Abstract
A new affinity chromatography method was developed by modifying a zonal elution method. The new method targets transient protein-protein interactions, such as those encountered during direct ligand transfer between the ligand transporter and its cognate receptor. A ligand-loaded transport protein is immobilized on the solid support, and a plug containing a putative receptor is flowed through the column. Elution profiles of proteins not interacting with the immobilized transporter can be approximated with a simple Gaussian curve, while the elution profiles of cognate receptors show significant delay and exhibit complex shape. Ligand transfer from the immobilized transporter molecules to the receptors is verified by both UV absorbance measurements and mass spectrometry. The sensitivity of the method is demonstrated using retinoic acid (RA) transfer from various isoforms of cellular RA binding proteins (CRABPs) and RA receptor γ (RARγ). Although these interactions have been hypothesized long ago to proceed via direct mechanism (i.e., via transient docking of the receptor and the transporter), the existing biophysical techniques failed to detect the presence of the transporter-receptor complexes. However, the modified zonal elution method provides unequivocal evidence of direct interaction between RARγ and one of the CRABP isoforms (CRABP II) during the ligand transfer to the receptor.
INTRODUCTION
Protein-protein interactions are essential elements in all biochemical processes, and play particularly important roles in regulation of signaling cascades both inside and outside the cells. While a large number of such encounters involve direct interactions, such as binding of co-repressors or co-modulators in gene expression pathways, many others involve either transient or indirect interactions. One particularly interesting example is gene activation by retinoic acid (RA), which controls expression of dozens of genes in mammals1 Since RA is a hydrophobic molecule, it must be solubilized in the cellular environment prior to binding to its cognate receptor. This task is accomplished in mammalian cells by cellular retinoic acid binding proteins (CRABP) I and II, with at least one of them (CRABP II) playing a transporter function, i.e. ferrying RA from the cytosol to the nuclear receptor site2 RA transfer from CRABP II to the retinoic acid receptor (RAR) was postulated to proceed via direct mechanism involving docking of the transporter and the receptor3 however, this complex has never been isolated leading to the conclusion that it is transient and the binding is weak.
There is a variety of other examples of transient complexes, which are usually considered to be a sub-class of the so-called non-obligate interactions in vivo.4-7 Transient interactions can be further divided into weak and strong interactions, where the weak interactions are characterized by a dissociation constant (KD) in the micromolar range and a lifetime of seconds and where the strong interactions last longer and have a lower KD in the nanomolar range. Since transient protein interactions frequently evade detection by classical biophysical tools, they have been studied traditionally by yeast two hybrid or mammalian two hybrid technologies, which are very labor intensive8. Alternative approaches include FRET9, SPR10, affinity-based capture11, and other methods, which are also labor intensive or else run the risk of losing weak and/or transiently interacting proteins during long incubation or washing steps. NMR, chemical cross-linking,12 size exclusion chromatography, 13 native electrospray ionization mass spectrometry (ESI-MS),14 are methods that can allow for the detection for protein interactions that do not suffer as much from washing and incubation steps, can be very sensitive and give stoichiometry data in the case of ESI-MS.15 In this work, we approached the task of detecting transient CRABP-RAR interactions by modifying the well-established technique of zonal elution chromatography, which has been used successfully in the past mostly for drug-protein binding studies16 or protein-protein interactions.17 Unlike classical zonal elution technique, the small ligand is only playing a role of the interaction mediator between the two proteins, one of which is immobilized on a solid support, and another is flowed over it. This modification of the method, where a ligand mediates the interaction between two or more proteins is assessed has already been utilized,18-20 however, those protein/ligand/protein interactions were long lived enough to allow “fishing” out of interacting proteins or in the other case is a long lived DNA/ligand/protein interaction that was assessed for function upon different ligand binding. Here, the ligand binding allows for a very transient interaction that can't be detected by any other method.
MATERIALS AND METHODS
CRABP II, CRABP I and its mutants, one containing a double cysteine mutation in its portal region (A35C/T57C),21 the other the double cysteine mutation in the portal region and a triple mutation (E75K/K81P/E102K) in the Ω-loop (CRAPBP Iε) were expressed on the backbone of CRABP I and CRABP II plasmids generously provided by Prof. Lila M. Gierasch (UMass-Amherst). All three proteins contained identical N-terminal His-tags (amino acid sequences are presented in Supporting Information). The ligand binding domain the RAR isoform γ (RARγ) was generously provided by Prof. Michael I. Schimerlik (Oregon State University, Corvallis, OR). Carbonic anhydrase II (CA II) and all-trans retinoic acid (RA) were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO).
Electrospray ionization mass spectra of a mixture of holo-CRABP II and RARγ (2:1 ratio) were acquired with a QStar XL (ABI Sciex, Toronto, Canada) hybrid quadrupole time-of-flight mass spectrometer. Solutions of the two proteins (10 μM in 10 mM ammonium acetate) were mixed immediately prior to data acquisition, and the mass spectra were acquired continuously for 15 min at a flow rate of 5μL/min at a declustering potential of 30V and a focusing potential of 165 V. Chromatographic measurements were carried out by immobilizing CRABP isoforms on a home-made Ni-NTA column (the Ni-NTA resin was purchased from Qiagen, Valencia, CA). Depending on a particular experiment, 1mL of either 10 μM apo- or holo-forms of the transporters were immobilized on a 275 μL bed volume of Ni-NTA. The column was then washed with 5 mL of 20 mM imidazole 500 mM NaCl and fractions of the wash were checked by UV to ascertain that no CRABP was being unbound from the resin. Either 1 mL of 10 μM apo-RARγ or CA II (negative control) was flowed through the column, using the same wash solution as previously with fraction collection at 30 sec time intervals. The amounts of the protein and RA in each fraction were determined by acquiring the absorbance spectrum for each fraction from 240nm to 450nm to measure protein elution (280 nm) and RA transfer (350 nm). Following each experiment the Ni-NTA column was stripped of all immobilized CRABP molecules using imidazole gradient and the CRABP protein elute absorbance was measured at 350 nm to determine any remaining RA that did not transfer to RAR.
RESULTS AND DISCUSSION
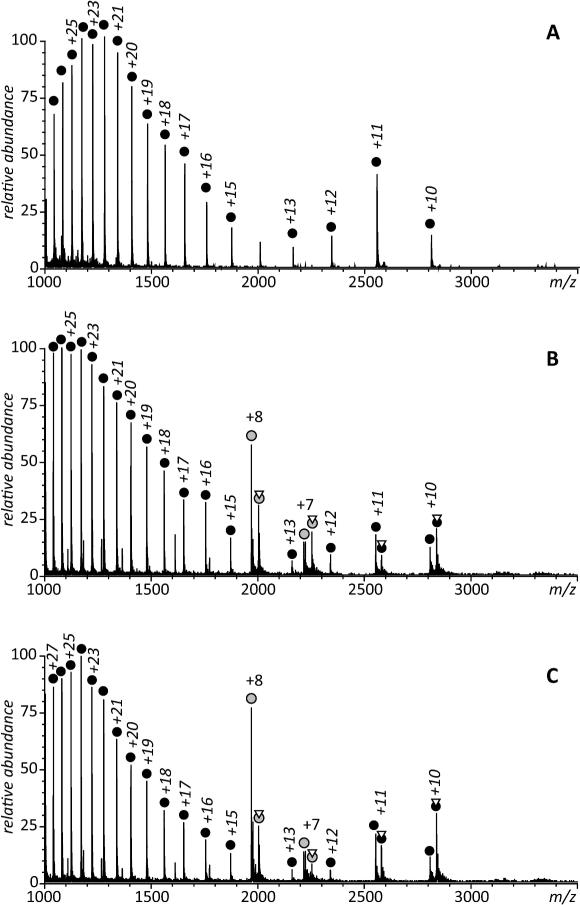
Analytical methods that are commonly used to study protein/receptor interactions, such as chemical cross-linking22 and size exclusion chromatography,13 have not been successful in detecting transient complexes formed by CRABPs and RARγ (data not shown). Native electrospray ionization mass spectrometry (ESI MS) has recently enjoyed popularity as a technique of choice in the studies of protein complexes. It has been particularly useful in the studies of protein-receptor interactions, allowing even complexes of moderate stability to be detected and their binding affinities estimated.14 Furthermore, native ESI-MS measurements can also be used to monitor small ligand exchange between two proteins.23, 24 Initially, we used native ESI-MS to monitor the exchange of ligand from holo-CRABP II to apo-RARγ since the change in mass can be easily followed in real time as the protein mixture in injected in the mass spectrometer, and to try and detect the formation of a CRABPII/RARγ/RA complex during the transfer process, provided the complex is strong enough not to be dissociated once in the gas phase and long lived enough to make it to the detector. However, no association between the two proteins was detected (Figure 1), although these experiments clearly demonstrate a decrease of the population of ions representing holo-CRABP II and the growing abundance of the holo-RARγ ions over time (compare panels A, B and C in Figure 1) showing transfer of the ligand.
Figure 1.
ESI mass spectra of RARγ alone (A) and mixed with RA·CRABP II complex accumulated during first three minutes following mixing of the protein solutions (A) and between 10-14 min after the mixing (B). Concentration of the two proteins (in 10 mM ammonium acetate) were 10 and 20 μM, respectively. CRABP II ions are labeled with gray circles and RARγ ions are labeled with black circles; triangles indicate attachment of RA to a protein ion.
A very peculiar feature of the mass spectra shown in Figure 1 is the presence of high charge density RARγ ions (but not CRABP II ions) in the low m/z region of the spectra (< 2,000 u). These ions usually represent protein molecules that fail to maintain compact, well-defined structures in solution.25 and their presence in ESI mass spectra acquired under near-native conditions is usually a manifestation of intrinsic structural disorder in the entire protein or some parts of it.26 Conformational plasticity has been suggested in the past to be an important factor facilitating interactions of nuclear receptors with their multiple physiological partners27; therefore, observing conformational disorder within RARγ is not surprising. In general, nuclear receptors (a large family of proteins that includes RARγ) are flexible proteins, and the classic lock-and-key model is inadequate to explain their behavior. Regions of intrinsic disorder (including those located in the ligand-binding domain) are known to be functionally important and ligand binding is thought to stabilizing the protein structure at varying degrees28.
Although such partially unfolded RARγ molecules may interact with other proteins and small ligands in solution, the resulting complexes are unlikely to survive the transition to the gas phase in the ESI interface, hence the absence of the RARγ·RA complexes in the low m/z region of the spectra shown in Figure 1. The putative RARγ·RA·CRABP II complexes are not observed in the entire m/z region (the ionic signal for a complex formed by completely folded CRABP II and RAR γ would be expected to be seen in the m/z region between 3,000 and 4,000 u based on the correlation between the protein physical size and the average ionic charge in ESI MS;29 lower m/z values would be expected if one of the partners is partially unfolded30).
The inability of classical analytical techniques to detect CRABP·RARγ complexes prompted us to explore the utility of affinity chromatography. Selection of the affinity ligand immobilization method is a critical step in designing a successful affinity separation or purification protocol. While the covalent methods of affinity ligand attachment to the support resin remain to be the most popular choice in many cases, they are known to be able to generate artifacts due to a variety of factors, such as multisite attachment, improper orientation and steric hindrance.31 Such artifacts can be avoided by using biospecific absorption methods to immobilize the affinity ligand to the support resin non-covalently. However, these methods either target a specific but rather limited set of analytes (e.g., constant regions of antibodies targeted by protein A and protein G), or else require introduction of biotin tags to enable interaction with avidin and streptavidin.31
In order to minimize possible artifacts in the studies of the interaction between various forms of CRABP and RARγ during the transfer of RA, we took advantage of the N-terminal His-tags present in all CRABP constructs (whose initial purpose was to facilitate protein purification following its expression21). Previous studies showed that the presence of the His-tag had no influence on the protein conformation, or its stability.32 Since the His-tag itself contains 10 histidine residues and is 21 amino acid residues long, both the extent of native structure distortion and interference with the receptor binding are expected to be minimal following its immobilization on Ni-NTA resin. This is in contrast to commonly used cross-linking methods of protein immobilization (usually targeting primary amines), which results in a heterogeneous population and may impact protein conformation and function. Another advantage of the His-tag over more classical cross-linking reagents is that the protein can easily be stripped from the column by using a high concentration of imidazole without any deleterious effect to it. A highly efficient interaction between the protein and its cognate receptor immobilized on the metal column via a His-tag had previously been demonstrated using human serum transferrin and transferrin receptor, in which case the interaction between the two proteins was strong enough that the transferrin receptor could be pulled down with the immobilized transferring using an imidazole gradient.33
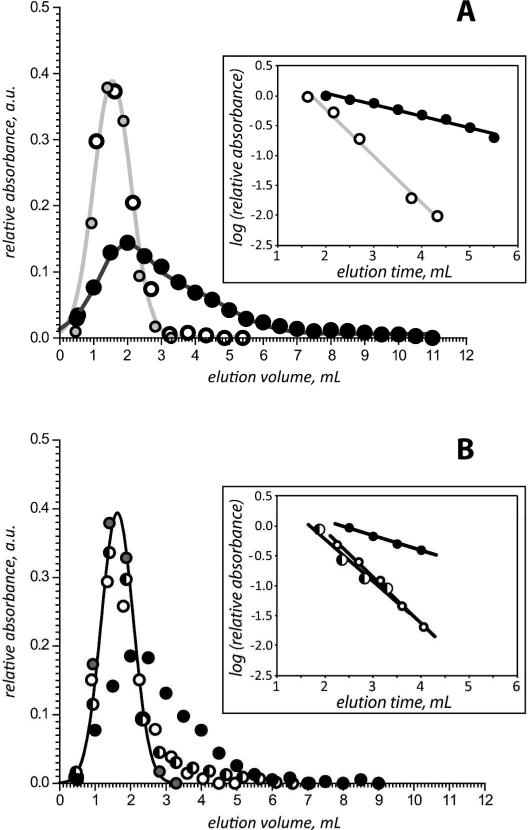
Carbonic anhydrase II was used as a negative control for binding to immobilized CRABPs, as both RARγ and CA II have a similar mass (ca. 30 kDa) and a similar size based (a gyration radius of RARα, a protein that is highly homologous to RARγ, is 18.6 Å,34 while the same parameter of CA II is 19.0 Å).35 CA II eluted at 1.6 mL when flowed over the 4 immobilized CRABPs (CRABPI, CRABPI double and triple mutant and CRABPII), irrespective of the presence of RA (Figure 2A). This provides a reference point for elution volume of proteins not interacting with CRABPs.
Figure 2.
A: elution profiles of RARγ flowed over apo-CRABP II (open circles) and holo-CRABP II (black circles) immobilized on a Ni-NTA column. Gray-shaded circles represent an elution profile of CA II flown over apo-CRABP II. B: elution profiles of RARγ flowed over apo-CRABP Iε (open circles), reduced holo-CRABP Iε (half-shaded circles), and oxidized holo-CRABP Iε (black circles). Gray-shaded circles represent an elution profile of CA II flowed over apo-CRABP II (same as in panel A). All elution profiles have been normalized to have identical areas under the curve. Insets in both panels show descending slopes of the elution profiles plotted on the log scale.
Injection of a plug of apo-RARγ to that of the immobilized apo-CRABP II: the elution profile of RARγ was nearly identical to that of CA II, with the peak elution volume being 1.6 mL. This suggests that, not surprisingly, there is no interaction between apo-CRABP II and apo-RARγ (Figure 2A). However, when an equimolar amount of apo-RARγ was flowed over immobilized holo-CRABP II, the elution of RARγ was shifted towards longer retention time (peak elution volume is 2.0 mL), and the profile changed from that of a Gaussian to an asymmetric peak with significant tailing (Figure 2A). This suggests that the two proteins (apo-RARγ and holo-CRABP II) interact with each other, and that the presence of RA is critical for this interaction. The ability of RARγ to remove RA from CRABP II is known, and can be elegantly demonstrated using native ESI MS (Figure 1). However, the transient RARγ/CRABP II complex (which had been postulated to be formed as an obligatory step during RA transfer from its transporter to the receptor2) has not been detected (vide supra); the transient affinity chromatography data presented in this work provides conclusive evidence for the existence of this complex. It is noteworthy that there are very little conformational differences between the apo- and holo-forms of CRABP II, and yet RAR only interacts with the holo-form of CRABP II. One hypothesis for ligand transfer is that CRABP II and RAR interact continuously with each other and if the ligand is present, a transfer occurs. However, our results provide clear indication that this view of the ligand transfer process is incorrect, and gives credibility to an alternative hypothesis, which views the interaction between the two proteins as being driven primarily by electrostatic forces.3 It is the change in the surface electrostatic potential of CRABP II due to ligand binding, particularly on the protrusion above the entrance for the ligand into the binding cavity of CRABPII (rather than a conformational change), that guides its transient association with and, ultimately, ligand transfer to the receptor.3
CRABP I, unlike CRABP II does not interact directly with RARγ; instead, RA transfer from CRABP I to RAR occurs passively (i.e., dissociation of RA from CRABP I is followed by its pick-up from solution by RARγ2). A triple mutation (E75K/K81P/E102K) in the Ω-loop of CRABP I was shown to create surface charge distribution mimicking that of CRABP II; as a result, the mutant CRABP I transfers RA to RAR directly, just as CRABP II does.3 We took advantage of this feature and designed a CRABPI mutant (CRABP Iε) containing 5 mutations, the triple mutation in the Ω-loop creating the RAR recognition site on the CRABP I surface, and the double mutation in the portal region to lock the RA inside the binding cavity. The latter structural feature was introduced in an attempt to increase the lifetime of the protein-receptor complex by locking RA inside the transport protein21 to prevent the possibility of its dissociation from the protein in the absence of the receptor.
The elution profile of RARγ flowed over immobilized apo-CRABP Iε is similar to that of CA II (Figure 2B), indicating that there is no interaction between the two proteins. However, elution profile of RARγ changed dramatically when flowed over immobilized holo-CRABP Iε with intact disulfide bridge in the portal region (Figure 1B). The average elution volume of RARγ increases to 2.3 mL, and the elution profile of the receptor becomes highly asymmetric with significant peak tailing. At the same time, the elution profile of RARγ flowed over immobilized holo-CRABP Iε in the presence of 10 mM β-mercaptoethanol is similar to that of CA II (Figure 2B). Reduction of the disulfide bond in the portal region was shown previously to allow full transfer of RA to lipid vesicles from a CRABP I mutant containing two engineered cysteine residues in the portal region.21 The remarkable sensitivity of the elution behavior of RARg to the presence of the disulfide bond in the portal region of CRAPBI confirms our hypothesis that the lifetime of the transient /RA/RARγ complex can be increased by creating barriers to ligand transfer.
One thing that may seem surprising is the similarity of the elution profile of RARγ flowed over immobilized apo-CRABP Iε and the reduced form of holo- CRABP Iε, which contains the RAR “recognition” patch and, therefore, is expected to interact with the receptor directly during the ligand transfer process. However, we note that even though the elution volume alone is not sufficient to detect the transient protein interaction in this case, the peak does become asymmetric, and the tailing is indicative of the receptor interaction with the protein. The peak decay method36 is used to get access to the dissociation constant of the analytes and has been shown not to need a competing analyte in the case of weak affinity systems.37 Applied to our system, the log plot of the tailing slope of the elution profile (insets in Figure 2) allows for further distinction between pairs of interacting and non-interacting proteins. The average logarithm of the slope of the elution profile of RAR flowed on the non interacting CRABPs (apo-CRABP II, and apo-CRABP Ιε) is 0.776 ± 0.005. The logarithm of the slope for holo-CRABP II is 0.193, that of holo-CRABP Iε is 0.250 and that of reduced holo-CRABP Iε is 0.699 which is significantly different than the result for the apo-CRABP. The logarithm of the slope of reduced holo-CRABPIε is much closer to that of the apo-CRABPs than that of holo-CRABPII, but still statistically different than that of the non interacting CRABPs, which can explain why by the elution volume alone, one cannot tell whether reduced holo-CRABPIε interacts with RAR. As the logarithm of the slope is proportional to the dissociation constant of the CRABP/RAR pair, we can conclude that the strongest interaction is that between holo-CRABPII and RAR followed by oxidized holo-CRABPIε and finally reduced holo-CRABPIε showing again that the introduction of the disulfide bond increases the lifetime of the CRABPIε/RA/RARγ complex. We did not design a CRABPII mutant with a disulfide bond in its portal region (which could lead to a stronger complex than with WT holo-CRABPII) as it doesn't have the same dynamics in its portal region as CRABPI and the putative disulfide could form in the apo-form of the protein. Since the aim of this work was only to establish the fact of transient binding of the ligand receptor with the ligand transporter (rather than measure the ligand/protein dissociation constants), no attempt was made to keep the protein concentrations at their physiological levels. Monitoring of the absorbance at 350 nm in the elution fraction of RARγ correlated with the absorbance at 280 nm indicating efficient transfer of RA from immobilized CRABP to RARγ and the absorbance at 350 nm of the stripped CRABP once transfer had occurred was null, indicating full transfer of RA from CRABP to RARγ (1:1) ratio (data not shown).
CONCLUSIONS
We have demonstrated that by immobilizing a protein to a Ni-NTA column and monitoring the elution profile of transiently interacting partners, conclusions can be made whether the protein interacts with the immobilized protein directly (i.e., via docking). Both the peak elution volume and the logarithm of the slope of the elution profile allow conclusions to be made whether two proteins are interacting partners. To our knowledge, this is the first time that the zonal elution method has been modified to be applied to proteins and not only to small molecules. This method allows the detection of the CRABP/RA/RARγ interactions that are too short lived to be detected by mass spectrometry. This method can be complementary or an initial step to surface Plasmon resonance (SPR) to detect whether two proteins interact before using SPR to get kinetic constants.
Many diseases, including some cancers are treated by RA derivates and analogues, and there is a continuous search for synthetic retinoids that will bind to both CRABP and RAR to exert their effect.38 This method can be adapted to be a first screen for retinoid-based therapies39 to identify synthetic retinoids levels that can compete effectively with endogenous levels of retinoids. Above and beyond the retinoid work, the method presented in this work can be adapted to any protein-protein interacting pair where both proteins can be purified and one has a remaining tag such as a His-tag or GST-tag. One of the screens that could be developed is an assay of kinases and phosphatases and their putative substrates. Many signaling pathways are not known in detail, all the proteins involved in a particular node for the signal to propagate are not always identified and this method could be another means to validate systems results. In this case, the system would be adapted not to two interacting proteins, but to a multi-protein complex.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a grant R01 GM061666 from the National Institutes of Health. RARγ was a generous gift from Dr. Schimerlik (Oregon State University) and the CRABP I and CRABP II plasmids were provided by Dr. Gierasch (University of Massachusetts-Amherst).
REFERENCE
- 1.Balmer JE, Blomhoff R. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 2.Dong D, Ruuska SE, Levinthal DJ, Noy N. J Biol Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- 3.Budhu A, Gillilan R, Noy N. J Mol Biol. 2001;305:939–949. doi: 10.1006/jmbi.2000.4340. [DOI] [PubMed] [Google Scholar]
- 4.Levy ED, Pereira-Leal JB. Curr Opin Struct Biol. 2008;18:349–357. doi: 10.1016/j.sbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Nooren IM, Thornton JM. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 6.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Nooren IM, Thornton JM. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 9.Gau D, Ding Z, Baty C, Roy P. Cell Mol Bioeng. 2011;4:1–8. doi: 10.1007/s12195-010-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beattie J, Phillips K, Shand JH, Szymanowska M, Flint DJ, Allan GJ. Mol Cell Biochem. 2008;307:221–236. doi: 10.1007/s11010-007-9601-8. [DOI] [PubMed] [Google Scholar]
- 11.Collins MO, Choudhary JS. Curr Opin Biotechnol. 2008;19:324–330. doi: 10.1016/j.copbio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Fabris D, Yu ET. J. Mass Spectrom. 2010;45:841–860. doi: 10.1002/jms.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Yang WC, Gao Q, Regnier F. J Chromatogr A. 2008;1178:24–32. doi: 10.1016/j.chroma.2007.10.067. [DOI] [PubMed] [Google Scholar]
- 14.Leverence R, Mason AB, Kaltashov IA. Proc Natl Acad Sci U S A. 2010;107:8123–8128. doi: 10.1073/pnas.0914898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebong IO, Morgner N, Zhou M, Saraiva MA, Daturpalli S, Jackson SE, Robinson CV. Proc Natl Acad Sci U S A. 2011;108:17939–17944. doi: 10.1073/pnas.1106261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David S, Hage JAA, Abby J. Jackson, Ryan Matsuda, Efthimia Papastavros, Erika Pfaunmiller, Zenghan Tong, John Vargas-Badilla, Yoo Michelle J., Zheng Xiwei. Analytical Methods. 2011;3:1449–1460. [Google Scholar]
- 17.Takekawa N, Li N, Kojima S, Homma M. J Bacteriol. 2012 doi: 10.1128/JB.06552-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marszall MP, Moaddel R, Kole S, Gandhari M, Bernier M, Wainer IW. Anal Chem. 2008;80:7571–7575. doi: 10.1021/ac801153h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFadden MJ, Junop MS, Brennan JD. Anal Chem. 2010;82:9850–9857. doi: 10.1021/ac102164d. [DOI] [PubMed] [Google Scholar]
- 20.Moaddel R, Price GB, Juteau JM, Leffak M, Wainer IW. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;820:197–203. doi: 10.1016/j.jchromb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Sjoelund V, Kaltashov IA. Biochemistry. 2007;46:13382–13390. doi: 10.1021/bi700867c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinz A. J Mass Spectrom. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 23.Zaia J, Fabris D, Wei D, Karpel RL, Fenselau C. Protein Sci. 1998;7:2398–2404. doi: 10.1002/pro.5560071117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hathout Y, Fabris D, Fenselau C. Int. J. Mass Spectrom. 2001;204:1–6. [Google Scholar]
- 25.Kaltashov IA, Abzalimov RR. J. Am. Soc. Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Frimpong A,K, Abzalimov R,R, Uversky V,N, Kaltashov I,A. Proteins. 2010;78:714–722. doi: 10.1002/prot.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. PLoS Comp. Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilser VJ, Thompson EB. J. Biol. Chem. 2011;286:39675–39682. doi: 10.1074/jbc.R111.278929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaltashov IA, Mohimen A. Anal. Chem. 2005;77:5370–5379. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa L, Brocca S, Grandori R. Anal. Chem. 2011;83:6459–6463. doi: 10.1021/ac201740z. [DOI] [PubMed] [Google Scholar]
- 31.Kim HS, Hage DS. In: Handbook of affinity chromatography. 2 ed. Hage DS, editor. Vol. 92. Taylot and Francis; Boca Raton, London, New York, Singapore: 2006. pp. 35–78. [Google Scholar]
- 32.Clark PL, Weston BF, Gierasch LM. Fold Des. 1998;3:401–412. doi: 10.1016/s1359-0278(98)00053-4. [DOI] [PubMed] [Google Scholar]
- 33.Byrne SL, Leverence R, Klein JS, Giannetti AM, Smith VC, MacGillivray RTA, Kaltashov IA, Mason AB. Biochemistry. 2006;45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 34.Egea PF, Rochel N, Birck C, Vachette P, Timmins PA, Moras D. J Mol Biol. 2001;307:557–576. doi: 10.1006/jmbi.2000.4409. [DOI] [PubMed] [Google Scholar]
- 35.Semisotnov GV, Kihara H, Kotova NV, Kimura K, Amemiya Y, Wakabayashi K, Serdyuk IN, Timchenko AA, Chiba K, Nikaido K, Ikura T, Kuwajima K. J Mol Biol. 1996;262:559–574. doi: 10.1006/jmbi.1996.0535. [DOI] [PubMed] [Google Scholar]
- 36.Moore RM, Walters RR. Journal of Chromatography A. 1987;384:91–103. [Google Scholar]
- 37.Schiel JE, Hage DS. J Sep Sci. 2009;32:1507–1522. doi: 10.1002/jssc.200800685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez S, Bourguet W, Gronemeyer H, de Lera AR. Expert Opin Ther Pat. 2011;21:55–63. doi: 10.1517/13543776.2011.536531. [DOI] [PubMed] [Google Scholar]
- 39.Moise AR, Noy N, Palczewski K, Blaner WS. Biochemistry. 2007;46:4449–4458. doi: 10.1021/bi7003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.