPI stained nuclei per image. Fluorescent nuclei images were overlaid with the bright-field images to determine the number of ALP positive cells per well.
Visual inspection Analysis
The image of a representative positive control was kept open for comparison while visually scanning the images of wells that had compounds added. Wells were rated categorically on a four-point scale: (1) clear augmenter of ALP expression, (2) potential augmenter that appeared to have ALP expression above the representative positive control, (3) changes in the morphology of the cells, and (4) did not change or suppressed ALP expression. Duplicate wells that were independently confirmed by eye for increased ALP expression were compared with the other two search strategies.
Quantitative RT-PCR
Total RNA was extracted by the TRIzol method as recommended by the manufacturer (Invitrogen). Isolated RNA was cleaned up using the Rneasy Kit (Qiagen) and 500ng of RNA was reverse-transcribed using the High Capacity cDNA Synthesis Kit (Applied Biosystems #4368813). Taqman expression assays were used for detection. The expression of 18S (Applied Biosystems #4319413E) was used for normalization of gene expression values. Real-time primers Runx2, Sp7 (Osx), Bglap1 (Ocn) and Smad7 were obtained from Applied Biosystems (Mm00501580_m1, Mm00504574_m1, Mm03413826_m1 and Mm00484742_m1 respectively). Quantification was determined using the ΔΔCT method and normalized to the untreated sample.
Western Blot
MC3T3 cells were lysed, on ice, for 20 minutes using lysis buffer containing 10mM Tris, pH 7.6, 150mM NaCl, 2mM EDTA, 1% Triton, 0.1% SDS, 0,1g deoxycholic acid, 1X protease inhibitor cocktail (Roche), 500mM sodium fluoride, 100mM sodium pyrophosphate, and 400mM β-glycerophosphate and centrifuged at 16,000rpM for 10min at 4°C. Protein concentrations were determined using the BCA protein assay kit (Pierce). Equal amounts of protein (20ug) were resolved by SDS polyacrylamide gel electrophoresis. Gels were transferred to Trans-Blot Transfer Medium Pure Nitrocellulose Membrane (Bio-Rad #162-0115) and probed with either Phospho-Smad 1/5/8 (Cell Signaling 9511S) or Smad-1/5/8 (Santa Cruz SC-6031R), as primary antibodies overnight at 4°C in 5% milk. After washing, membranes were probed with the corresponding secondary anti-rabbit HRP-conjugated (Cell Signaling 7074S) for 1 hour at room temperature. Bands were visualized by chemiluminescence using Amersham ECL Western Blotting Detection Reagents (GE Healthcare RPN2106). To observe correlating amounts of total protein and phosphoprotein, the same blot was stripped for 30 minutes at 65°C in 62.5mM Tris pH 6.8, 2%SDS and 0.6% β-mercaptoethanol. Stripped blots were washed for 30 minutes and then re-blocked before the primary antibody. Bands were analyzed for density with ImageJ and normalized to loading control Smad 1/5/8. Values represent fold change compared to untreated samples.
Alizarin Red Staining
Cells were fixed with 2.5% glutaraldehyde after 21 days of stimulation and washed with PBS adjusted to a pH of 4.2. They were stained with 2% Alizarin Red S (Sigma-Aldrich A5533-25G) for 20 minutes at 37°C. After being washed with PBS four images were captured for each well.
RESULTS
High-throughput screen identifies osteoblast-inducing compounds
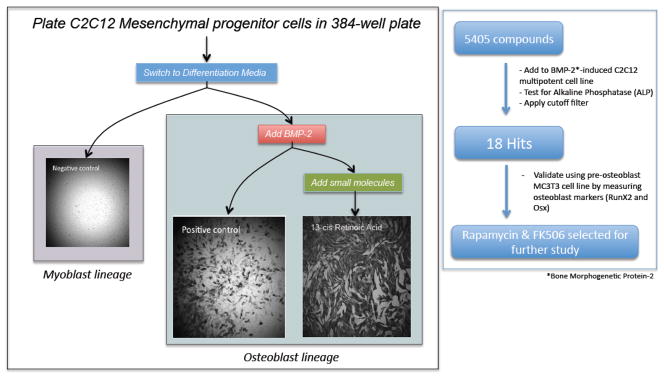
In the course of our studies on muscle differentiation using the C2C12 cell line, we observed incomplete BMP-2-mediated conversion of C2C12 cells to osteoblasts based on ALP staining with virtually no background in untreated cells, which prompted us to use this system to screen a library of compounds that augment BMP-2 mediated osteoblast conversion. C2C12 cells were plated at 750 cells per well in a 384-square well plate and were allowed to adhere for 24 hours in Growth Media (GM). The GM was replaced with either low serum Differentiation Media (DM) (Negative control), DM + BMP-2 (Positive control), or DM + BMP-2 + small molecules (Figure 1a). After 72 hours, cells were fixed and stained for Alkaline Phosphatase (ALP) and DAPI for nuclei visualization. We found that the negative control wells expressed close to zero ALP, reinforcing the specificity of this assay. Positive control wells consistently had ALP expression in 20–30% of the cells. Initially, we performed a pilot study of 240 FDA approved compounds on our system; a number of the small molecules added in combination with BMP-2 strikingly enhanced conversion to osteoblast (ALP+) cells. Notably, there were several phenotypes observed including: no obvious effect on conversion, attenuated conversion to osteoblasts, morphological changes and enhancement or suppression of proliferation of cells. Encouraged by these initial findings, we were interested in screening a larger set of small molecules from more diverse backgrounds to identify enhancement of osteoblast conversion.
Figure 1. Protocol for high-throughput screening of small molecules to investigate BMP-2 induced promotion of osteoblast formation.
A: Overview of C2C12 cells switched to DM media (low serum) Media and that received either no BMP-2 (Negative Control), BMP-2 only (Positive Control), or BMP-2 + Compound*. The level of alkaline phosphatase (ALP) activity measured colorimetrically was scored in all wells using three criteria (see methods). Compounds that enhanced ALP expression were considered for further analysis. B: Schematic of 5405 compounds tested on C2C12 cells in a 384-well plate format, in duplicate for BMP-2 induced ALP expression*. ALP intensity images were acquired using a Digilab Plate reader. Images were analyzed using three independent criteria and considered for secondary screening and validation in the MC3T3 pre-osteoblast cell line. Enhancement of the mRNA and protein levels of the mature osteoblast markers RunX2 and Osterix was tested. DM = differentiation media. *Plates were run in duplicate.
Results from screen of 5405 compounds
We were most interested in those compounds that enhanced osteoblast formation beyond BMP-2 alone due to their therapeutic potential to promote bone formation. To ensure a robust detection of augmenters (i.e., those that promoted osteoblast differentiation), we applied three search strategies to measure the effects of added compounds’ on ALP expression. After screening 5405 compounds in duplicate from five different chemical libraries, we then applied a cutoff filter that revealed 45 compounds with enhanced ALP expression. From these 45 hits (18 strongest hits and 27 potential hits, Table 1 and Supplementary Figure 1) we decided to further evaluate two unexpected compounds, rapamycin and FK-506 (FK-520 was also identified but was not among the top hits), for their effect on the pre-osteoblast mouse cell line MC3T3 (Figure 1b). We chose rapamycin and FK-506 to validate our screen because they are not widely recognized as osteoblast potentiators, and because, based on prior studies, their role in osteoblasts is mixed. Collectively, we found that screening a large number of compounds allowed us to discover several expected and unexpected compound hits among the libraries tested.
Table 1. Small compound libraries.
5 different chemical libraries were used and the number of compounds in each library is indicated in the table. The number of hits per library is broken down into “strongest hits” and “potential hits.” Strongest hits were characterized as compounds that were positive in all three analyses, whereas potential hits were characterized as compounds that were positive in two of the three approaches. The BU-CMLD is composed of stereochemically and structurally complex chemical libraries (sprioketal, epoxyquinol, oxime, macrodiolide, diketopiperazine, cyclic ether). This library uniquely probes three-dimensional space by employing stereochemical and positional variation within the molecular framework as diversity elements for library design. The NIH and FDA approved drug library, and ICCB are comprised of small molecules that are all known bioactives. These collections were assembled to affect a wide variety of biological pathways. The Chembridge represent drug-like small molecules, rationally selected based on 3D pharmacophore analysis to cover the broadest part of biologically relevant pharmacophore diversity space.
| Library Name | Number of Compounds | Strongest Hits | Potential Hits |
|---|---|---|---|
| NIH | 446 | 1 | 6 |
| ICCB | 480 | 13 | 13 |
| FDA | 640 | 4 | 3 |
| BUCMLD | 1839 | 0 | 1 |
| ChemBridge | 2000 | 0 | 4 |
| Total: | 5405 | 18 | 27 |
Comparison and characteristics of results from three different search strategies (ImageJ, Digilab, visual inspection)
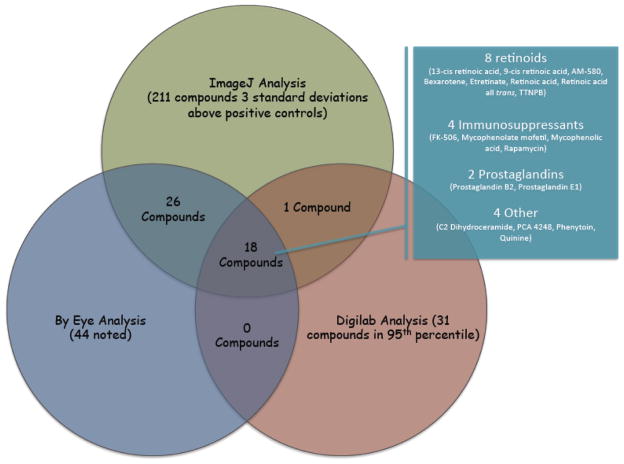
To ensure true positivity, images of wells from the initial screen of chemical libraries were processed using three different search strategies based on method of analysis (ImageJ, Digilab and visual inspection) to ensure reproducibility and reduce false positives. Resultant images were analyzed using ImageJ software, Digilab eaZYX Image Analyzer software, and systematically scanned by eye. ImageJ: Analysis using ImageJ software (see methods section) indicated 211 compounds with ALP expression > three standard deviations above the average (99%) of positive controls. Digilab: Analysis using the eaZYX Image Analysis software from Digilab indicated 31 compounds that had ALP expression in the 95th percentile above the positive controls (see methods). Visual inspection: Image scanning by eye revealed 44 compounds that appeared to enhance ALP expression above representative positive control wells. There were also 32 compounds that were noted by eye to have morphologic changes as compared to positive controls (data not shown). If a compound well was detected as an augmenter of ALP expression in at least two of the three analysis search strategies, the compound was considered for further analysis. Cross validation of the results from the three analyses revealed thirty-one compounds that appeared in only two out of three searches and eighteen compounds that were identified in all three searches (Figure 2 and Table 1). Therefore, using three different methods of analysis became the most stringent way to determine the strongest positive hits.
Figure 2. Venn diagram depiction of the 3 analysis approaches.
ImageJ analysis was used to find compounds that were three standard deviations above the positive controls. 211 compounds were identified with ImageJ. Digilab analysis was used to find compounds that were in the 95th percentile and 31 compounds were identified under this category. The compounds were also analyzed by visual inspection with 44 noted. Of these, 18 compounds were common to all three analyses. Functional categories of the 18 compounds are indicated in the inset box.
We were also interested in the functional class types of compounds represented in the top 18 hits and whether they included known and/or novel effectors. Therefore, we conducted a literature search on the eighteen compounds that were identified in all three analyses and organized them into groups based on common features. Of the eighteen compounds, eight were retinoid derivatives/analogs (13-cis retinoic acid, bexarotene, TTNPB, etc.), four known immunosuppressant drugs (FK-506, mycophenolate mofetil, mycophenolic acid, and rapamycin), two prostaglandins (prostaglandin B2 and prostaglandin E1), one fatty acid (C2 dihydroceramide), one platelet-activating factor (PAF) receptor antagonist (PCA 4248), one anticonvulsant medication (phenytoin), and one antimalarial medication (quinine) (Figure 2). There were also notable hits among the top 45 compounds including Reseveratrol, which has been implicated in pro-osteogenic activity [32, 33] and supplementary Figure 1.
Validation of Rapamycin and FK-506 induced osteoblastogenesis using MC3T3 pre-osteoblast cells
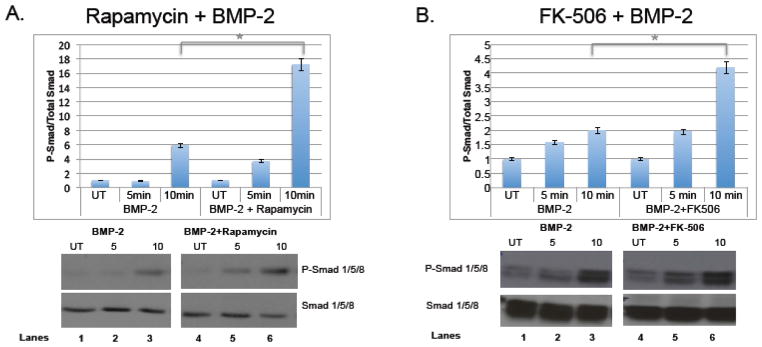
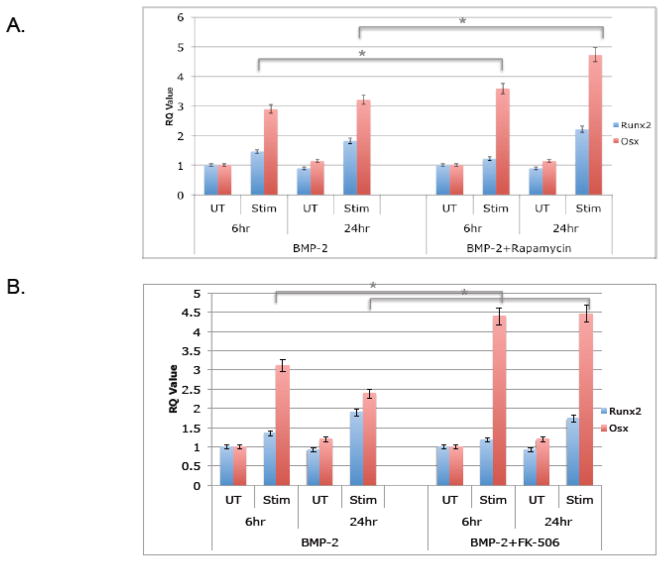
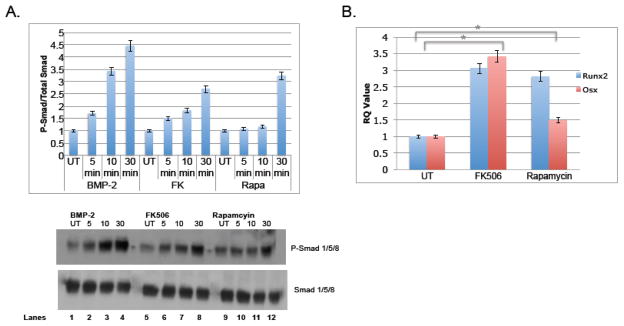
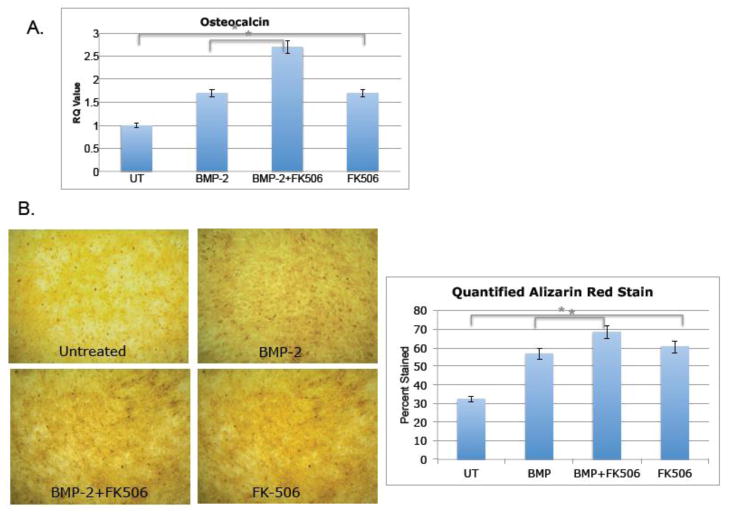
To validate our screening assay with functional assays, we utilized an osteoblast precursor cell line, MC3T3, with the two compounds rapamycin and FK506. These compounds were initially added in addition to BMP-2, which upon engagement with BMPR-I/II promotes phosphorylation of Smad 1/5/8 (P-Smad 1/5/8) and downstream activation of osteoblastogenesis such as Runx2 and Osx [4, 10, 14]. As shown in figure 3a, rapamycin was added to MC3T3-E1 cells in addition to BMP-2 and protein was collected 5 and 10 minutes after stimulation. Phospho-Smad 1/5/8 levels were measured via western blot and compared to total Smad 1/5/8 levels. Rapamycin significantly increased phosphorylation (lanes 3 and 6). The same experiment was carried out with FK-506 and an increase in phosphorylation was also observed (Figure 3b, lanes 3 and 6), indicating that both immunosuppressants increase signaling of BMP-2. To determine whether or not osteoblast specific genes were increased with rapamycin and FK-506, the compounds were added to the cells with BMP-2 and then RNA was collected 6 hours and 24 hours after stimulation to quantify Runx-2 and Osx transcripts using quantitative realtime-PCR (qRT-PCR). Both compounds significantly increased Runx2 and Osx transcripts, although Osx had a much more robust change, especially at 24 hours, both transcription factors were up regulated in reproducible experiments (Figure 4). This result is somewhat expected since Runx2 levels are tightly regulated and must be shut down at certain time points soon to allow for differentiation to continue and therefore it is possible that at 6 hours there is less of a change compared to 24 hours. We were also interested in whether rapamycin and FK-506 require BMP-2 to augment osteoblastogenesis. To address this, we evaluated bone markers in response to rapamycin and FK-506 alone. As shown in Figure 5, both FK506 and rapamcyin were sufficient to increase P-Smad 1/5/8 ratios (Figure 5a, lanes 4,8,12) and Osx and Runx2 mRNA levels (Figure 5b). However, the level of induction was not as dramatic as it was in the presence of exogenous BMP-2 but did reach levels similar to BMP-2 alone. Since the compounds had an effect on early differentiation capability, we wanted to assess their influence on late differentiation. Osteocalcin (Ocn) is an osteoblast-specific marker of late differentiation and is often observed at later time points, e.g., 14 days and later. Similar to previous experiments, FK-506 and rapamycin were added to the cells with BMP-2, with new media and compounds added every two days. We observed that Ocn transcripts were significantly increased when FK-506 was present, with or without BMP-2 (Figure 6a), but not with rapamycin (data not shown). To assess late differentiation Alizarin-red staining was conducted to measure mineralization. Staining was done after 21days and images were captured demonstrating an increase in the level of mineralization with FK-506 treatment, with or without BMP-2 (Figure 6b).
Figure 3. Rapamycin and FK-506 increase BMP-2 induced phosphorylation of Smad 1/5/8.
MC3T3 pre-osteoblast cells were plated at a density of 8×105 cells per 9.6cm2 well. BMP-2 was added to differentiate the cells with rapamycin (A) or FK-506 (B) at a concentration of 100ng/mL. Total protein was collected at 5 and 10 minutes and analyzed by western blot with antibodies to phospho-Smad 1/5/8 and total Smad 1/5/8. Graphs are shown as a ratio of phosphorylated and total Smad 1/5/8 and compared to untreated samples. Shown are representative results (average of duplicates) of at least three independent experiments. (* indicates p-value=0.001)
Figure 4. Rapamycin and FK-506 increase BMP-2 induced.
Runx2 and Osx transcripts. MC3T3 pre-osteoblast cells were plated at a density of 8×105 cells per 9.6cm2 well. BMP-2 was added to differentiate the cells with rapamycin (A) or FK-506 (B) at a concentration of 100ng/mL. Cells were harvested at 6hr and 24hr time points, RNA was purified and target transcripts Runx-2 and Osx were analyzed by qRT-PCR. UT=untreated and Stim=stimulated with the indicated compound. Values represent fold change compared to untreated samples after normalization using the ΔΔCT method. Shown are representative results (average of duplicates) of at least three independent experiments. (* Indicates p-value=0.001 for both Runx2 and Osx)
Figure 5. Rapamcyin and FK-506 induce osteoblast differentiation independently of BMP-2.
The same experiments were done as shown in figures 3 & 4, except rapamycin and FK-506 were added to the MC-3T3 cells without BMP-2. (A) Phosphorylation of Smad 1/5/8 was observed at 5, 10 and 30 minutes after stimulation and compared to total Smad 1/5/8 levels via western blot. The graph represents the ratio of P-Smad to total Smad and is representative of 3 independent experiments. (B) RNA was collected 24 hours after simulation and analyzed by qRT-PCR for Runx2 and Osx levels. The values for the graph were determine by ΔΔCT and compared to the untreated sample and were normalized to 18S. This was representative of three independent experiments (* indicates p-value=0.02 for both Runx2 and Osx).
Figure 6. FK-506 induces late differentiation markers.
MC-3T3 cells were plated at a density of 8×105 cells per 9.6cm2 well and treated with 100ng/ml FK-506 in the presence or absence of 100ng/ml BMP-2. (A) Media was replaced with fresh compound stimulation every two days and then RNA was collected and purified for qRT-PCR of Ocn mRNA transcripts on day 14. Values in the graph represent fold change compared to untreated samples after normalization using the ΔΔCT method. Shown are representative results (average of duplicates) of at least three independent experiments. (* indicates p-value=0.001). (B). Media was replaced with fresh compound simulation every two days and then stained on day 21 with Alizarin-red. Pictures are representative of three independent experiments. Quantification was done using ImageJ (* indicates p-values=0.02).
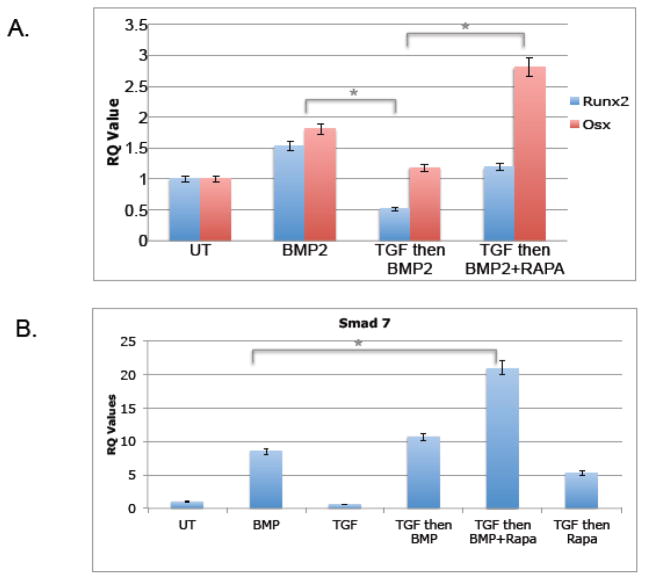
Rapamycin but not FK506 rescues TGFβ1 mediated inhibition of BMP-2 induced Runx2 and Osx and augments Smad 7 mRNA induction via BMP-2
We were then interested in exploring whether rapamycin and FK506 were equivalently capable of attenuating TGFβ1 mediated decline in osteoblastogenesis, an antagonistic effect that has been well documented [7, 25, 34]. When TGFβ1 was added to MC3T3 cells, it reduces basal levels of Runx2 and Osx mRNA as well as preventing the induction of these transcripts by BMP-2, as expected (Figure 4). However, when rapamycin was added with BMP-2 to cells after 24hours of TGFβ1 pre-treatment, the ability of TGFβ1 to inhibit osteoblast formation was attenuated (Figure 7a). Interestingly, by contrast, when FK506 was added after TGFβ1 stimulation, there was no apparent rescue of TGFβ1 mediated suppression (data not shown). These data suggest that rapamycin interferes with the TGFβ1 consistent with these compounds having different mechanisms of action.
Figure 7.
TGFβ inhibits osteoblast differentiation and Rapamycin rescues this inhibition while increasing induction of Smad7 transcripts. MC3T3 cells were treated with media containing 1ng/mL of TGFβ1 for 24 hours and then the media was replaced with BMP-2 or BMP-2 with 100ng/ml rapamycin. RNA was collected 24 hours afterwards and analyzed by qRT-PCR and the ΔΔCT method. (A) Values of Runx2 and Osx transcripts are represented as fold change compared with untreated samples (* indicates p-value=0.02). (B) Values of Smad 7 transcripts. Shown are representative results (average of duplicates) of at least three independent experiments. (* indicates p-value=0.03)
We were then interested in whether Smad-7, a suppressor of Smad-2/3 downstream of TGFβ1, might be a target of rapamycin and/or FK-506. As shown in Figure 7b, rapamycin augmented Smad 7 expression in the context of BMP-2 and TGFβ1; whereas TGFβ1 had no apparent affect on Smad-7 levels (similar results were seen with FK-506, data not shown). Because BMP-2 alone induces Smad-7, BMP-2 signaling may involve activation of protective factors that negatively regulate Smad-2/3, further ensuring expression of bone markers, and augmented by rapamycin/FK-506. Collectively, these data suggest that while both immunosuppressants are capable of inducing bone markers, they nevertheless differ in detail.
DISCUSSION
In this study, we describe a novel high-throughput screening approach to identify bone-promoting compounds that induce osteoblastogenesis in vitro. Among over 5000 compounds from several chemical libraries that were screened, 45 compounds were identified and cross-validated using three different criteria. Two compounds were of particular interest to us due to their notable role as immunosuppressive drugs (i.e., rapamycin and FK506, [35]). Our results support prior but mixed studies suggesting an osteogenic role for rapamycin and FK506. After we were able to validate the use of FK506 and rapamycin in promoting osteoblastogenesis, we evaluated the capacity for these compounds to attenuate bone growth antagonists. Because TGFβ1 has a prominent role in bone loss studies and has been previously shown to inhibit osteoblastogenesis, this antagonist was chosen. Our data support a capacity for rapamycin but not FK-506 to rescue TGFβ1 mediated inhibition. By studying these compounds in the context of antagonist perturbation that share features with common bone disease models, we were able to gain further insights into the pathways involved in osteoblast differentiation. Notably, TGFβ1 can promote osteoblastogenesis, often in the very early stages of commitment [36]. In this study, however, we chose to focus on the inhibitory role due to previous studies linking increased TGFβ1 levels with decreased bone mass [37].
Among the most potent compounds identified in this screen were the retinoids, metabolically active forms of Vitamin A. Reports of in vivo experiments clearly show enhanced bone healing after injury in mice with increased Vitamin A in their diet and those studies have suggested that retinoic acid enhances osteoblast differentiation by increasing BMP2 mRNA expression [38].
Notable compounds among the strongest hits and potential hits (Table 1, Supplementary Figure 1) included five commonly used immunosuppressant drugs (rapamycin, FK-506, FK-520, mycophenolate mofetil and mycophenolic acid) that are reported to function differently but ultimately suppress B and/or T cell proliferative responses [39–41]. We also identified four different DNA topoisomerase inhibitors as well as two actin polymerization inhibitors. Another group of interest that enhanced ALP expression were four prostaglandins (prostaglandins B1, E1, E2 and 13,14-Dihydro-prostaglandin E1). It is interesting to note that our high-throughput screen identified different compounds that fell into specific functional classes, suggesting common mechanisms of action affecting osteoblast lineage commitment. An unexpected observation for the majority of compound hits that enhanced ALP expression in our screen was their effect on cell proliferation. A general model for an inverse relationship between proliferation and differentiation has been proposed, possibly explaining why our hits would induce differentiation at the same time they prevent proliferation [42].
It has been observed that different immunosuppressant regimens used for organ transplant have varying effects post-transplantation bone loss [43, 44]. In vitro and in vivo data show conflicting results, possibly due to the wide variation of conditions and concentrations of immunosuppressant drugs used [45]. In vitro, FK-506 has been shown to enhance osteoblast differentiation at low concentrations (10nM-1uM) [46]. At higher concentrations (>25uM), FK-506 has been shown to inhibit osteoblast differentiation [47]. Interestingly, rat studies have indicated that FK-506 treatment was shown to decrease bone mineral density whereas rapamycin was shown to be bone sparing [48]. However, one study shows that FK-506 increased bone formation in alveolar bone of rats [49]. Recently, several studies have shown novel therapeutic roles for rapamycin, making it important to note that it should no longer just be thought of as an irrelevant immunosuppressive drug. One study showed that rapamycin was able to increase longevity in mice [50], whereas another recent study has shown that rapamycin can reverse the phenotype of Hutchinson-Gilford Progeria Syndrome cells [51]. For our screen, the immunosuppressant drugs were tested at relatively low doses (i.e. <1uM).
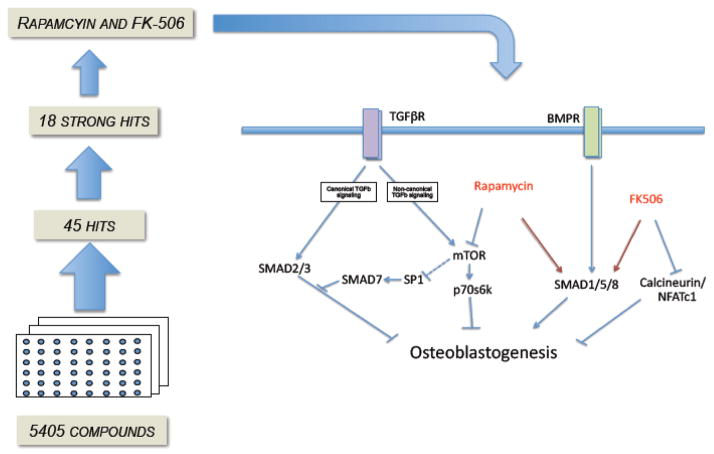
Although rapamycin and FK-506 share common signaling targets (i.e., Smad-1/5/8), they also have compound specific effects. FK-506 is a well-recognized calcineurin inhibitor and rapamycin inhibits the mammalian target of rapamycin (mTOR). Both appear to act through FKBP12 and are indicated in our graphical model (Figure 8). Calcineurin is a phosphatase that acts upon nuclear factor of activated T cells (NFAT) allowing it to translocate into the nucleus and act as a transcription factor [52]. Constitutively active NFATc1 has been shown to inhibit osteoblast differentiation and function [53]. mTOR’s role in osteoblastogenesis is not as clear but it has been shown to be critical in the differentiation of mesenchymal stem cells [18, 20, 54]. Whether rapamycin’s suppression of mTOR contributes to the rescue from TGFβ1 suppression will require further study.
Figure 8.
Graphical model of role for Rapamycin and FK-506 promoting osteoblastogenesis.
The capacity for both rapamycin and FK-506 to enhance osteoblastogenesis independently of BMP-2 may be due to low but detectable levels of BMP-2 present in unstimulated cells (see Figure 3). A model describing the augmentation of BMP-2 consistent with our observations is shown in figure 8. In this model, both FK-506 and rapamycin promote phosphorylation of Smad 1/5/8. This, then, allows for the downstream signaling leading to activation of osteoblast specific genes. Either alone or in the presence of exogenous BMP-2, these compounds, or more potent derivatives, show promise for promoting osteoblast differentiation. Interestingly, only FK-506 was observed to enhance late differentiation, indicating that FK-506 and rapamycin signaling are non-redundant. One possible explanation may be differential effects on Runx2 levels during late differentiation, explaining why rapamycin did not enhance Ocn and mineralization. Although FK-506 also enhances Runx2, it is likely that it enhances osteoblastogenesis though a Runx2-independent mechanism as well.
TGFβ1 has been demonstrated to decrease osteoblast differentiation [27, 55] and in vivo experiments have shown that blockade of TGFβ1 results in increased bone mineral density accompanied by increased osteoblast numbers [24]. Elevated TGFβ1 has been implicated in several disease states. For example, TGFβ1 is increased in the serum of HIV positive patients compared to HIV negative patients. These HIV positive patients also have increased loss of bone density compared to their HIV negative age matched controls [56, 57]. Although FK-506 and rapamycin both acted to increase osteoblast formation in our system, they were not equally efficacious in attenuating TGFβ1 mediated decline in osteogenic signaling. Only rapamycin was able to attenuate the loss of differentiation. Figure 8 shows a hypothetical model for TGFβ1 inhibition and the ability of rapamycin to rescue. Previous studies indicate that TGFβ1 can signal through two different pathways; a canonical pathway that induces Smad 2/3, which blocks osteogenesis, and a non-canonical pathway that induces mTOR, also blocking osteogenesis [58, 59]. We hypothesize that when rapamycin is introduced to pre-osteoblasts blockage of mTOR leads to downstream effects on the non-canonical TGFβ1 signaling pathway. The rapamycin-FKBP12 complex is known to bind mTOR and, thereby, block p70s6kinase (p70s6K) activation [60]. Rapamycin has been shown to potentiate osteoblast differentiation via a p70s6K dependent manner [61]. It has also been proposed that mTOR signaling affects Sp1 transcriptional activity [62]. Sp1 has previously been implicated in TGFβ1 signaling [63] and overexpression of Sp1 resulted in six-fold increase of basal Smad7 promoter activity [64], indirectly enhancing Smad7 activity, a TGFβ1 inducible antagonist [65, 66]. Therefore, Smad7 upregulation may assist but is insufficient for rapamycin to attenuate TGFβ1 induced repression of osteoblast differentiation, since FK-506 also induces Smad-7. FK-506 did not appear to significantly rescue TGFβ1 mediated decline in osteoblast differentiation. Further studies are clearly needed to dissociate the effects of rapamycin and FK-506 on TGFβ1 signalling.
Although this screen was focused on the identification of bone promoting compounds, an added advantage of our high-throughput approach is the utility for discovery of novel pathways with desirable outcomes. For example, our screen identified a platelet activation factor (PAF) receptor antagonist as a hit that increases osteoblast formation. Having this hit allows for the further study of the role of PAF receptors in the mechanism through which bone is formed. An isolated study has reported a potential role of PAF in bone metabolism [67] but there has not been follow up on this observation, making the need for pathway analysis even more clear. High-throughput screens may also allow for the identification of multiple alternatives for use in patient oriented approaches by providing multiple alternative pathways that may overcome host specific deficits, such as genetic diseases in one pathway. Thus, high throughput screening is a powerful technology in translational medical research (i.e., targeted therapeutics). The more obvious benefit to high throughput screens is the ability to discover new drugs quickly and efficiently. An additional benefit is the ability to use the compound hits to explore mechanisms. Through these benefits, we were able to identify an uncommon role for two common immunosuppressants. Further studies will need to be done to understand either their true potential, or derivatives thereof, as potential therapeutics in mitigating bone loss.
Highlights.
High-throughput screen identifies osteogenic compounds.
Two immunosuppressors among many selected for secondary analysis.
Rapamycin and FK-506 promote osteogenesis.
Pathway analysis supports different role.
Novel approach for identifying potential factors mitigating bone loss.
Supplementary Material
List of top compounds in this screen detected with at least two search strategies.
Acknowledgments
The authors would like to acknowledge the help of Dr. Sarah Haigh Molina and Ada Kane in the Boston University High Throughput Screening Core, as well as David Drapcho from Digilab. This research was in part funded by the Boston OAIC Pepper Center, the Boston University Ignition Award Program (M.M.) and RO1 AR055115 from the National Institutes of Health (M.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arora S, Agrawal M, Sun L, Duffoo F, Zaidi M, Iqbal J. HIV and bone loss. Curr Osteoporos Rep. 8:219–26. doi: 10.1007/s11914-010-0036-x. [DOI] [PubMed] [Google Scholar]
- 2.Wongdee K, Charoenphandhu N. Osteoporosis in diabetes mellitus: Possible cellular and molecular mechanisms. World J Diabetes. 2:41–8. doi: 10.4239/wjd.v2.i3.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto M, Murai J, Imai Y, Ikegami D, Kamiya N, Kato S, Mishina Y, Yoshikawa H, Tsumaki N. Conditional deletion of Bmpr1a in differentiated osteoclasts increases osteoblastic bone formation, increasing volume of remodeling bone in mice. J Bone Miner Res. doi: 10.1002/jbmr.477. [DOI] [PubMed] [Google Scholar]
- 4.Eriksen EF. Cellular mechanisms of bone remodeling. Rev Endocr Metab Disord. 2010;11:219–27. doi: 10.1007/s11154-010-9153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raggatt LJ, Partridge NC. Cellular and molecular mechanisms of bone remodeling. J Biol Chem. 285:25103–8. doi: 10.1074/jbc.R109.041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Yeo LS, Vidal C, McCorquodale T, Herrmann M, Fatkin D, Duque G. Decreased bone formation and osteopenia in lamin a/c-deficient mice. PLoS One. 6:e19313. doi: 10.1371/journal.pone.0019313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rufo A, Del Fattore A, Capulli M, Carvello F, De Pasquale L, Ferrari S, Pierroz D, Morandi L, De Simone M, Rucci N, Bertini E, Bianchi ML, De Benedetti F, Teti A. Mechanisms inducing low bone density in duchenne muscular dystrophy in mice and humans. J Bone Miner Res. 26:1891–903. doi: 10.1002/jbmr.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings SR, San Martin J, McClung MR, Siris ES, Eastell R, Reid IR, Delmas P, Zoog HB, Austin M, Wang A, Kutilek S, Adami S, Zanchetta J, Libanati C, Siddhanti S, Christiansen C. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N Engl J Med. 2009;361:756–65. doi: 10.1056/NEJMoa0809493. [DOI] [PubMed] [Google Scholar]
- 9.Giddings AM, Maitra R. A disease-relevant high-content screening assay to identify anti-inflammatory compounds for use in cystic fibrosis. J Biomol Screen. 2010;15:1204–10. doi: 10.1177/1087057110384612. [DOI] [PubMed] [Google Scholar]
- 10.Dallas S. Dynamics of the transition from osteoblast to osteocyte. Ann NY Acad Sci. 2010;1192:437–443. doi: 10.1111/j.1749-6632.2009.05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aubin JE. Osteoblast and Chondroblast Differentiation. Bone. 1995;17:77S–83S. doi: 10.1016/8756-3282(95)00183-e. [DOI] [PubMed] [Google Scholar]
- 12.Stein GS. Transcriptional control of osteoblast growth and differentation. Physiol Rev. 1996;76:593–618. doi: 10.1152/physrev.1996.76.2.593. [DOI] [PubMed] [Google Scholar]
- 13.Rosen V. BMP2 signaling in bone development and repair. Cytokine and Growth Fac Rev. 2009;20:475–480. doi: 10.1016/j.cytogfr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Nakashima K. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi A. Regulation of osteoblast differentiation mediated by none morphogenetic proteins, hedgehogs, and cbfa1. Endocr Rev. 2000;21:393–411. doi: 10.1210/edrv.21.4.0403. [DOI] [PubMed] [Google Scholar]
- 16.Hoemann CD, El-Gabalawy H, McKee MD. In vitro osteogenesis assays: influence of the primary cell source on alkaline phosphatase activity and mineralization. Pathol Biol (Paris) 2009;57:318–23. doi: 10.1016/j.patbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–66. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singha UK, Jiang Y, Yu S, Luo M, Lu Y, Zhang J, Xiao G. Rapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cells. J Cell Biochem. 2008;103:434–46. doi: 10.1002/jcb.21411. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa T. Osteoblastic differentiation is enhanced by rapamycin in rat osteoblast-like osteosarcoma (ROS 17/2. 8) cells. Biochem Biophys Res Commun. 1998;249:226–230. doi: 10.1006/bbrc.1998.9118. [DOI] [PubMed] [Google Scholar]
- 20.Lee K. Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR pathway and stimulating the BMP/Smad pathway. Stem Cells and Dev. 2010;19:557–568. doi: 10.1089/scd.2009.0147. [DOI] [PubMed] [Google Scholar]
- 21.Kaihara S. Simple and effective osteoinductive gene therapy by local injection of a bone morphogenetic protein-2-expressing recombinant adenoviral vector and FK506 mixture in rats. Gene Therapy. 2004;11:439–447. doi: 10.1038/sj.gt.3302176. [DOI] [PubMed] [Google Scholar]
- 22.Kugimiya F. Mechanism of osteogenic induction by FK506 via BMP/Smad pathways. Biochem Biophys Res Commun. 2005;338:872–879. doi: 10.1016/j.bbrc.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Tang L. FK506 enhanced osteoblastic differentiation in mesenchymal cells. Cell Biol International. 2001;26:75–84. doi: 10.1006/cbir.2001.0812. [DOI] [PubMed] [Google Scholar]
- 24.Edwards Inhibition of TGF-beta Signaling by 1D11 Antibody Treatment Increases Bone Mass and Quality In Vivo. JBMR. 2010 doi: 10.1002/jbmr.139. [DOI] [PubMed] [Google Scholar]
- 25.Ehnert S. TGF-b1 as possible link between loss of bone mineral density and chronic inflammation. PLOS One. 2010;5:e14073. doi: 10.1371/journal.pone.0014073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang JS. Repression of Runx2 function by TGF-b through recruitment of class II histone deacetylase by Smad3. European Molecular Biology Organization. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chambers TJ. Regulation of the differentiation and function of osteoclasts. J Pathol. 2000;192:4–13. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH645>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 28.Centrella M. Transforming growth factor beta is a bifunctional regulator of replication and collagen synthesis in steoblast enriched cell cultures from fetal rat bone. J Cell Biochem. 1987;262:2869–2874. [PubMed] [Google Scholar]
- 29.Centrella M. Multiple regulatory effects by transforming growth factorbeta on type I collagen levels in osteoblast-enriched cultures from fetal rat bone. Endocrinology. 1992;131:2863–2872. doi: 10.1210/endo.131.6.1446624. [DOI] [PubMed] [Google Scholar]
- 30.Wrana JL. Diferential effects of transforming growth factor-beta on the synthesis of extracellular matrix proteins by normal fetal rat calvarial bone cell populations. J Cell Biol. 1988;106:915–924. doi: 10.1083/jcb.106.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takuwa Y, Ohse C, Wang EA, Wozney JM, Yamashita K. Bone morphogenetic protein-2 stimulates alkaline phosphatase activity and collagen synthesis in cultured osteoblastic cells, MC3T3-E1. Biochem Biophys Res Commun. 1991;174:96–101. doi: 10.1016/0006-291x(91)90490-x. [DOI] [PubMed] [Google Scholar]
- 32.Rayalam S, Della-Fera MA, Baile CA. Synergism between resveratrol and other phytochemicals: Implications for obesity and osteoporosis. Mol Nutr Food Res. 2011;55:1177–85. doi: 10.1002/mnfr.201000616. [DOI] [PubMed] [Google Scholar]
- 33.Uysal T, Gorgulu S, Yagci A, Karslioglu Y, Gunhan O, Sagdic D. Effect of resveratrol on bone formation in the expanded inter-premaxillary suture: early bone changes. Orthod Craniofac Res. 2011;14:80–7. doi: 10.1111/j.1601-6343.2011.01511.x. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto T. TGF-b-related mechanisms of bone destruction in multiple myeloma. Bone. 2011;48:129–134. doi: 10.1016/j.bone.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 35.Taylor AL, Watson CJ, Bradley JA. Immunosuppressive agents in solid organ transplantation: Mechanisms of action and therapeutic efficacy. Crit Rev Oncol Hematol. 2005;56:23–46. doi: 10.1016/j.critrevonc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z, Zhao L, Nagy TR, Peng X, Hu J, Feng X, Van Hul W, Wan M, Cao X. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–65. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004;23:552–63. doi: 10.1038/sj.emboj.7600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka K, Tanaka S, Sakai A, Ninomiya T, Arai Y, Nakamura T. Deficiency of vitamin A delays bone healing process in association with reduced BMP2 expression after drill-hole injury in mice. Bone. 47:1006–12. doi: 10.1016/j.bone.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Ohdan H. Quantification of T-cell proliferation for individualizing immunosuppressive therapy for transplantation patients. Clin Pharmacol Ther. 87:23–6. doi: 10.1038/clpt.2009.171. [DOI] [PubMed] [Google Scholar]
- 40.Sehgal SN. Rapamune (RAPA, rapamycin, sirolimus): mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin Biochem. 1998;31:335–40. doi: 10.1016/s0009-9120(98)00045-9. [DOI] [PubMed] [Google Scholar]
- 41.Hamawy MM. Molecular actions of calcineurin inhibitors. Drug News Perspect. 2003;16:277–82. doi: 10.1358/dnp.2003.16.5.829315. [DOI] [PubMed] [Google Scholar]
- 42.Zhu L, Skoultchi AI. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev. 2001;11:91–7. doi: 10.1016/s0959-437x(00)00162-3. [DOI] [PubMed] [Google Scholar]
- 43.Tamler R, Epstein S. Nonsteroid immune modulators and bone disease. Ann N Y Acad Sci. 2006;1068:284–96. doi: 10.1196/annals.1346.032. [DOI] [PubMed] [Google Scholar]
- 44.Epstein S, Shane E, Bilezikian JP. Organ transplantation and osteoporosis. Curr Opin Rheumatol. 1995;7:255–61. doi: 10.1097/00002281-199505000-00018. [DOI] [PubMed] [Google Scholar]
- 45.Yeo H, Beck LH, McDonald JM, Zayzafoon M. Cyclosporin A elicits dose-dependent biphasic effects on osteoblast differentiation and bone formation. Bone. 2007;40:1502–16. doi: 10.1016/j.bone.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang L, Ebara S, Kawasaki S, Wakabayashi S, Nikaido T, Takaoka K. FK506 enhanced osteoblastic differentiation in mesenchymal cells. Cell Biol Int. 2002;26:75–84. doi: 10.1006/cbir.2001.0812. [DOI] [PubMed] [Google Scholar]
- 47.Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11:880–5. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- 48.Romero DF, Buchinsky FJ, Rucinski B, Cvetkovic M, Bryer HP, Liang XG, Ma YF, Jee WS, Epstein S. Rapamycin: a bone sparing immunosuppressant? J Bone Miner Res. 1995;10:760–8. doi: 10.1002/jbmr.5650100513. [DOI] [PubMed] [Google Scholar]
- 49.Andia DC, Nassar CA, Nassar PO, Guimaraes MR, Cerri PS, Spolidorio LC. Treatment with tacrolimus enhances alveolar bone formation and decreases osteoclast number in the maxillae: a histomorphometric and ultrastructural study in rats. Histol Histopathol. 2008;23:1177–84. doi: 10.14670/HH-23.1177. [DOI] [PubMed] [Google Scholar]
- 50.Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, Orihuela CJ, Pletcher S, Sharp ZD, Sinclair D, Starnes JW, Wilkinson JE, Nadon NL, Strong R. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao K. Rapamycin reverses cellular phenotypes and enhanes mutant protein clearance in Hutchinson-Gilford Progeria Syndrom cells. Science Translational Medicine. 2011;3:1–11. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- 52.Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 53.Choo MK, Yeo H, Zayzafoon M. NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation. Bone. 2009;45:579–89. doi: 10.1016/j.bone.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang X, Zhao J, Xu G, Li Y, Zhang W. mTOR and the differentiation of mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai) 43:501–10. doi: 10.1093/abbs/gmr041. [DOI] [PubMed] [Google Scholar]
- 55.Ehnert S, Baur J, Schmitt A, Neumaier M, Lucke M, Dooley S, Vester H, Wildemann B, Stockle U, Nussler AK. TGF-beta1 as possible link between loss of bone mineral density and chronic inflammation. PLoS One. 5:e14073. doi: 10.1371/journal.pone.0014073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cazanave C, Dupon M, Lavignolle-Aurillac V, Barthe N, Lawson-Ayayi S, Mehsen N, Mercie P, Morlat P, Thiebaut R, Dabis F. Reduced bone mineral density in HIV-infected patients: prevalence and associated factors. AIDS. 2008;22:395–402. doi: 10.1097/QAD.0b013e3282f423dd. [DOI] [PubMed] [Google Scholar]
- 57.Dolan SE, Kanter JR, Grinspoon S. Longitudinal analysis of bone density in human immunodeficiency virus-infected women. J Clin Endocrinol Metab. 2006;91:2938–45. doi: 10.1210/jc.2006-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–78. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 59.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–39. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dumont FJ, Su Q. Mechanism of action of the immunosuppressant rapamycin. Life Sci. 1996;58:373–95. doi: 10.1016/0024-3205(95)02233-3. [DOI] [PubMed] [Google Scholar]
- 61.Vinals F, Lopez-Rovira T, Rosa JL, Ventura F. Inhibition of PI3K/p70 S6K and p38 MAPK cascades increases osteoblastic differentiation induced by BMP-2. FEBS Lett. 2002;510:99–104. doi: 10.1016/s0014-5793(01)03236-7. [DOI] [PubMed] [Google Scholar]
- 62.Astrinidis A, Kim J, Kelly CM, Olofsson BA, Torabi B, Sorokina EM, Azizkhan-Clifford J. The transcription factor SP1 regulates centriole function and chromosomal stability through a functional interaction with the mammalian target of rapamycin/raptor complex. Genes Chromosomes Cancer. 49:282–97. doi: 10.1002/gcc.20739. [DOI] [PubMed] [Google Scholar]
- 63.Lai CF, Feng X, Nishimura R, Teitelbaum SL, Avioli LV, Ross FP, Cheng SL. Transforming growth factor-beta up-regulates the beta 5 integrin subunit expression via Sp1 and Smad signaling. J Biol Chem. 2000;275:36400–6. doi: 10.1074/jbc.M002131200. [DOI] [PubMed] [Google Scholar]
- 64.Brodin G, Ahgren A, ten Dijke P, Heldin CH, Heuchel R. Efficient TGF-beta induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J Biol Chem. 2000;275:29023–30. doi: 10.1074/jbc.M002815200. [DOI] [PubMed] [Google Scholar]
- 65.Itoh S, Landstrom M, Hermansson A, Itoh F, Heldin CH, Heldin NE, ten Dijke P. Transforming growth factor beta1 induces nuclear export of inhibitory Smad7. J Biol Chem. 1998;273:29195–201. doi: 10.1074/jbc.273.44.29195. [DOI] [PubMed] [Google Scholar]
- 66.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 67.Shibata Y, Ogura N, Moriya Y, Abiko Y, Izumi H, Takiguchi H. Platelet-activating factor stimulates production of prostaglandin E2 in murine osteoblast-like cell line MC3T3-E1. Life Sci. 1991;49:1103–9. doi: 10.1016/0024-3205(91)90598-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of top compounds in this screen detected with at least two search strategies.