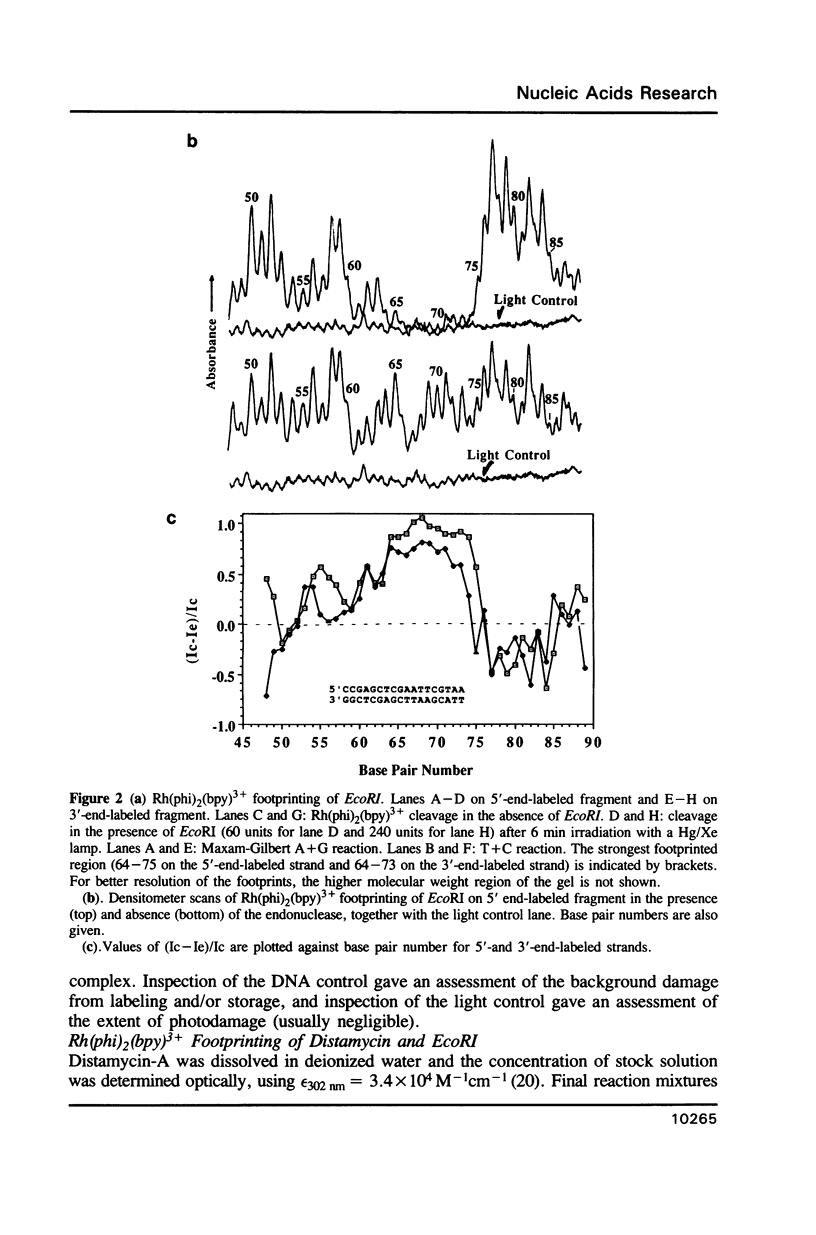
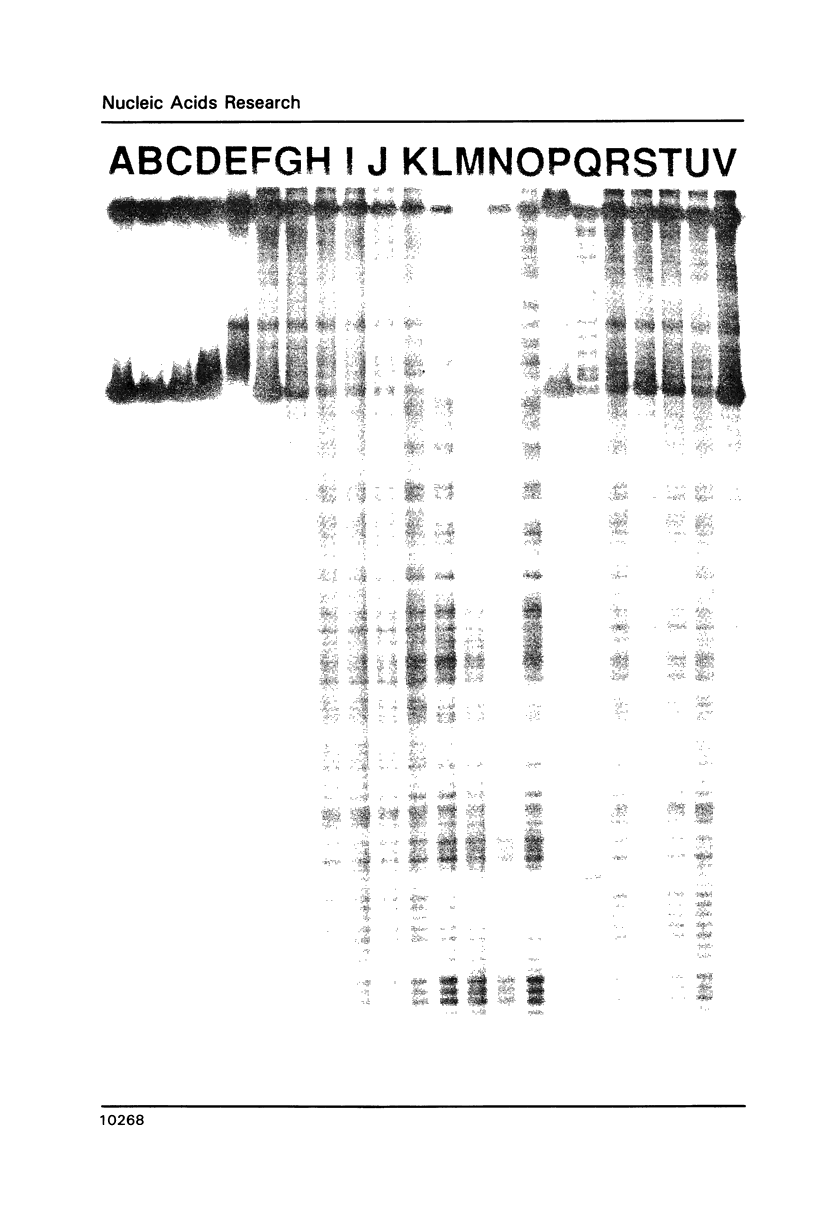
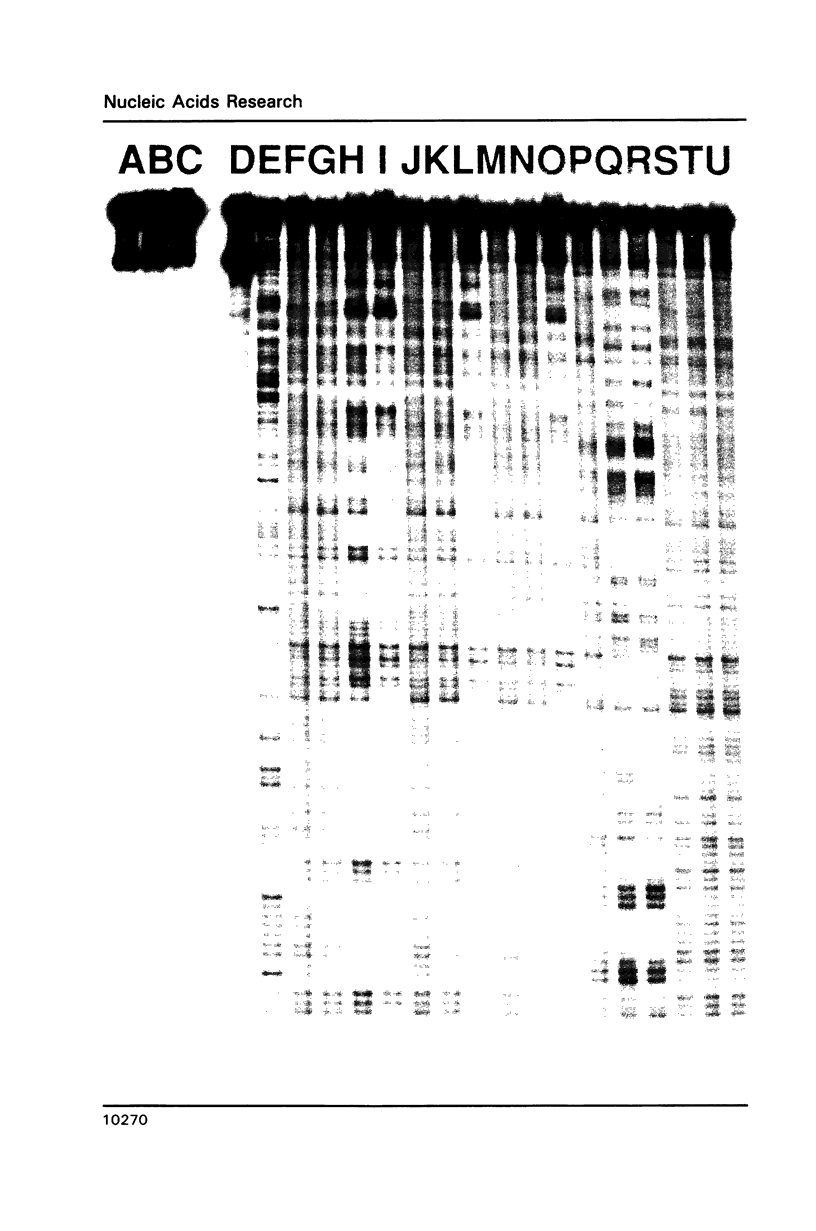
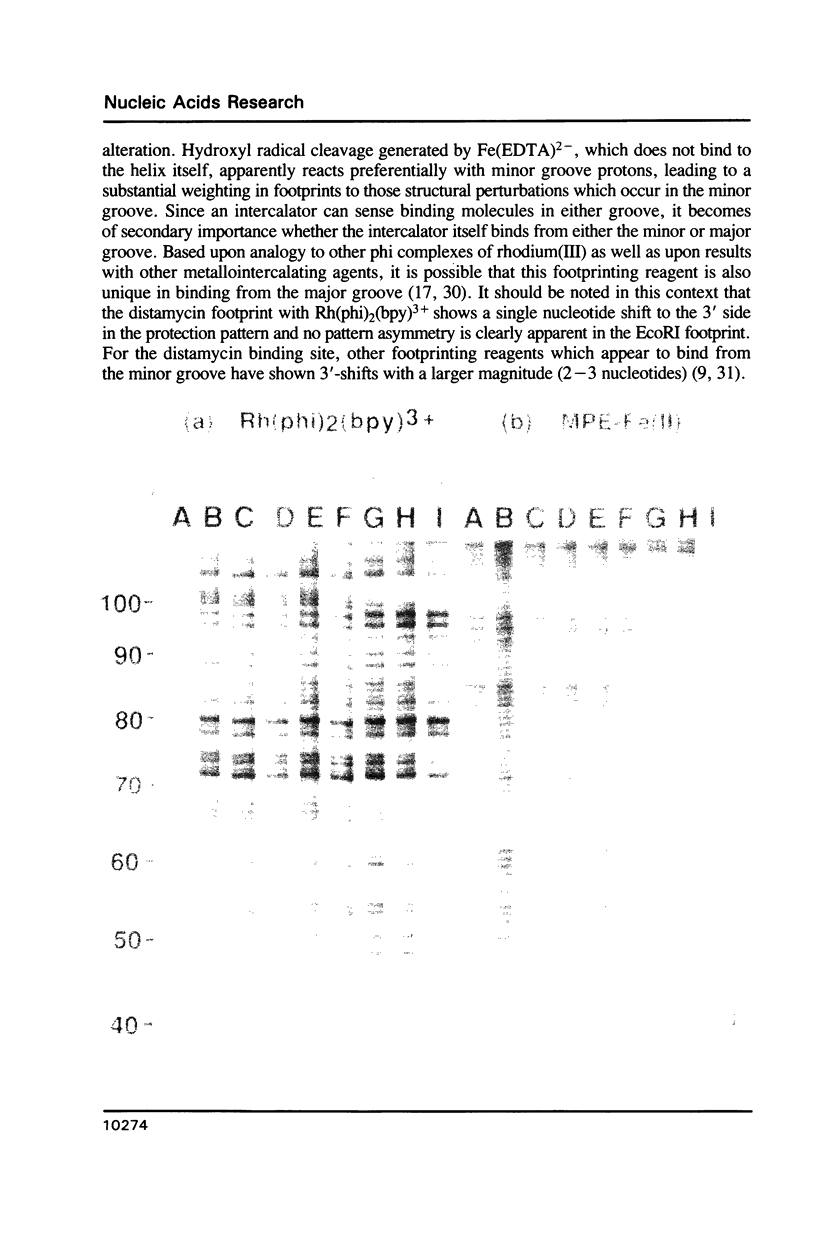
Abstract
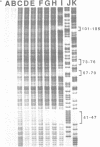
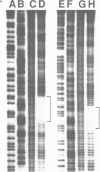
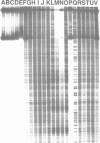
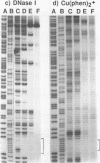
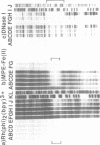
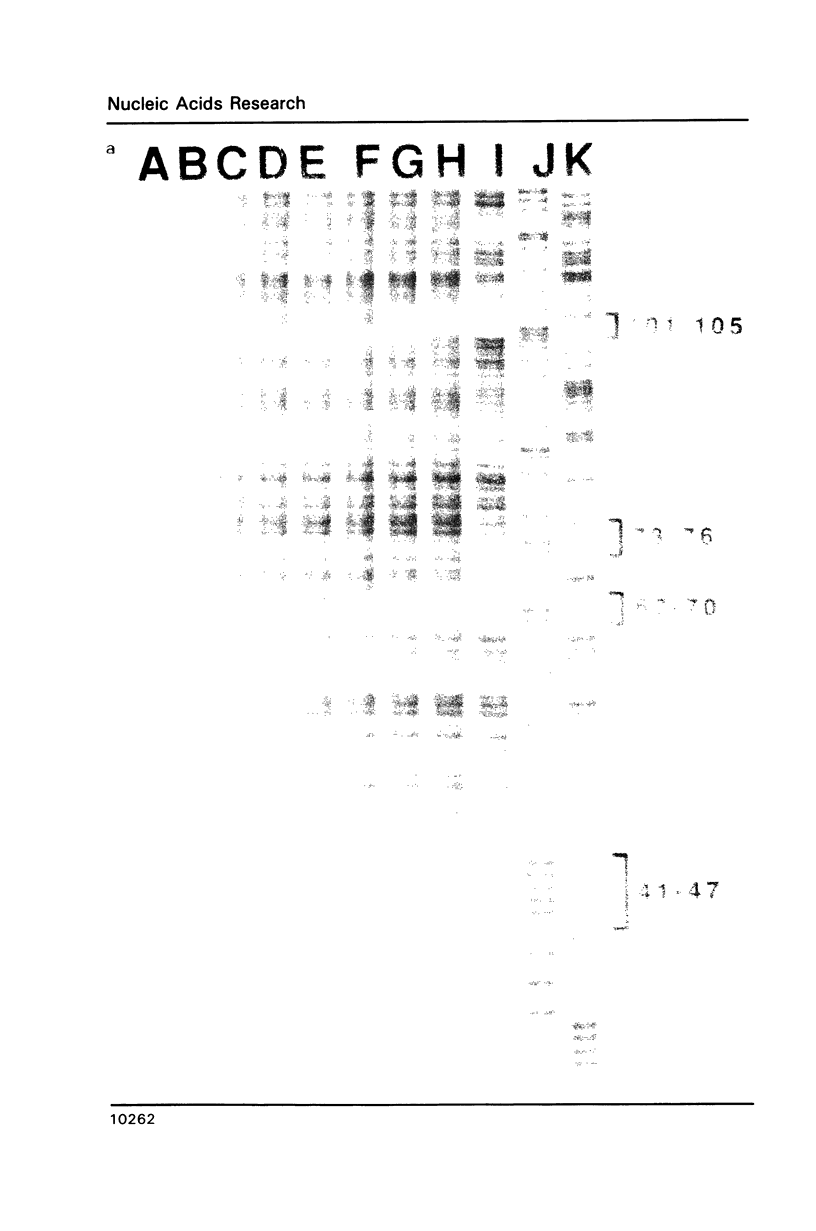
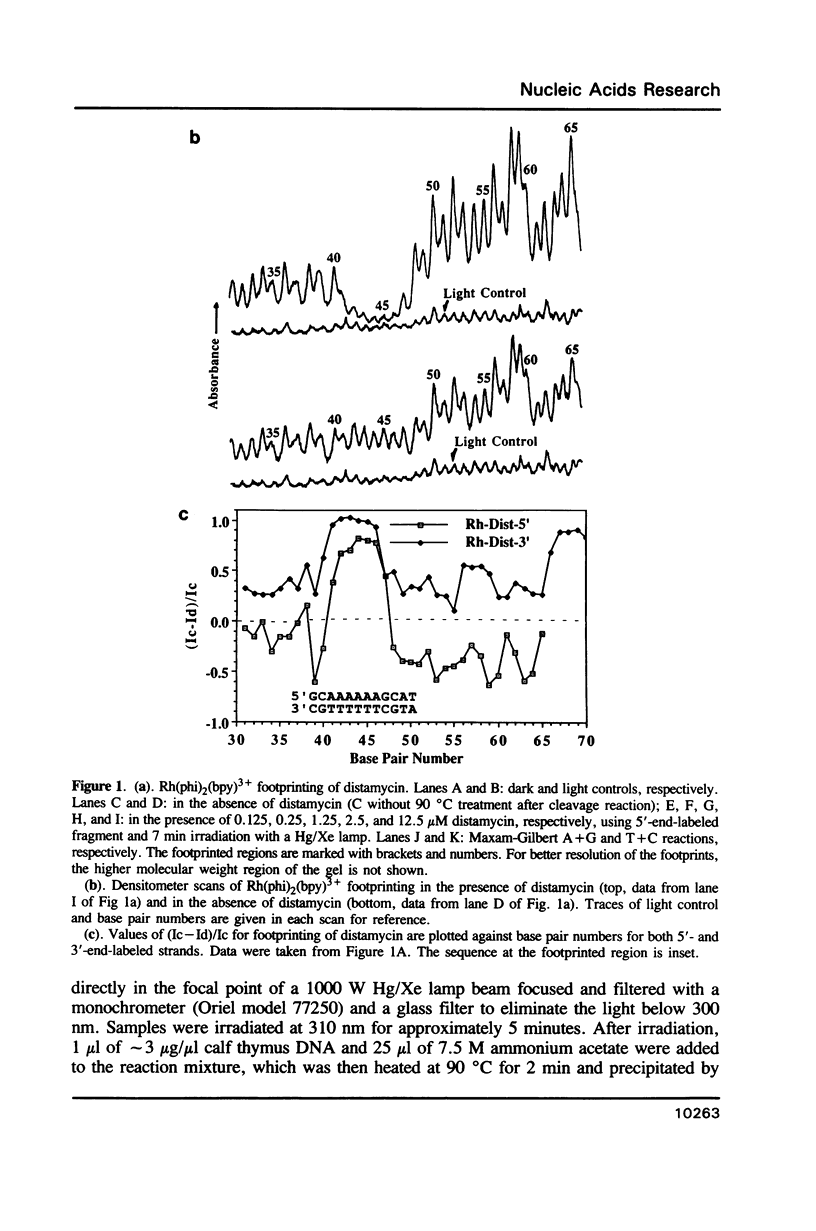
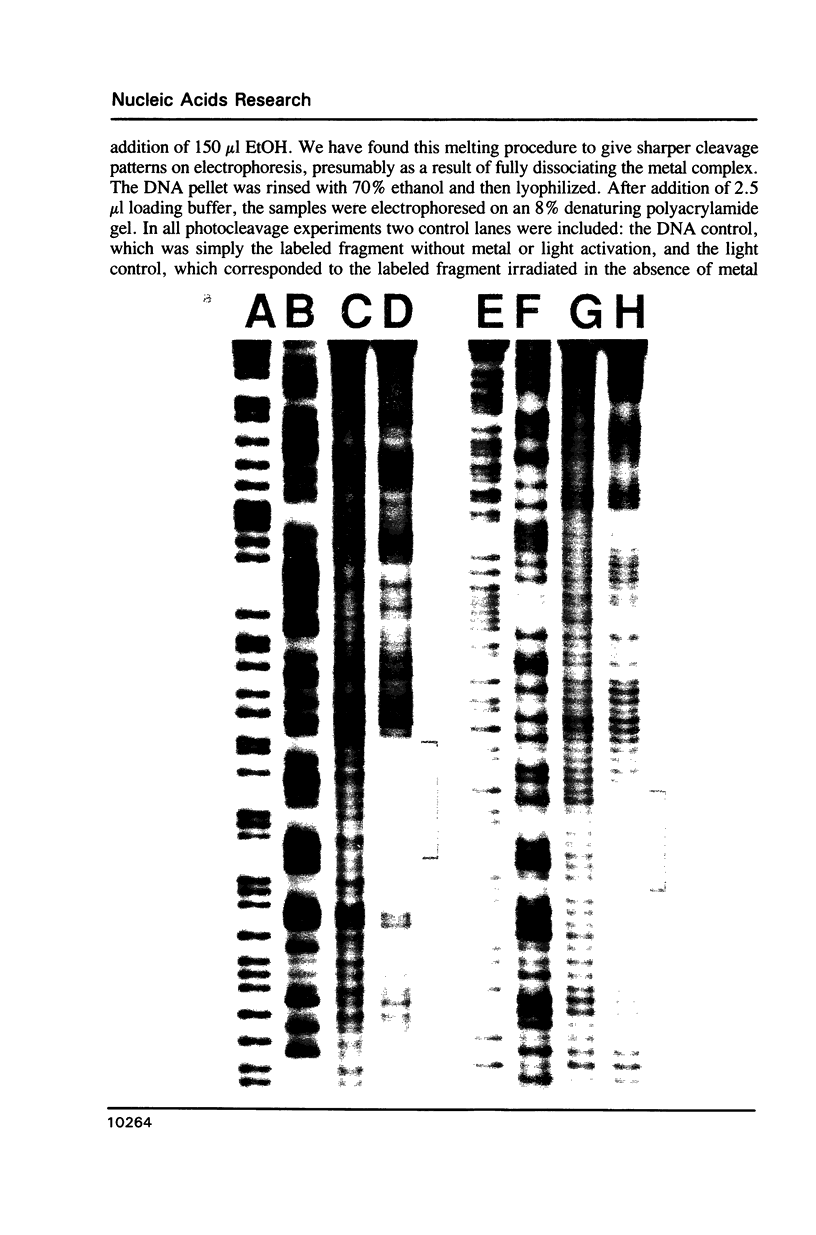
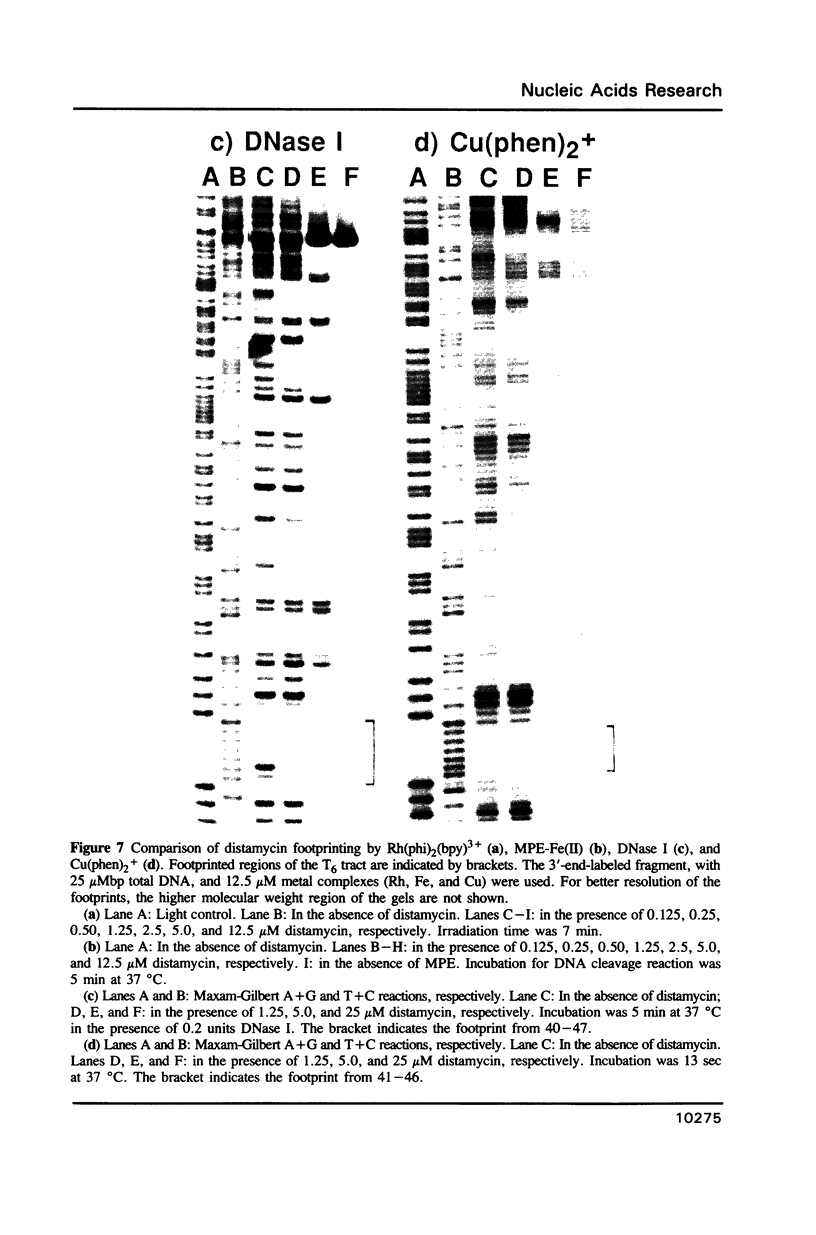
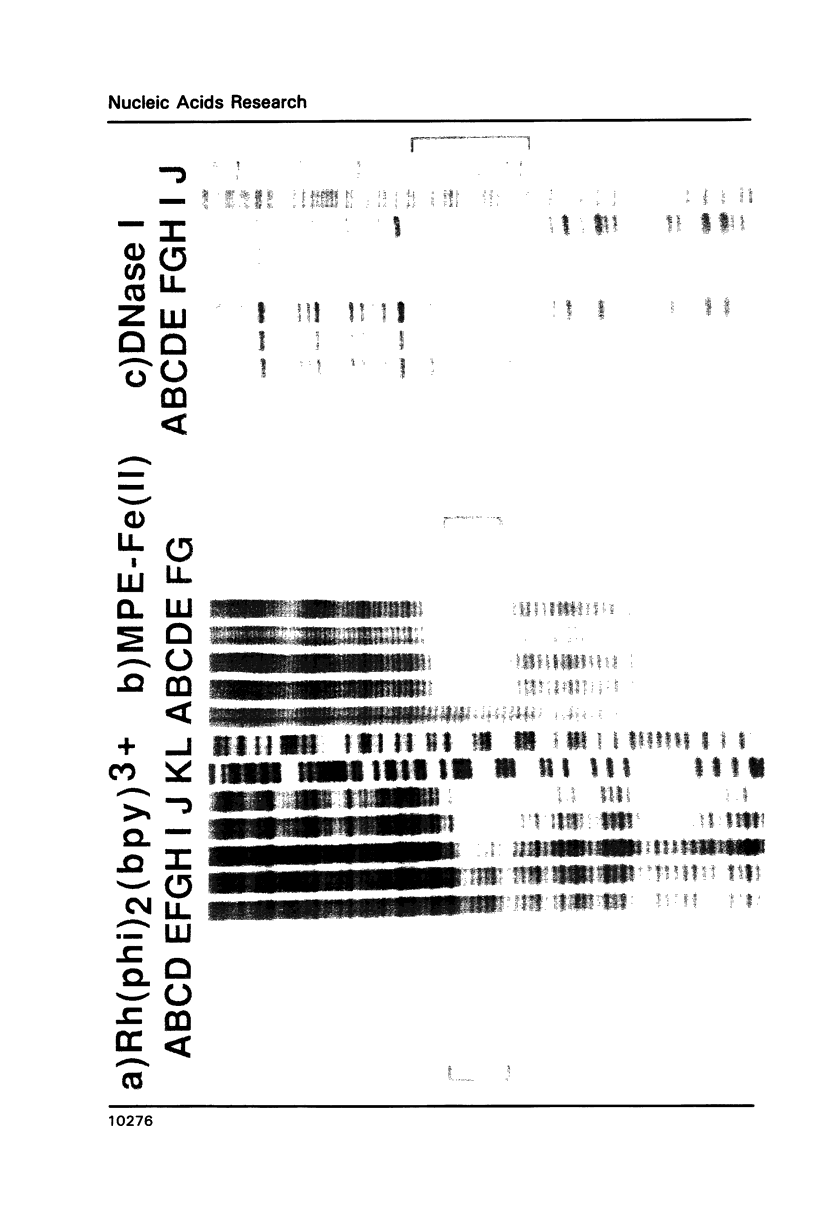
The complex bis(phenanthrenequinone diimine)(bipyridyl)rhodium(III), Rh(phi)2(bpy)3+, cleaves DNA efficiently in a sequence-neutral fashion upon photoactivation so as to provide a novel, high resolution, chemical photofootpring reagent. Photofootprinting of two crystallographically characterized DNA-binding agents, distamycin, a small natural product which binds to DNA in the minor groove, and the endonuclease EcoRI, which binds in the major groove, gave respectively a 5-7 base pair footprint for the drug at its A6 binding site and a 10-12 base pair footprint for the enzyme centered at its recognition site (5'-GAATTC-3'). Both footprints agree closely with the crystallographic results. The photocleavage reaction can be performed using either a high intensity lamp or, conveniently, a simple transilluminator box, and the photoreaction is not inhibited by moderate concentrations of reagents which are sometimes required for examining interactions of molecules with DNA. When compared with other popular footprinting agents, the rhodium complex shows a number of distinct advantages: sequence-neutrality, high resolution, ability to footprint major as well as minor groove-binding ligands, applicability in the presence of additives such as Mg2+ or glycerol, ease of handling, and a sharply footprinted pattern. Light activated footprinting reactions furthermore offer the possibility of examining DNA-binding interactions with time resolution and within the cell.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker M. M., Lesser D., Kurpiewski M., Baranger A., Jen-Jacobson L. "Ultraviolet footprinting" accurately maps sequence-specific contacts and DNA kinking in the EcoRI endonuclease-DNA complex. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6247–6251. doi: 10.1073/pnas.85.17.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowiak J. C., Ward B., Goodisman J. Quantitative footprinting analysis using a DNA-cleaving metalloporphyrin complex. Biochemistry. 1989 Apr 18;28(8):3314–3322. doi: 10.1021/bi00434a029. [DOI] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Fox K. R. DNAase I footprinting of restriction enzymes. Biochem Biophys Res Commun. 1988 Sep 15;155(2):779–785. doi: 10.1016/s0006-291x(88)80563-1. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman K. D., Dervan P. B. Molecular recognition of B-DNA by Hoechst 33258. Nucleic Acids Res. 1985 Jul 11;13(13):4825–4835. doi: 10.1093/nar/13.13.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Hurley L. H., Lee C. S., McGovren J. P., Warpehoski M. A., Mitchell M. A., Kelly R. C., Aristoff P. A. Molecular basis for sequence-specific DNA alkylation by CC-1065. Biochemistry. 1988 May 17;27(10):3886–3892. doi: 10.1021/bi00410a054. [DOI] [PubMed] [Google Scholar]
- Jeppesen C., Buchardt O., Henriksen U., Nielsen P. E. Photocleavage of DNA and photofootprinting of E. coli RNA polymerase bound to promoter DNA by azido-9-acridinylamines. Nucleic Acids Res. 1988 Jul 11;16(13):5755–5770. doi: 10.1093/nar/16.13.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessee B., Gargiulo G., Razvi F., Worcel A. Analogous cleavage of DNA by micrococcal nuclease and a 1-10-phenanthroline-cuprous complex. Nucleic Acids Res. 1982 Oct 11;10(19):5823–5834. doi: 10.1093/nar/10.19.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985 Jun 25;183(4):553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Kuwabara M., Yoon C., Goyne T., Thederahn T., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion: reaction with CGCGAATTCGCG and its complexes with netropsin and EcoRI. Biochemistry. 1986 Nov 18;25(23):7401–7408. doi: 10.1021/bi00371a023. [DOI] [PubMed] [Google Scholar]
- Luck G., Zimmer C., Reinert K. E., Arcamone F. Specific interactions of distamycin A and its analogs with (A-T) rich and (G-C) rich duplex regions of DNA and deoxypolynucleotides. Nucleic Acids Res. 1977 Aug;4(8):2655–2670. doi: 10.1093/nar/4.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Mei H. Y., Barton J. K. Tris(tetramethylphenanthroline)ruthenium(II): a chiral probe that cleaves A-DNA conformations. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1339–1343. doi: 10.1073/pnas.85.5.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J Biol Chem. 1976 Oct 10;251(19):5866–5874. [PubMed] [Google Scholar]
- Nielsen P. E., Jeppesen C., Buchardt O. Uranyl salts as photochemical agents for cleavage of DNA and probing of protein-DNA contacts. FEBS Lett. 1988 Aug 1;235(1-2):122–124. doi: 10.1016/0014-5793(88)81245-6. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. In vivo "photofootprint" changes at sequences between the yeast GAL1 upstream activating sequence and "TATA" element require activated GAL4 protein but not a functional TATA element. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5399–5403. doi: 10.1073/pnas.85.15.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Bruist M. F., Simon M. I., Dervan P. B. Synthesis of a sequence-specific DNA-cleaving peptide. Science. 1987 Nov 20;238(4830):1129–1132. doi: 10.1126/science.3120311. [DOI] [PubMed] [Google Scholar]
- Spassky A., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion. Conformational analysis and footprinting of the lac operon. Biochemistry. 1985 Dec 31;24(27):8050–8056. doi: 10.1021/bi00348a032. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Suggs J. W., Wagner R. W. Nuclease recognition of an alternating structure in a d(AT)14 plasmid insert. Nucleic Acids Res. 1986 May 12;14(9):3703–3716. doi: 10.1093/nar/14.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G., Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988 Sep 9;241(4871):1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- Veal J. M., Rill R. L. Sequence specificity of DNA cleavage by bis(1,10-phenanthroline)copper(I): effects of single base pair transitions on the cleavage of preferred pyrimidine-purine-pyrimidine triplets. Biochemistry. 1989 Apr 18;28(8):3243–3250. doi: 10.1021/bi00434a019. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Nathans J., van der Marel G., van Boom J. H., Rich A. Molecular structure of a double helical DNA fragment intercalator complex between deoxy CpG and a terpyridine platinum compound. Nature. 1978 Nov 30;276(5687):471–474. doi: 10.1038/276471a0. [DOI] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Goodisman J., Dabrowiak J. C. Determination of netropsin-DNA binding constants from footprinting data. Biochemistry. 1988 Feb 23;27(4):1198–1205. doi: 10.1021/bi00404a020. [DOI] [PubMed] [Google Scholar]
- Ward B., Skorobogaty A., Dabrowiak J. C. DNA binding specificity of a series of cationic metalloporphyrin complexes. Biochemistry. 1986 Dec 2;25(24):7827–7833. doi: 10.1021/bi00372a007. [DOI] [PubMed] [Google Scholar]
- Yoon C., Kuwabara M. D., Law R., Wall R., Sigman D. S. Sequence-dependent variability of DNA structure. Influence of flanking sequences and fragment length on digestion by conformationally sensitive nucleases. J Biol Chem. 1988 Jun 15;263(17):8458–8463. [PubMed] [Google Scholar]
- Zhen W. P., Jeppesen C., Nielsen P. E. Psoralen photofootprinting of protein-binding sites on DNA. FEBS Lett. 1988 Feb 29;229(1):73–76. doi: 10.1016/0014-5793(88)80800-7. [DOI] [PubMed] [Google Scholar]