Abstract
Conventional cell separation against multiple markers typically requires the attachment of antibody tags, typically fluorescent or magnetic, to selected cell types in a heterogeneous suspension. This work describes how such separation can be accomplished in a series of microfluidic systems without the need for such tags. Two capture stages containing antibody-functionalized alginate hydrogels are utilized for the isolation of CD34+ and Flk1+ cells from untreated, whole human blood, respectively. The capture-release capability of these degradable coatings is harnessed by a mixing chamber and a simple valving system such that the suspension emerging from the first capture stage is prepared for the second capture stage for further enrichment. With this configuration we demonstrate the isolation of CD34+/Flk1+ endothelial progenitor cells from blood enabled by the depletion of CD34+/Flk1-hematopoietic stem cells population. This ability to achieve isolation of cells against multiple markers in an untagged separation method is of particular significance in applications involving cell implantation-based therapeutics including tissue engineering, and molecular analysis.
Introduction
The isolation of particular cell types from heterogeneous suspensions such as blood or digested tissue is an essential first step in many clinically-relevant protocols. Examples include cell-based diagnostics, molecular analysis of cells via proteomics and genomics, and tissue engineering and cell-based therapeutics. For example, the ability to isolate endothelial progenitor cells (EPCs) from whole blood is desired for vascular tissue engineering and cell-based therapeutic applications.1–3 EPCs are typically isolated from blood using a multi-cycle method of centrifugation and plating,1 which is a highly time-intensive process spanning days or weeks. Alternatively, these and other cells can be isolated at very high purity using the well-established techniques of fluorescence- and magnet-activated cell sorting (FACS and MACS, respectively). FACS and MACS are currently the gold standard methods for cell isolation. However, techniques require the attachment of antibody tags in the form of fluorescent dyes or magnetic beads, respectively, to one or more cell types in the sample. Such pre-processing tagging requires additional time and may be undesirable for cell-based therapeutic applications as well as in downstream molecular analysis. The approach of adhesion-based microfluidic cell separation, which has seen significant progress in the last decade, aims to overcome this limitation via the use of a variety of immobilized capture molecules4–9 and methods to release captured cells, including deformable cell capture monoliths5,10–12 A key challenge in this mode of separation is the ability to isolate target cells that do not have one unique surface marker that distinguishes them from the non-target cells in the sample. For instance progenitor cell markers expressed by EPCs such as CD34 are also expressed by other cell types present in blood, such as hematopoietic stem cells.13, 14 Furthermore, endothelial markers expressed by EPCs, such as Flk1 (also known as VEGFR-2 and KDR), are also expressed by mature endothelial cells.15, 16
We recently described how alginate hydrogels co-functionalized with capture antibodies and poly(ethylene glycol) (PEG) are capable of accomplishing high purity capture of CD34+ cells from whole, untreated blood in microfluidic devices followed by efficient release.10 The release capability of these hydrogels arises from the ability to remove the divalent cations that physically-crosslink the alginate molecules to form the hydrogel using a chelator. The significance of the present work lies in the demonstration of the ability to separatecells expressing two different surface antigens, CD34 and Flk1. The scale-up from single to dual marker-based separation using the functionalized alginate hydrogels is not a trivial extension due to the need to achieve chelator based release in the first separation device without compromising the hydrogel coating in the second separation device and the need to neutralize the chelator molecules after cell release in the first separation stage. We term the approach described herein as “tag-free” as opposed to “label-free” because both stages of capture still require the identification of a capture molecule to bind to a known cell surface receptor.
Experimental Section
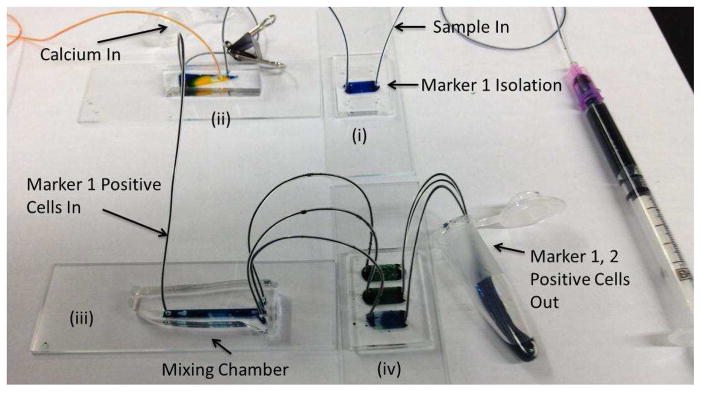
The configuration of microfluidic devices for the dual marker separation described above is shown in Fig. 1. Here, a sample is injected via a syringe pump into the first alginate-based capture stage (“Marker 1 isolation”/stage (i) in Fig. 1). This stage is connected to stage (ii), which is a diverter valve chip.17 In its “closed” configuration, this valve lets the waste from stage (i) pass through to a collection tube. After all of the waste has gone through, the waste stream is closed using a clip.17 Next a solution of ethylene diamine tetraacetic acid (EDTA) is injected into stage (i) to release captured cells while simultaneously a 100 mM calcium chloride solution in [2-(N-morpholino)ethanesulfonic acid] (MES) buffer is injected into stage (ii). The purpose of the calcium chloride is to neutralize the EDTA in the cell suspension emerging from stage (i). To ensure mixing of the calcium chloride solution with this cell suspension, the combined output (which is in laminar flow) is sent into a mixing chamber (stage (iii)) containing herringbone features.18 The mixed solution then enters stage (iv) where the cells expressing receptors for the second capture molecule are captured. The final step in the separation process is the injection of an EDTA solution into the stage (i) inlet, which releases the captured cells from stage (ii). This solution is collected in a tube containing an excess of culture medium in order to minimize any deleterious effect of the EDTA on the cells.
Figure 1.
Sequence of devices for adhesion-based microfluidic separation of cells against multiple surface markers. Following capture and release from device (i), cells expressing CD34 enter device (ii) where a calcium chloride solution is co-injected to neutralize the EDTA present in the cell suspension. Device (iii) mixes the calcium chloride solution and cell suspension. Device (iv) captures cells against the second marker, Flk1, which are eluted out by EDTA solution introduced at the inlet to device (i).
All of the chips shown in Fig. 1 were fabricated using standard poly(dimethylsiloxane) (PDMS)-based soft lithography.19 Each of the capture devices is a post-array fabricated as described previously.10 Herringbone devices were fabricated with a single inlet and three outlets with an overall channel height of approximately 55 μm and herringbone features of approximately 20 μm in height (details in Supporting Information).
Two sets of dual-marker separation experiments were carried out. Preliminary experiments were carried out with ovine EPCs isolated as described by Kaushal et al.1 Cell suspensions were prepared by removing cells from a culture flask with trypsin and diluting to a concentration of 100,000 cells per mL in serum-free EBM-2 medium (Lonza). For experiments, 100 μL volumes of these suspensions were injected into the devices at a flow rate of 5 μL/min. The results of these experiments are shown in Supporting Information.
The second set of experiments was performed with untreated whole human blood. Blood samples were drawn from healthy volunteers in heparin-coated Vacutainer collection tubes under a protocol approved by the Northeastern University Institutional Review Board. The blood samples were injected at a flow rate of 5 μL/min for 60 min.
Following the injection of homogeneous EPC suspensions or whole blood into the capture chip functionalized with anti-CD34, the clip on the valve device (stage (ii) in Fig. 1) was moved to send all effluent from stage (i) into the herringbone chip and the alginate coating within stage (i) was dissolved by introduction of the chelating agent, EDTA, at 10 μL/min for 10 min. The flow of EDTA into stage (i) was accompanied by the injection of calcium chloride solution into the second inlet of the valve chip at 5 μL/min. The EDTA and calcium chloride solution were then mixed in stage (iii). In order to maintain the same flow rate in the second capture device (stage (iv)), the flow exiting the herringbone mixer was divided into three streams and passed through three post array devices containing alginate conjugated with anti-Flk. Cells were released from stage (iv) by introduction of EDTA through the entire system at a flow rate of 30 μL/min (10 μL/min per stage (iv) device) for 10 min.
Characterization of the cells output from the multistage sequence was performed by conjugation of the recovered cells with anti-CD31-PE, anti-Flk1-APC, and anti-CD45-FITC (eBioscience) and enumeration using a Beckman Coulter Quanta SC flow cytometer. The viability of cells emerging from the second capture stage was measured using a calcein/ethidium bromide live/dead staining kit (Invitrogen). For evaluation of capture after the anti-CD34-functionalized device, separate experiments were carried out with both sample types with only one capture device in addition to the multistage experiments.
Results and Discussion
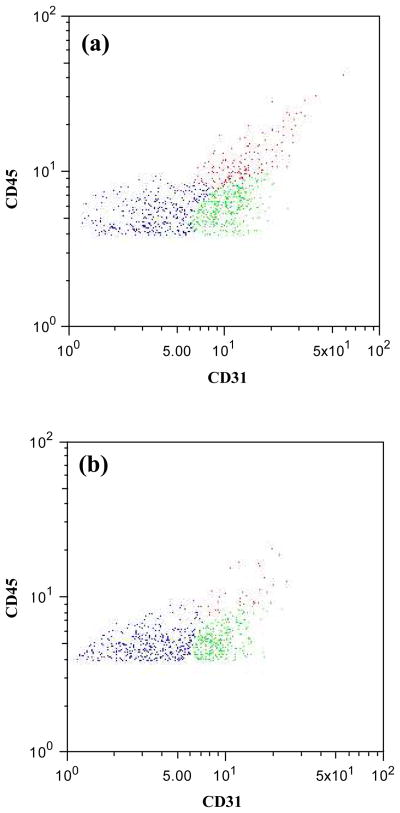
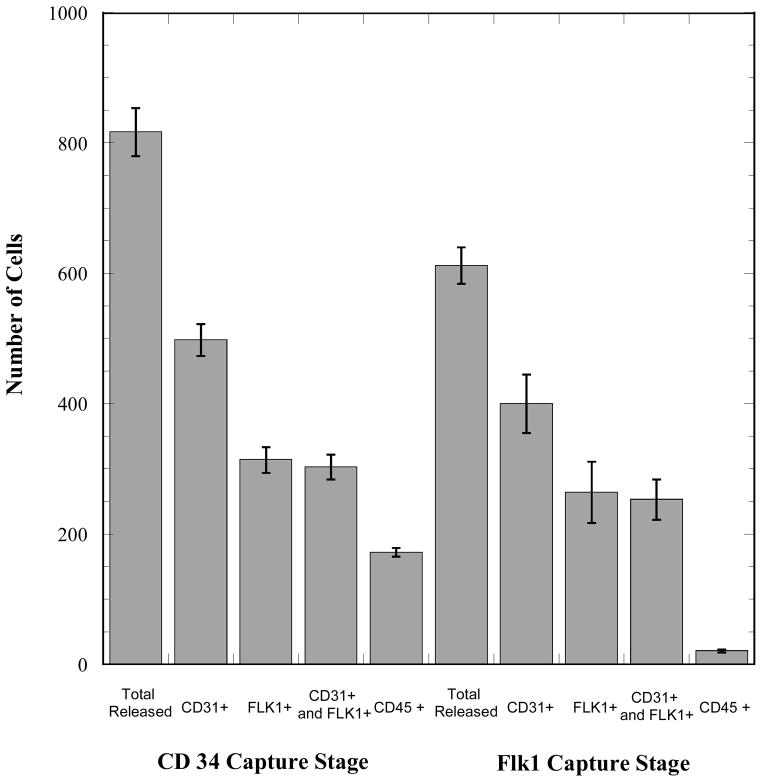
The output cell content from the single and dual capture configurations for cell capture from whole blood are shown in Figs. 2 and 3. The CD34 receptor was targeted for the first stage of cell isolation based on the high expression level of this marker by the target population of EPCs.10, 20 However, within the CD34+ population are CD31+ EPCs21 and CD45+ HSCs.22 The EPC content (CD31+/Flk1+) of the CD34 capture stage output is 37 ± 2% and the HSC content is 21 ± 1% (tabulated composition data in Supporting Information). This suspension is processed by the valve chip and the herringbone mixer before flowing into the Flk1 capture stage. The objective of this second capture stage is to enrich the Flk1+ subset of cells from the CD34+ fraction emerging from the first capture stage. In the suspension recovered from the Flk1 capture stage, which has an overall viability of approximately 75%, a major reduction in the number of CD45+ cells is readily apparent (Figs. 2 and 3; contour plot in Supporting Information, Fig. S.3). The EPC content of the Flk1 capture stage output is approximately 41 ± 4%. The output from this second capture stage also contains non-endothelial CD31+ cells (22 ± 11%), as evidenced by the greater number of CD31+ cells relative to the number of Flk1+ cells. Flk1 is expressed by EPCs as well as by more mature circulating endothelial cells and both of these populations are CD31+ in addition to being CD34+.
Figure 2.
Flow cytometry dot plots representing cells released from (a) the first stage and (b) the second stage. The y-axis represents CD45 expression and the x-axis CD31 expression. Blue dots represent red blood cells, red dots represent CD31+ and CD45+ positive cells (hematopoietic stem cells), green dots represent CD31+ and CD45− cells (EPCs).
Figure 3.
Performance of the multistage capture-release device system in dual-marker separation of EPCs from whole, untreated human blood. The target cells for these experiments were EPCs, which are CD34+ and Flk1+. The first capture stage is designed to selectively capture CD34+ cells. The second stage is designed to further enrich this population and remove CD34+ hematopoietic stem cells. The sample volume for each experiment was 300 μL of whole blood and numbers of cells in this figure are reported without any normalization. Cell counts were determined via flow cytometry. Error bars represent standard deviations based on 3 replicates.
It is known that CD31 is expressed by some subsets of leukocytes which are Flk1−.23, 24 CD31 is also expressed by HSCs,25 which are also Flk1− and hence the total content of the CD31+/Flk1− leukocytes is 19 ± 11%.
The relatively large content of CD31+ leukocytes in the Flk1 capture stage output is surprising given their lack of expression of Flk1 as shown in Fig. 3. While the antibody-functionalized alginate hydrogel coating in the Flk1 capture stage is quite successful in suppressing the adhesion of CD45+ HSCs, the data indicate that is susceptible to the adhesion of these CD31+ and Flk− leukocytes. For tissue engineering applications, recovered cells will typically be placed in culture for expansion prior to seeding on scaffolds; in this situation these leukocytes can be easily removed via a medium change because they are non-adherent.
Lastly, 35 ± 9 % of the Flk1 capture stage output consists of red blood cells (RBCs). The presence of RBCs in this instance is attributable to physical trapping of these cells within the pillar arrays as opposed to extracellular matrix or surface receptor-mediated binding to device surfaces. As with the CD31+/Flk1− leukocytes described above, these cells are non-adherent and can be removed when the recovered cells are placed in culture. Alternatively, mixing the recovered cell suspension with a lysis buffer is another means of eliminating these cells.
As shown in the Supporting Information, when the CD31+/Flk1− leukocytes and RBCs are excluded, the remaining cells consist of 92 ± 11% EPCs and 8 ± 1% HSCs. These values describe the composition of the adhered cells when the output of the Flk1 capture stage is placed in culture.
The overall yield of the capture devices (defined as total cells released versus total cells injected) utilized for CD34 was approximately 60%. This value, which provides a measure of the binding capacity of the hydrogel coating, is consistent between the two sample types examined, namely homogeneous ovine EPC suspensions (Supporting Information) and whole blood. The overall yield of the second capture stage, by contrast, was higher, approximately 75%. This higher value is likely due to lower concentrations of cells entering the second capture stage relative to the starting sample that enters the first capture stage. In general, these levels of overall yield and efficiency of selective capture can be improved by optimizing the pillar array configurations to provide a greater likelihood of capture and by minimizing areas of non-homogeneous coating with the hydrogels. Still, the current work demonstrates reasonable levels of EPC recovery from the 300 μL volumes of the whole blood samples.
The throughput of the two-stage system can be significantly increased by simply adding devices in parallel. Due to the low cost and simplicity of the overall configuration of devices, such parallel operation is relatively straightforward. For example, a parallel configuration of ten 2-stage arrangements set up with two ten-port syringe pumps could process a total volume of 3 mL of whole blood per hour. The significance of the separation platform described herein is the ability to carry out an affinity separation of cells against two different surface markers from a complex sample without any tagging of target cells a priori. The entire sequence of marker 1-based capture, release, mixing, and marker 2-based capture is accomplished simply by flowing a sequence of solutions in the following order (a) sample, (b) rinse solution (MES buffer), (c) EDTA solution which is mixed with calcium chloride solution in the mixing device and carries the recovered cells to the second capture stage, and finally (d) an EDTA solution to recover cells from the second capture stage and carry them into a container with excess culture medium to suppress any deleterious effects of the EDTA on the recovered cells. With the exception of the calcium chloride solution injected into the valve chip, all solutions are injected into the first capture chip, making the entire protocol easy to execute and automate.
Conclusions
Microfluidic devices containing alginate hydrogels functionalized with antibodies can be employed for sequential enrichment of a target cell population against two surface markers. Such enrichment can be accomplished by a serial array of microfluidic devices for capture and intermediate stage devices to neutralize the chelator utilized to release cells from the first stage. The strength of the method lies in the relative simplicity of the layout and the reasonable levels of cell recovery from a complex sample.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge financial support from the National Institutes of Health through grant R01-EB009327 and thank Dr. Juan Melero-Martin for providing the ovine EPCs.
Footnotes
Supporting Information Available: Schematic diagram showing herringbone feature dimensions for mixer stage (Fig. S.1); cell counts from each capture stage for experiments with ovine EPCs (Fig. S.2); flow cytometry contour plot (Fig. S.3); tabulated composition data for output from each stage for experiments with whole blood (Table S.1).
References
- 1.Kaushal S, Amiel GE, Guleserian KJ, Shapira OM, Perry T, Sutherland FW, Rabkin E, Moran AM, Schoen FJ, Atala A, Soker S, Bischoff J, Mayer JE. Nature Medicine. 2001;7(9):1035–1040. doi: 10.1038/nm0901-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eunju O, Lee BH, Ahn HY, Shin JC, Kim HK, Kim M, Park IY, Park YG, Joe YA. FASEB J. 2011;25(1):159–169. doi: 10.1096/fj.10-162040. [DOI] [PubMed] [Google Scholar]
- 3.Kolvenbach R, Kreissig C, Ludwig E, Cagiannos C. Journal of Cardiovascular Surgery. 2007;48(1):39–44. [PubMed] [Google Scholar]
- 4.Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Chemical Engineering Science. 2011;66(7):1508–1522. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dainiak MB, Kumar A, Galaev IY, Mattiasson B. Proc Natl Acad Sci U S A. 2006;103(4):849–854. doi: 10.1073/pnas.0508432103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P, Gao Y, Pappas D. Anal Chem. 2011;83(20):7863–7869. doi: 10.1021/ac201752s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan WH. Anal Chem. 2009;81(3):1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Marshall MK, Garza G, Pappas D. Anal Chem. 2008;80(6):2118–2124. doi: 10.1021/ac702553w. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Phillips JA, Yan JL, Li QG, Fan ZH, Tan WH. Anal Chem. 2009;81(17):7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatch AHA, Hansmann G, Murthy SK. Langmuir. 2011;27(7):4257–4264. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Anal Chem. 2012 doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurkan UA, Anand T, Tas H, Elkan D, Akay A, Keles HO, Demirci U. Lab Chip. 2011;11(23):3979–3989. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim I, Kim YJ, Metais JY, Dunbar CE, Larochelle A. Experimental Hematology. 2012;40(1):84–91. doi: 10.1016/j.exphem.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghaedi M, Soleimani M, Shabani I, Duan YY, Lotfi AS. Cell Mol Biol Lett. 2012;17(1):89–106. doi: 10.2478/s11658-011-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa S, Hirashima M, Matsuyoshi N, Kodama H. Development. 1998;125(9):1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- 16.Chung YS, Zhang WJ, Arentson E, Kingsley PD, Palis J, Choi K. Development. 2002;129(23):5511–5520. doi: 10.1242/dev.00149. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Radisic M, Murthy SK. Lab on a Chip, Chips & Tips online article. published 17 October 2011.
- 18.Stroock AD, Dertinger SK, Whitesides GM, Ajdari A. Anal Chem. 2002;74(20):5306–5312. doi: 10.1021/ac0257389. [DOI] [PubMed] [Google Scholar]
- 19.Xia YN, Whitesides GM. Angewandte Chemie-International Edition. 1998;37(5):551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Plouffe BD, Kniazeva T, Mayer JE, Murthy SK, Sales VL. FASEB J. 2009;23(10):3309–3314. doi: 10.1096/fj.09-130260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timmermans F, Plum J, Yoder MC, Ingram DA, Vandekerckhove B, Case J. J Cell Mol Med. 2009;13(1):87–102. doi: 10.1111/j.1582-4934.2008.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Zeng L, Ding S, Xu K. Transplant Proc. 2010;42(9):3745–3749. doi: 10.1016/j.transproceed.2010.07.094. [DOI] [PubMed] [Google Scholar]
- 23.Newman PJ. J Clin Investig. 1997;99(1):3–7. doi: 10.1172/JCI119129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi TP, Dumont DJ, Conlon RA, Breitman ML, Rossant J. Development. 1993;118(2):489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- 25.Baumann CI, Bailey AS, Li WM, Ferkowicz MJ, Yoder MC, Fleming WH. Blood. 2004;104(4):1010–1016. doi: 10.1182/blood-2004-03-0989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.