Abstract
Although liquid chromatography/mass spectrometry using selected reaction monitoring (LC/SRM-MS) holds great promise for targeted protein analysis, quantification of therapeutic monoclonal antibody (mAb) in tissues represents a daunting challenge due to the extremely-low tissue levels, complexity of tissue matrices, and the absence of an efficient strategy to develop an optimal LC/SRM-MS method. Here we describe a high-throughput, streamlined strategy for the development of sensitive, selective and reliable quantitative methods of mAb in tissue matrices. A sensitive nano-LC/nanospray-MS method was employed to achieve a low lower limit of quantification (LOQ). For selection of signature peptides (SP), the SP candidates were identified by a high-resolution Orbitrap and then optimal SRM conditions for each candidate were obtained using a high-throughput, on-the-fly orthogonal array optimization (OAO) strategy, which is capable of optimizing a large set of SP candidates within a single nano-LC/SRM-MS run. Using the optimized conditions, the candidates were experimentally evaluated for both sensitivity and stability in the target matrices and SP selection was based on the results of the evaluation. Two unique SPs, respectively from the light and heavy chain, were chosen for quantification of each mAb. The use of two SP improves the quantitative reliability by gauging possible degradation/modification of the mAb. Standard mAb proteins with verified purities were utilized for calibration curves, to prevent the quantitative biases that may otherwise occur when synthesized peptides were used as calibrators. We showed a proof of concept by rapidly developing sensitive nano-LC/SRM-MS methods for quantifying two mAb (8c2 and cT84.66) in multiple preclinical tissues. High sensitivity was achieved for both mAb with LOQ ranged from 0.156 to 0.312 μg/g across different tissues, and the overall procedure showed a wide dynamic range (≥500 fold), good accuracy (RE<18.8%) and precision (inter-batch RSD<18.1%, intra-batch RSD<17.2%). The quantitative method was applied to a comprehensive investigation of the steady-state tissue distribution of 8c2 in wild-type mice vs. those deficient in FcRn α-chain, FcγIIb, and FcγRI/FcγRIII, following a chronic dosing regimen. This work represents the first extensive quantification of mAb in tissues by an LC/MS-based method.
INTRODUCTION
Due to their expanding role in the treatment of a variety of refractory diseases such as cancer, autoimmunity and inflammation and neurological disorders, therapeutic monoclonal antibodies (mAb) have attracted growing interest in recent years 1-5. Comparing with small-molecule agents, mAb exhibits high targeting specificity, low off-target toxicity, low clinical risk and prolonged efficacy. Therefore mAb engineering has been predicted as one of the most promising fields in drug development over the next decade 5-7.
Despite of the significant advances2, 4, 8, the exact mechanisms of the absorption, distribution and elimination of mAb, have yet to be fully elucidated. For instance, there has been considerable debate as to whether Fc receptor should be responsible for the low exposure of mAbs in mouse brain 9. In order to investigate the factors that may regulate the tissue exposure and alter the clearance of mAb, a method capable of determining both systematic (e.g. in plasma) and local (e.g. in tissues) levels is critical. Such a method is also valuable for the development and preclinical/clinical evaluation of candidate mAb.
Enzyme-linked immunosorbent assay (ELISA) is the most commonly employed approach for the mAb quantification. Nevertheless, the quantitative accuracy, specificity and reproducibility are often compromised by the interferences from endogenous IgGs, mAb degradation/modification, and by the occurrence of anti-mAb antibody10, 11. Moreover, development of an industry-grade ELISA method for each new mAb is both time-consuming and costly, representing a prominent disadvantage in the drug discovery phase. Finally, ELISA is matrix-dependent and usually could not be transferred across different species/matrices (e.g. from plasma to tissues) 10, 12. By comparison, radiolabeled mAb, which are often used for the semi-quantitative pre-clinical investigation of mAb pharmacokinetics (PK), may be used to track mAb in multiple matrices. Nonetheless, the use of radiolabeled mAb falls short in that its reliability may be compromised by the low stability of the labeled antibody, and that the incorporated tag may cause immunoreactivity and/or alter the characteristics of mAb binding and distribution 10, 13.
Liquid chromatography/mass spectrometry (LC/MS) based methods have emerged as a promising alternative for protein quantification in biological matrices, because these methods provide high specificity, high sensitivity and multiplexing capability, and are often not matrix-selective 14. Recently, several LC/MS methods have been developed for the pre-clinical/clinical investigation of mAb. These methods provide important benefits over the immunoassay methods and therefore could markedly advance the PK research on therapeutic mAbs 15-19. Nevertheless, there are several challenges associated with the development of LC/MS-based methods. First, to achieve a sensitive, selective and accurate analysis, it is critical to choose the optimal signature peptides (SP) for quantification 14. Current methods for selecting SP, such as these via in silico predication or from previous proteomic data, may not be able accurately predict the most sensitive proteolytic peptides and the optimal matrix-dependent parameters such as chemical interferences in samples20. Second, most strategies employ the selected reactions monitoring (SRM) on a triple-quadrupole MS 14. It is desirable to optimize the SRM conditions (e.g. the optimal transitions and the de-cluster/collision energy) for a number of potential SP candidates from a digestion mixture. Many current approaches rely on the synthesis of multiple peptide candidates, which are often costly and time-consuming 21. Third, most methods use a lone SP for the quantification of a mAb, which may carry a significant risk of error where the mAb could be truncated biologically outside the SP domain or certain residues within the SP domain could be biologically modified. Furthermore, the quantitative accuracy could be compromised by the instability of SP, which is often poorly predictable22. Unfortunately, many of the above problems are often overlooked. Moreover, it was estimated that mAb levels in tissues are usually orders-of-magnitude lower than these in plasma 23. Such extremely low concentrations, in conjunction with the high complexity of tissue matrices, pose a daunting challenge for accurate quantification of mAbs in tissues. To our knowledge, so far the available LC/MS-based methods for mAb quantification were exclusively developed for plasma/serum and none is for tissues.
To address these fundamental challenges, we describe a generally applicable strategy that enables high-throughput, streamlined method development for sensitive, selective and reliable quantification of mAb in multiple tissue matrices and plasma. A highly sensitive nano-LC/nanospray-MS method was employed to achieve a low lower limit of quantification (LOQ). A suite of technical advances were employed, which include i) the identification of SP candidates by a high resolution/accuracy Orbitrap analyzer, ii) high-throughput, on-the-fly optimization of SRM conditions for many SP candidates using an orthogonal array optimization (OAO) strategy, iii) the evaluation of stability of SP candidates in the target tissue digests, iv) the use of two unique SP to enhance the quantitative reliability and v) the utilization of pure mAb proteins as calibrators to prevent the quantitative biases that may otherwise occur when synthesized peptides were solely used for calibration 20.
The strategy was developed and then evaluated using the quantification of an anti-topotecan antibody (8c2) and a chimerical anti-CEA antibody (cT84.66) in tissue samples as the model systems. As a proof-of-concept, we applied this method to a comprehensive investigation of the tissue distributions of 8c2 at the steady state after repetitive dosing in different animal models, where the mAb deposition in brain is of particular interest but the levels are expected to be very low6.
EXPERIMENTAL
Reagents and antibody production and purification
cf. SI Experimental.
Animal model and design of animal study
Four different strains of mice were used to investigate the possible role of FcRn and FcγR on the tissue distribution of 8c2. These strains include I) B6.129P2-Fcer1gtm1Rav N12: a dual knockout strain for the activating FcγRI and FcγRIII. II) B6.129S4-Fcgr2btm1TtK N12: a knockout strain for the inhibitory FcγRIIb. III) C57BL/6 strain: used as control/wild type/genetic background strain. Each mouse model is available from Taconic Laboratories. IV) B6.129X1-Fcgrttm1DCR/DCRJ: a knockout for FcRn α-chain. This mouse model is available from the Jackson Laboratory (Bar Harbor, ME). 8c2 (1 mg/kg) was administered by intra-peritoneal injection every 2 days for 73 days. On Day 75, each mouse was perfused using 80 mL heparinized saline followed by 15 ml of freshly prepared protease cocktail inhibitors (Roche diagnostic, IN) to efficiently remove the blood. Briefly, mice were anesthetized with intra-peritoneal injection of ketamine (150 mg/kg) and xylazine (15 mg/kg). Under anesthesia, the abdomen and the thorax area were opened to expose both the heart and the inferior vena cava: a 25.5-gauge cannula was inserted into the apex of the heart to access the left ventricle. The inferior vena cava was cut and heparinized saline was then allowed to flow through the heart and to exit from the cut in the vena cava. The extent of removal of residual blood was quantified by infusing 100 μL 51Cr-labeled red blood cells and then compare the radioactivity counts in the perfused vs. non-perfused tissues. Tissue samples (brain, spleen, liver, lung, heart, kidney) were collected, and frozen in liquid nitrogen until transferred to −80°C. A xenograft model was developed to investigate the distribution of cT84.66 in tumor tissues. LS174T human colon cancer cells (ATCC# CL-188, Manassas, VA, USA), known to express human CEA were used to establish xenografts in male athymic Foxnu mice (Harlan, Indianapolis, IN, USA). All animal procedures were performed in accordance with the protocols approved by the Institutional Animal Use and Care Committee of the University at Buffalo.
Development of nanoLC/SRM-MS method by OAO
mAb proteins were spiked into the extracts of blank tissues at two levels (tissue conc. 1559 μg/g and 78.0 μg/g), then processed and digested following an efficient acetone precipitation/on-pellet digestion procedure (SI Experimental). Accurate peptide identification was performed by analyzing the 1559 μg/g spiked sample on a shallow gradient nano-LC coupled with a high resolution/accuracy Orbitrap-MS. A set of stringent criteria was used to filter out the peptides that are non-specific or with observed or potential modifications. The detailed procedure was described in SI Experimental. Peptide sequence, retention time and fragmentation data (e.g. m/z of precursors and abundant product ions) were recorded for each eligible peptide (Supplementary Table S1).
The 78.0 μg/g spiked sample was analyzed on a TSQ Quantum triple quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with the same nano-LC configuration and gradient condition used for peptide identification. Given the high reproducibility of the nano-LC separation, the peptide candidates were grouped according into retention time windows, and one group was monitored in each window. A predefined L25 (35) orthogonal array design was applied to evaluate the critical SRM parameters including product ion, collision energy (CE) and tube-lens voltage. The ranges of tube-lens voltage and CE were predefined based on the empirical values we established before20. The optimization was conducted by programming 25 independent SRM trials with strategically varied parameters. For each SRM trial, the dwell time was set to 25 ms. The peak area, peak height and S/N ratio were extracted from each SRM channel and exported to a statistic analysis module, which automatically calculate the effect curves for each parameter (c.f. Fig 2 and SI Experimental).
Figure 2.
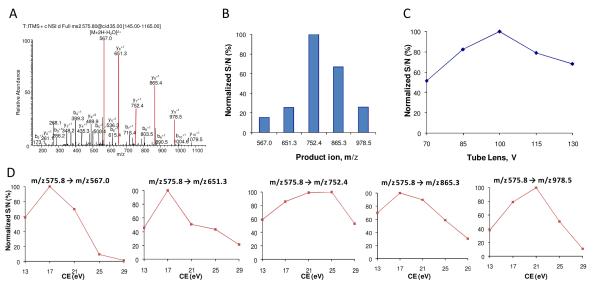
An illustration for OAO data processing and evaluation, using the SP candidate A67TIITDTSSNK77. (A) Product ion spectrum acquired by LTQ-Orbitrap for the doubly charged precursor at m/z 575.8. The five abundant product ions (in red) were selected for OAO procedure. (B) OAO evaluation in tissue revealed the rank of sensitivity achieved by the five products on a SRM-MS platform, which differs from that by the LTQ-Orbitrap. (C) Effect of tube lens offset. (D) Individual effect curves of collision energy for each product ion. Raw data and statistic methods were presented in Supplementary Figure S-2.
Tissue sample homogenization, extraction, cleanup and digestion
The frozen tissues were dissected and ground into a fine powder in the presence of liquid nitrogen. An aliquot of approx. 100 mg of tissue powder was suspended in 300 L PBS buffer (100 mM, pH7.4) containing a protease inhibitor cocktail and homogenized on ice using a Polytron homogenizer (Kinematica, Lucerne, Switzerland). The homogenates were centrifuged at 10,800g for 20min at 4°C and the supernatants were collected. The concentration of total proteins in the PBS extract was measured using the BCA assay (Pierce, Rockford, IL). An aliquot of 50 L PBS extract was mixed with 6 volumes of cold acetone in two steps and then incubated overnight at −20 °C. After centrifugation at 12,000 g for 20 min, the supernatant was removed carefully and the pellet was allowed to air dry. The on-pellet-digestion procedure consisted of two steps. In Step 1, 70 L of Tris buffer (50 mM, pH 8.2) containing the I.S. and trypsin was added to a final enzyme:substrate ratio of 1:30 (w/w). The solution was incubated at 37°C and vortexed at 120 rpm for 6 h to dissolve the pellet. In Step 2, the sample was reduced with 2 mM TCEP and then alkylated with 100 mM iodoacetamide at 37°C for 30 min in the dark. A second aliquot of trypsin was added at an enzyme:substrate ratio of 1:25 (w/w), and the mixture was incubated at 37 °C overnight to achieve complete digestion. Formic acid was added to a final concentration of 1% (v/v) in order to quench the digestion.
Identification of SP candidates using nanoLC-LTQ/Orbitrap
cf. SI Experimental.
NanoLC/SRM-MS
An Eksigent two-dimensional-LC system (Eksigent Technologies, Dublin, CA) equipped with a refrigerated nano-scale autosampler was employed. Solvents used were 0.1% formic acid in water (mobile phase A) and 85% acetonitrile/0.1% formic acid (mobile phase B). Samples were loaded onto a trap column (5 × 0.3 mm ID, packed with Zorbax 3- m C18 material) at an injection volume of 9.5 L, at the flow rate of 10 L/min with 3% B. For nano-LC analysis, the trap was switched online with a 25 cm × 75 m ID fused-silica column packed with 3- m Pepmap C18 particles. The column temperature was maintained at 55°C. The flow rate for separation was 350 nL/min. The gradient consisted of a linear increase of B from 3% to 10% in 5 min, followed by an increase to 22% B in 15 min, and then another increase to 97% B within the next 25 min, and 97% B was held for the final 5 minutes of the gradient. At the end of the run the trap column was back-flushed with 97%B and then equilibrated with 3% B at a flow rate of 10 L/min. The analytical column was re-equilibrated at 3% B at 350 nL/min for 10 min. The total cycle time was around 60 min per injection. The nano-LC system was connected to a TSQ Quantum triple quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a lab-made nanospray interface operated in the positive mode. The spray voltage was set at 2kV and the capillary temperature was set at 320°C. Two SP were simultaneously quantified for each target mAb: A67TIITDTSSNK77 and T156LADGVPSR164 for 8c2; Q58RPEQGLEWIGR69 and A75SNLESGIPVR86 for cT84.66. The quantification were carried out independently with each SP, and then the mean of the two data was reported provided that there was no significant discrepancy (<25% of the larger value) between the two values. Three SRM transitions were monitored for each SP and its IS: two for quantification and one for confirmation. The detailed SRM parameters for the selected SP and IS peptides, including precursor/product ion transitions, optimal collision energy, tube-lens voltages etc were shown in Table 1. The dwell time was 100 ms for each transition. Q1 and Q3 resolutions were both set at 0.7 FWHM (full width at half maximum).
Table 1.
Selected signiture peptides and the optimal SRM conditions for 8c2 and cT84.66
| Target mAb | Signature Peptide | Domain | Optimums of SRM conditions | Role | ||
|---|---|---|---|---|---|---|
| Transition | TL (V) | CE (eV) | ||||
| 8c2 | A67TIITDTSSNK77 | Heavy chain | 575.8→752.3 | 105 | 25 | Quantification |
| (variable region) | 575.8→865.3 | 105 | 17 | Confirmation | ||
| ATIITDTSSNK* | 579.8→760.5 | 105 | 25 | Internal standard | ||
| T156LADGVPSR164 | Light chain | 458.2→701.4 | 100 | 17 | Quantification | |
| (CDR L2) | 458.2→359.3 | 100 | 13 | Confirmation | ||
| TLADGVPSR* | 463.3→711.3 | 100 | 17 | Internal standard | ||
| cT84.66 | Q58RPEQGLEWIGR69 | Heavy chain | 490.3→660.4 | 115 | 21 | Quantification |
| (CDR H2) | 734.9→726.3 | 130 | 18 | Confirmation | ||
| QRPEQGLEWIGR* | 493.9→670.3 | 115 | 21 | Internal standard | ||
| A75SNLESGIPVR86 | Light chain | 571.8→371.7 | 85 | 19 | Quantification | |
| (CDR L2) | 571.8→757.5 | 85 | 25 | Confirmation | ||
| ASNLESGIPVR* | 576.8→381.6 | 85 | 19 | Internal standard | ||
Sythesized IS peptide with K or R coded by 13C and 15N;
Peptide stability assessment, calibrations and method evaluation
cf. SI Experimental.
Comparison of calibration curves for mAb using protein standards vs synthesized peptides
cf. SI Experimental.
Plasma sample analysis
cf. SI Experimental.
RESULTS AND DISCUSSION
1. Overall strategy for developing the quantitative method
The overall strategy consists the following steps: SP candidates were identified confidently by a nano-LC/LTQ/Orbitrap, followed by an on-the-fly OAO optimization to obtain rapidly and accurately the optimal SRM conditions for each candidates; with the optimal SRM conditions, all candidates were then evaluated for sensitivity and stability in the tissue matrix, to facilitate the final selection of the optimal SP. Finally, the sample preparation and nano-LC/SRM-MS procedures for high throughput quantification were developed. With the purpose of achieving high sensitivity and selectivity to the extent possible, each step was thoroughly optimized using 8c2 and cT84.66 as the model analytes.
1.1 Identification of SP candidates with high confidence by nano-LC/LTQ/Orbitrap
Due to the high sequence homology between therapeutic mAb and the endogenous IgG, only a limited number of unique tryptic peptides are eligible as the SP candidates. Therefore, an extensive identification of tryptic peptides is desirable to obtain a comprehensive pool of SP candidates. In this study, an exhaustive trypsin digestion procedure 24 was employed and peptides were resolved thoroughly with a shallow-gradient on a sensitive nano-LC system, followed by Orbitrap detection (Supplementary Experimental).
Under a set of stringent filters (Supplementary Experimental), 27 and 32 unique peptides were identified respectively for 8c2 and cT84.66 with high confidence. Peptides that are not specific to the target mAbs (as revealed by the interrogation of a murine protein database) or bearing either miss-cleavage(s) or known modifications (e.g. Glycosylation) were removed from the list. The peptides surviving this screening process were designated as the SP candidates, which were subjected to further evaluations for sensitivity, stability and interference in the target matrices.
For each SP candidate, important information such as the retention times and abundant product ions were also recorded, which are necessary to setup the OAO procedure. As the charge states of a peptide precursor may markedly affect the sensitivity of SRM 25-27, all identified charge states of the precursors were optimized by OAO and further evaluated (SI Table S1).
1.2 On-the-fly orthogonal array optimization (OAO) of SP candidates
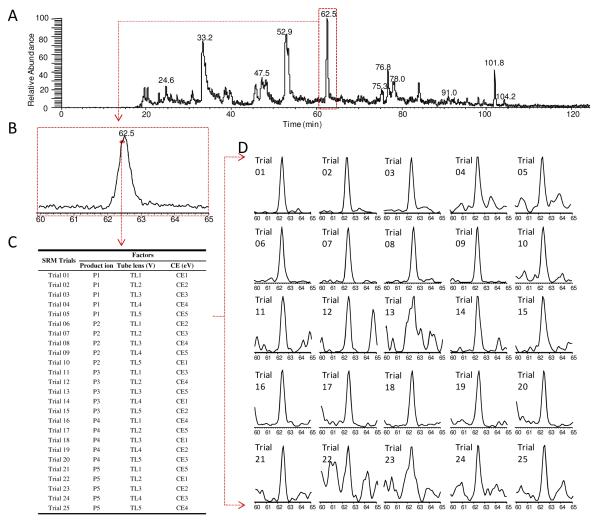
Experimentally optimizing the SRM parameters using target peptides provides highly accurate SRM optimization 14. Nonetheless currently such optimizations mostly employ synthesized peptides, which may be cost-prohibitive since it is often necessary to synthesize multiple candidate peptides (e.g. >5) for each protein in order to enhance the likelihood of obtaining an optimal SP for quantification 28-30. Here we implemented an orthogonal array optimization (OAO) strategy, which provides an accurate, rapid and on-the-fly optimization of multiple key SRM parameters for all candidate peptides in a single LC/MS run, without the need of synthesized peptides20, 31. Details of this technique can be found in a previous publication20. As outlined in Fig 1, OAO approach evaluates the collective effects of several primary factors on the SRM sensitivity. Given the highly reproducible chromatographic separation on the nano-LC system (data not shown), the candidate peptides were grouped into narrow retention time windows for optimization (Fig. 1); the collective effects of fragment ion, collision energy and declustering potential were systematically investigated for each precursor by performing a series of 25 iterative SRM events with parameters dictated by a predefined orthogonal array design (Fig 1C and 1D). Compared to the conventional one-factor-at-a-time approaches, the OAO strategy is advantageous in that it provides high throughput, high reproducibility, and the capacity to evaluate the collective effects of multiple factors. Additionally, the OAO procedure was carried out in the matrix of a target tissue, rendering the optimized conditions directly applicable for the analysis of biological samples. To accelerate the method development, an automated workflow was developed for the analysis of the large data sets acquired by OAO (an example is shown in Supplementary Fig S-1). Based on these results, the optimal SRM parameters for all candidate peptides were readily identified (Table 1). An example of the effect plots for an SP candidate is shown in Figure 2.
Figure 1.
An illustration of the on-the-fly OAO strategy. (A) Total ion chromatogram (TIC) of the 8c2 spiked-in brain extract followed by digestion and nano-LC/SRM-MS analysis. SRM events were grouped by retention time windows. In total 675 SRM transitions, corresponding to 15 peptide candidates for 8c2, were monitored with a 125-min gradient. The full list of candidate peptides and SRM transitions is presented in Supplementary Table S-1. (B) Chromatogram of the scheduled retention time window of 60-65 min, within which several peptides were optimized. The peak represents A67TIITDTSSNK77. Each data point on the profile marks one independent optimization cycle. (C) The orthogonal array design (L25) for SRM optimization of three key factors, where each was investigated at five levels. (D) 25 SRM trials were performed in sequence within a short period (<1s total per cycle, and >20 cycles within the peak elution ). The extracted ion current (XIC) peak was obtained to evaluate the S/N for each trial (c.f. Supplementary Figure S-2).
The OAO procedure enabled efficient method development and optimization. For an example, in this study, the entire development procedure took only one week for the 2 mAbs across 7 different tissues.
1.3 Assessment of the stability of SP candidates
Degradation/modification of SP may severely compromise the accuracy, sensitivity and reproducibility of protein quantification even in the presence of isotope-coded internal standards 20, 22. Since such risks are not readily predictable from the peptide sequence. Therefore, it is necessary to experimentally examine the peptide stability in the digested target matrices prior to SP selection. Using the nano-LC/SRM-MS methods developed by the OAO procedure, such evaluation can be easily performed. Aliquots of spiked target matrix samples (e.g. pooled brain homogenates) were subjected to the precipitation/on-pellet-digestion procedure, and then the peptides stability was evaluated under conditions respectively mimicking the environments to which the tissue digests are exposed during tryptic digestion or queuing in a cooled autosampler. Subsequent LC/MS measurements revealed that a substantial number of candidate peptides for 8c2 and cT84.66 were unstable in tissue digests (Supplementary Fig S-2), and the unstable candidates were disqualified. Among the unstable peptides, some exhibited high LC/MS response, which underscores the risk of selecting SP merely based on the sensitivity achieved. Detailed result and discussion are in the Supplemental Information.
1.4 Selection of two unique signature peptides for each mAb
Using only one SP for the quantification of a protein may carry a significant but “hidden” risk of error due to the possible degradations (e.g. dissociation of light and heavy chain for a mAb) and modifications of the target protein32-35. By comparison, the use of multiple SP to quantify each protein can gauge degradation/modification and thus provides enhanced reliability 20, 36. Nevertheless, as yet most studies employed a lone SP for protein quantification, probably due to the high cost and effort involved to optimize and select multiple SP, each carrying a high sensitivity and stability, for each target protein. In this work, the OAO strategy enabled the extensive optimization and evaluation of numerous candidates with minor efforts and high throughput. Two optimal SP, one from light chain and the other from heavy chain, were identified with ease for each mAb. The quantification results for each sample were validated by examining the discrepancy of the data obtained independently from two SP. When a significant discrepancy is discovered (i.e. > 25% of the higher value), the result will be labeled as “unreliable data” that deserves further investigation.
All SP are located within the variable regions of the two target mAb. Specifically, T156LADGVPSR164 was derived from the second complementarity-determining region on the light chain (CDR L2) of 8c2, while another SP for 8C2, A67TIITDTSSNK77, was from the loop sequence bridging CDR H2 and CDR H3. The two SP of cT84.66, Q58RPEQGLEWIGR69 and A75SNLESGIPVR86, were derived from CDR H2 and CDR L2, respectively.
It is important to note that in certain cases, such as when there is very high sequence homology between the mAb and the endogenous species, it may not be feasible to obtain two unique SP for quantification of a mAb. In these occasions, alternative means are required to control the data quality.
1.5 Quantitative sample preparation
Due to the fact that the mAb tissue levels are much lower than that in plasma, it is critical to sufficiently remove residual blood from tissues, which could otherwise severely compromise the quantitative accuracy for tissue-associated mAbs. To alleviate this problem, we performed a vigorous perfusion on each animal (cf. Experimental). As estimated by infusing 51Cr-labeled red blood cells, the efficiencies for blood removal were >97% for brain and >99.2% for the rest of the tissues. This efficient perfusion minimizes the possible contribution of circulating mAb to the measured levels of tissue-associated mAb. For example, it is estimated that the mAb in residual blood contributes to less than 13% of the measured brain levels, and less than 5% for the rest of tissues.
As most therapeutic mAbs are highly hydrophilic, it could be feasible to extract these proteins with high efficiency using aqueous buffers with minimal or no detergents. Four buffers with increasing strengths for protein extraction, were evaluated to determine the optimal conditions for mAb extraction from target tissues (Supplementary Experimental). The extraction recovery, reproducibility and level of interferences were investigated with the nano-LC/SRM-MS. Interestingly, although the total protein yields (measured by BCA method) using different buffers varied substantially, there was no significant difference in either the yields or the variations of yields of the target mAb (SI Figure S-3). The comparable recoveries of the mAb by a detergent-free buffer (PBS) and detergent-rich buffers were probably due to the high polarity of mAb and the fact that most therapeutic mAb are distributed in tissue interstitial fluid rather than intra-cellular compartments 6, 9, 23. Because using PBS for extraction provides the similar recovery as using stronger buffers while extracting much less tissue proteins, we selected the PBS for the extraction buffer. The completeness of PBS extraction was confirmed by the observation that no target mAb was detected when re-extracting of the residue tissue pellets by a detergent-rich buffer (data not shown).
The extracts were further cleaned and digested using a straightforward, efficient and quantitative precipitation/on-pellet-digestion protocol we described previously 24. The procedure achieved an efficient and complete tryptic digestion while effectively removing non-protein interferences (e.g. salts, lipids, small nucleic acids etc) that would otherwise compromise loading capacity, chromatographic separation and assay robustness. As demonstrated by the validation data in Table 2, the sample preparation procedure is quantitative, reproducible, and applicable to various tissue species.
Table 2. Method sensitivity, linearity , accuracy and precision in tissues.
| Tissue | Sensitivity (LOD, μg/g)a |
Linearity | Accuracy (%)b | Precision (%)b | ||
|---|---|---|---|---|---|---|
| Range (μg/g) | Coefficient (r2) | Intra-batch RSD | Inter-batch RSD | |||
| 8c2 | ||||||
| Brain | 0.062 | 0.156-78.0 | ≥ 0.9901 | 89; 92; 104 | 14; 9; 10 | 18; 10; 11 |
| Liver | 0.031 | 0.312-156 | ≥ 0.9927 | 111; 102; 91 | 10; 8; 9 | 9; 12; 7 |
| Spleen | 0.016 | 0.156-78.0 | ≥ 0.9935 | 109; 107; 94 | 17; 11; 13 | 16; 13; 8 |
| Kidney | 0.031 | 0.156-78.0 | ≥ 0.9903 | 81; 87; 93 | 15; 9; 10 | 14; 11; 9 |
| Heart | 0.031 | 0.156-78.0 | ≥ 0.9895 | 88; 112; 106 | 11; 7; 7 | 7; 12; 5 |
| Lung | 0.078 | 0.312-156 | ≥ 0.9911 | 113; 105; 89 | 13; 9; 6 | 9; 7; 8 |
| cT84.66 | ||||||
| Tumor | 0.061 | 0.307 - 153 | ≥ 0.9890 | 83; 87; 107 | 16; 12; 10 | 14; 9; 9 |
Limit of detection was defined as the tissue concentration for which as least one signature peptide was detectable (S/N > 3);
Accuracy and precision were calculated based on the average of protein concentrations obtained by the two signature peptides. Intrabatch and interbatch precision were measured by repeating five determinations of the QC samples at 3 concentration levels within an analytical batch and on three different analytical batches.
1.6 Nano-LC/SRM-MS strategy for large-scale quantification
A nano-LC/nano-spray configuration was employed to achieve the high sensitivity necessary to quantify mAb in tissues, which are anticipate to present at low levels 6. Several unique features of this system help to alleviate problems often associated with low-flow LC systems and thus enables robust quantification. First, we used a large-ID (300- m) trap column to maintain a linear relationship between the peak areas and injection volumes up to 9.5 g of total tryptic peptides, as compared to less than 1 g peptides on a smaller-ID trap regularly used for a nano-LC system (e.g. 50- m ID). The increase in loading capacity also significantly improved the concentration sensitivity of low-flow-rate LC/MS analysis 37, 38. Second, an elevated separation temperature (55°C) was employed to improve the peak resolution and separation reproducibility, and to reduce the column back-pressure 39. The gradient conditions were optimized to achieve a significant shorter separation than these used for identification and OAO, without compromising the sensitivity and selectivity for the SP. The analytical cycle was approximately 60 min per injection, affording a reasonable throughput for analysis of a large batch of samples.
2. Method evaluation and validation
2.1 Selectivity and sensitivity
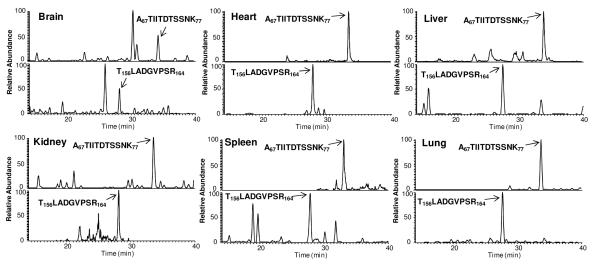
To evaluate the analytical selectivity, blank tissues from vehicle-treated animals (n=3) were analyzed experimentally. No interference was observed. This high degree of selectivity is attributable to the accurate optimization by OAO procedure and the sufficient chromatographic separation. Though the query of murine database resulted in a single hit of one of the chosen SP (T67LADGVPSR77, for 8c2), its endogenous level in target tissues, if expressed at all, was too low to be detected in any of the blank samples. Consequently, this SP is practically unique to 8c2 in the mouse tissues. Typical chromatographic profiles of the selected SP for 8c2 in various tissues are presented in Fig 3. The LOD (S/N=3) were in the range of 0.016~0.078 μg/g tissues for the two target mAbs across all tested tissues (Table 2). The practical LOQ, established based on the anticipated concentrations of 8c2 and cT84.66 in the proposed experimental sets, were significantly higher than the LOD (Table 2). The variation at LLOQ was within 19.6% and 18.5% for 8c2 and cT84.66 respectively (n=3). The good analytical reproducibility and the high S/N at LLOQ render this method highly robust and well-tolerant to possible compromised sensitivity that tends to occur in large-scale analysis.
Figure 3.
Representative nano-LC/SRM-MS chromatograms of the two signature peptides A67TIIDTSSNK77 (Heavy chain) and T156LADGVPSR164 (Light chain) in six different tissues acquired from an 8c2-dosed mouse (Mouse#04 in FcγRIIb(−/−) group). The unambiguous identification of each peak was confirmed by the co-elution of the isotope-labeled IS peptide and by monitoring the peak in qualification channel (Table 1). Nano-LC configuration and gradient settings are described in Experimental.
2.2 Calibration curves and the linear range
To prevent the risk of severe negative biases when use synthesized SP as calibrator20, we employed pure mAb proteins, with purities accurately measured by quantitative amino acid analysis (AAA), to prepare the calibration curve. Here all calibration curves in different tissue matrices were linear over at-least 500-fold concentration ranges, with regression coefficients ranged from 0.9890~0.9935 (Table 2). The wide dynamic range and good linearity can be attributed to the efficient mAb extraction, the quantitative recovery of SP and the minimized matrix effects by the optimized chromatographic separation.
2.3 Precision and accuracy
Precision and accuracy of 8c2 and cT84.66 quantification in target tissues are summarized in Table 2. For 8c2, the intra-assay and inter-assay precisions were respectively 5.3~10.2% and 9.4~18.1% across the six different mouse tissues. As for cT84.66, the intra-assay and inter-assay precisions were 10.1~15.8% and 8.9~14.4% in the xenograft tumor. The error ranged from - 18.8~13.3% for 8c2 in different tissues, and -17.3~7.1% for cT84.66 in tumor, respectively. The above data suggested that the overall sample preparation and analysis are highly quantitative and reproducible. The precision and accuracy were similar across all tissues evaluated, which suggested that tissue matrices showed no significant effect on the accuracy and reproducibility of the developed method.
2.4 Comparison of validation results using protein standards vs. synthesized peptides for calibration curves
In most methods for LC/MS-based protein quantification, synthesized SP (non-quality control samples (QC). In this study, we conducted a rigorous comparison of the accuracy and precision obtained respectively by using synthetic peptides and pure proteins for calibration curves (c.f. Supplementary Experimental). To avoid bias arising from the differences in the purities of these standards, the purities of both the peptide and protein standards were accurately measured by a quantitative amino acid analysis (AAA). The results demonstrated that both the calibration curves were linear over the concentration range investigated, while the peptide calibrations curve showed better regression co-efficiency and lower variability. This was expected because the peptide-based calibration method did not reflect the variations of most sample preparation steps, and thus tended to over-represent the linearity and reproducibility for protein quantification. Using these calibration curves, we attempted to measure the concentrations of 8c2 in QC samples prepared by spiking known concentrations of protein into blank brain tissue (Supplementary Experimental). The protein-based calibration curve provided good accuracy on all QC levels with either of the SP. By comparison, the peptide-derived calibration curves resulted in significant negative biases, and the quantitative data obtained independently with the two SP did not agree with each other (Supplementary Table S-2). The above observations suggest that using pure protein standards is preferable in order to enable an accurate SRM-based quantification of mAb.
3. Application of the developed method in a preclinical study of steady-state 8c2 distributions in various tissues
In order to direct therapeutic efforts, it is of high interest to evaluate the “steady-state” tissue distribution of mAb following long-term, multiple-dosing regimens, as mAb disposition in tissue is often influenced by concentration dependencies associated with receptor saturation (e.g. FcRn) and target-mediated elimination. To our knowledge, such effects have not been systemically investigated, largely due to the lack of a reliable method for the quantification of mAb levels in tissues following chronic dosing, e.g. radiolabeling approaches are not applicable because of the inadequate stability of the radio-labeled mAb. In this study, we employed the developed nano-LC/SRM-MS strategy to characterize the steady-state mAb distributions in tissues following a long-term dosing of 8c2 to mice. The steady-state was achieved by repetitive intraperitoneal injections of 8c2 at 1 mg/kg over a 73-day period (c.f. Experimental). Four strains of mice, including a wild type and three knocked-out strains (FcγRIIb (−/−); FcγRI/RIII (−/−) and FcRn α chain (−/−)), were used to investigate the roles of FcRn and Fcγ receptors on the exposure of 8c2 in tissues. For comparison, plasma samples were also collected (Supplementary Experimental) and analyzed using the same nano-LC/SRM-MS method developed here. To achieve a reliable quantification, the two SP were monitored and quantified in parallel and the mean of the two quantitative values was reported. Any inconsistent result obtained independently by the two SP may reflect quantitative inaccuracy arising from factors such as truncation or modification of the target mAb, and thus was excluded from the final data analysis. Approximately 6% of the results were excluded for this cause.
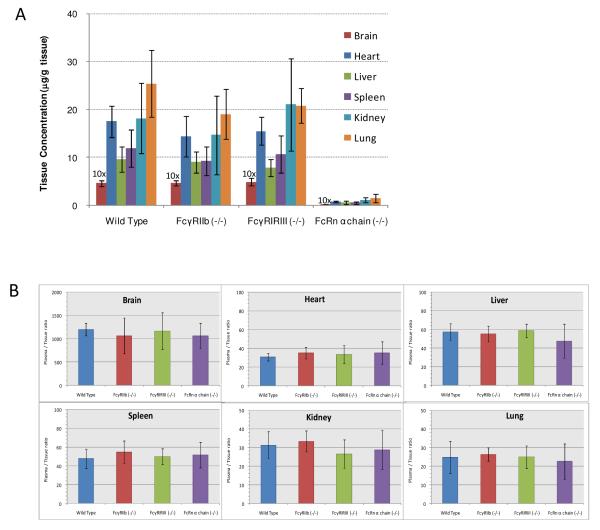
Due to the high sensitivity established in this work (Table 2), 8c2 could be quantified with good S/N in all tissue samples at the steady state. Representative chromatograms are shown in Figure 3. No evidence of accumulation was observed in any of the investigated tissues. For the wild-type group, the 8c2 concentrations in brain, heart, liver, spleen, kidney and lung were 0.466 ± 0.062 μg/g, 17.5 ± 3.2 μg/g, 9.65 ± 2.71 μg/g, 11.9 ± 3.9 μg/g, 18.2 ± 7.3 μg/g and 25.4 ± 7.0 μg/g, respectively (Figure 4A). The low brain distribution suggests that the blood-brain-barrier may limit mAb uptake into the central nervous system, which was consistent with the speculations in previous reports 9. Similar distribution characteristics was also observed in FcγRIIb (−/−) and FcγRI/RIII (−/−) groups. In terms of the levels of 8c2 in individual tissues, no statistically significant difference was observed among the FcγR knock-out groups (FcγRIIb (−/−) and FcγRI/RIII (−/−)) and wild-type group. However, the 8c2 tissue levels in the FcRn α-chain (−/−) group were significantly lower than that in wild-type group (p<0.05). Additionally, this group also showed much lower plasma levels compared to the other three groups (SI Figure S-4). These observations agree with our previous results on a shorter-term dosing study with FcRn-deficient mice9 and strongly suggested the low tissue exposure of 8c2 in FcRn (−/−) group was likely associated with the high rates of IgG catabolism, as expected in the absence of FcRn-mediated protection. Furthermore, comparison of the plasma-to-tissue ratios showed no significant differences across the four groups (Fig 4B), indicating that knocking out either FcγR or FcRn did not significantly affect the plasma-to-tissue ratios of 8c2 (possibly other mAb as well).
Figure 4.
Steady-state tissue distribution of 8c2 in different strains of mice. (A) steady state 8c2 tissue concentrations in wild type (control) , FcRn(−/−), FcγRIIb(−/−), FcγRI/RIII(−/−) and FcRn α chain groups after intraperitoneal injection of 1 mg/kg 8c2 for 73 days (n=4~6). 6% samples (9 out of 144 samples) showed discrepancy in the results by the two independent SPs and thus were not included. The brain concentrations were exaggerated by 10-fold for better visualization. (B) Plasma-to-tissue concentration ratios at the steady state. The density of plasma was assumed to be 1.0 g/mL. The plasma data, measured by the same nano-LC/SRM-MS strategy, were presented separately in Supplementary Figure S-4.
CONCLUSION
Measurement of therapeutic mAb in tissue is critical for evaluating the PK, immunogenicity and toxicology. Although several LC/SRM-MS-based methods have been developed for quantification of mAb in plasma 15-19, an efficient and high-throughput strategy for the development of a sensitive, selective and accurate method to determine tissue distributions remains elusive, due to the technical difficulties specified in the Introduction section. Here we described a streamlined procedure for high-throughput method development for sensitive and robust quantification of the mAb in plasma and various tissues. A sensitive nano-LC/SRM-MS was employed to achieve the LOQ sufficiently low for quantification in tissues. An on-the-fly OAO approach was employed for SRM optimization, which is superior to conventional optimization procedures because it is high-throughput and evaluates the collective effects of multiple factors in the target biological matrices. Prior to SP selection, all candidates were experimentally evaluated for stability and sensitivity in the target matrices, using the optimal SRM conditions obtained by OAO. Two unique SP respectively from the light and heavy chains, were selected for the quantification of each mAb. Using this procedure, it took only one week to develop nano-LC/SRM methods for 8c2 and cT84.66 in tissues and plasma. The extraction and digestion steps were extensively optimized and the quantitative methods were validated. The developed method is substantially more sensitive than those reported previously 11, and thus allowed measurement of the extremely-low levels of mAb in tissues such as the brain.
Moreover, several interesting observations in this study are i) many candidate peptides showed poor stability in tissue digests, underscoring the importance to evaluate peptide stability under the experimental conditions, prior to SP selection; ii) comparison of the quantitative results using pure mAb protein vs synthetic peptides as calibrators revealed that the use of protein standard provided much better quantitative accuracy; and iii) the use of two unique SP enhances the reliability for mAb quantification in tissues.
We applied the developed method to a comprehensive investigation of steady-state mAb tissue distribution in different mouse models that are deficient in various Fc receptors. Understanding of the impact of individual Fc receptors on mAb distribution is highly valuable but remains challenging to date due to the difficulties in analyzing mAb in tissues. This study presented, for the first time, a comprehensive comparison of mAb tissues distribution in various Fc-deficient models following a long-term dosing. A number of novel discoveries were made on the effects of different Fc receptors, which are of essential value for on-going modeling efforts on mAb PK/PD.
Overall, the method developed here provides superior accuracy, the ability to analyze different tissues with one method, and LOQs (0.156 to 0.312 μg/g tissue) compared favorably to immunoassays. Despite ELISA, once successfully developed, may offer higher throughput in sample analysis, the method developed here is more specific, and requires much less time and effort to develop. Though it has not been demonstrated in this study, the streamlined procedure has the potential to develop quantitative methods for a large number of mAbs in a short time period. The procedure is also adaptable to high-throughput development of sensitive and accurate methods for the quantification of mAb and other therapeutic proteins in various pharmaceutical and clinical matrices.
Supplementary Material
Acknowledgements
This work was mainly supported by an industrial consortium grant sponsored by the Center of Protein Therapeutics, SUNY-Buffalo to JQ and JPB, and also was supported principally by NIH grants CA118213 to JPB and DA027528 to JQ, and an AHA award 12SDG9450036 to JQ.
References
- (1).Nelson AL, Dhimolea E, Reichert JM. Nat Rev Drug Discov. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- (2).Chan AC, Carter PJ. Nat Rev Immunol. 2010;10:301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- (3).Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J. J Pharm Sci. 2008;97:2426–2447. doi: 10.1002/jps.21180. [DOI] [PubMed] [Google Scholar]
- (4).Weiner LM, Surana R, Wang S. Nat Rev Immunol. 2010;10:317–327. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC. Nat Biotechnol. 2005;23:1073–1078. doi: 10.1038/nbt0905-1073. [DOI] [PubMed] [Google Scholar]
- (6).Wang W, Wang EQ, Balthasar JP. Clin Pharmacol Ther. 2008;84:548–558. doi: 10.1038/clpt.2008.170. [DOI] [PubMed] [Google Scholar]
- (7).Reichert JM. MAbs. 2011;3:76–99. doi: 10.4161/mabs.3.1.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Carter PJ. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- (9).Garg A, Balthasar JP. AAPS J. 2009;11:553–557. doi: 10.1208/s12248-009-9129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hoofnagle AN, Wener MH. J Immunol Methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Damen CW, Schellens JH, Beijnen JH. Hum Antibodies. 2009;18:47–73. doi: 10.3233/HAB-2009-0206. [DOI] [PubMed] [Google Scholar]
- (12).Ezan E, Dubois M, Becher F. Analyst. 2009;134:825–834. doi: 10.1039/b819706g. [DOI] [PubMed] [Google Scholar]
- (13).Lee JW, Kelley M, King LE, Yang J, Salimi-Moosavi H, Tang MT, Lu JF, Kamerud J, Ahene A, Myler H, Rogers C. AAPS J. 2011;13:99–110. doi: 10.1208/s12248-011-9251-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA. J Proteome Res. 2009;8:787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dubois M, Fenaille F, Clement G, Lechmann M, Tabet JC, Ezan E, Becher F. Anal Chem. 2008;80:1737–1745. doi: 10.1021/ac7021234. [DOI] [PubMed] [Google Scholar]
- (16).Ji C, Sadagopan N, Zhang Y, Lepsy C. Anal Chem. 2009;81:9321–9328. doi: 10.1021/ac901800f. [DOI] [PubMed] [Google Scholar]
- (17).Heudi O, Barteau S, Zimmer D, Schmidt J, Bill K, Lehmann N, Bauer C, Kretz O. Anal Chem. 2008;80:4200–4207. doi: 10.1021/ac800205s. [DOI] [PubMed] [Google Scholar]
- (18).Lesur A, Varesio E, Hopfgartner G. Anal Chem. 2010;82:5227–5237. doi: 10.1021/ac100602d. [DOI] [PubMed] [Google Scholar]
- (19).Hagman C, Ricke D, Ewert S, Bek S, Falchetto R, Bitsch F. Anal Chem. 2008;80:1290–1296. doi: 10.1021/ac702115b. [DOI] [PubMed] [Google Scholar]
- (20).Cao J, Gonzalez-Covarrubias V, Straubinger RM, Wang H, Duan X, Yu H, Qu J, Blanco JG. Anal Chem. 2010;82:2680–2689. doi: 10.1021/ac902314m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, Carr SA, Maccoss MJ. Anal Chem. 2010;82:10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Arsene CG, Ohlendorf R, Burkitt W, Pritchard C, Henrion A, O’Connor G, Bunk DM, Guttler B. Anal Chem. 2008;80:4154–4160. doi: 10.1021/ac7024738. [DOI] [PubMed] [Google Scholar]
- (23).Garg A, Balthasar JP. J Pharmacokinet Pharmacodyn. 2007;34:687–709. doi: 10.1007/s10928-007-9065-1. [DOI] [PubMed] [Google Scholar]
- (24).Duan X, Young R, Straubinger RM, Page B, Cao J, Wang H, Yu H, Canty JM, Qu J. J Proteome Res. 2009;8:2838–2850. doi: 10.1021/pr900001t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Qu J, Straubinger RM. Rapid Commun Mass Spectrom. 2005;19:2857–2864. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- (26).Tu CJ, Li J, Young R, Page BJ, Engler F, Halfon MS, Canty JM, Qu J. Anal Chem. 2011;83:4802–4813. doi: 10.1021/ac200376m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang H, Straubinger RM, Aletta JM, Cao J, Duan X, Yu H, Qu J. J Am Soc Mass Spectrom. 2009;20:507–519. doi: 10.1016/j.jasms.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- (29).Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Fusaro VA, Mani DR, Mesirov JP, Carr SA. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Walker SH, Papas BN, Comins DL, Muddiman DC. Anal Chem. 2010;82:6636–6642. doi: 10.1021/ac101227a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ionescu R, Vlasak J. Anal Chem. 2010;82:3198–3206. doi: 10.1021/ac902752e. [DOI] [PubMed] [Google Scholar]
- (33).Correia IR. MAbs. 2010;2:221–232. doi: 10.4161/mabs.2.3.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hambly DM, Banks DD, Scavezze JL, Siska CC, Gadgil HS. Anal Chem. 2009;81:7454–7459. doi: 10.1021/ac901258g. [DOI] [PubMed] [Google Scholar]
- (35).Brady LJ, Martinez T, Balland A. Anal Chem. 2007;79:9403–9413. doi: 10.1021/ac7017469. [DOI] [PubMed] [Google Scholar]
- (36).Gallien S, Duriez E, Domon B. J Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- (37).Duan X, Weinstock-Guttman B, Wang H, Bang E, Li J, Ramanathan M, Qu J. Anal Chem. 2010;82:2488–2497. doi: 10.1021/ac902869y. [DOI] [PubMed] [Google Scholar]
- (38).Qu J, Qu Y, Straubinger RM. Anal Chem. 2007;79:3786–3793. doi: 10.1021/ac062184r. [DOI] [PubMed] [Google Scholar]
- (39).Farias SE, Kline KG, Klepacki J, Wu CC. Anal Chem. 2010;82:3435–3440. doi: 10.1021/ac100359p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.