Abstract
A rare pathway of HIV-1 resistance to small molecule CCR5 inhibitors such as Vicriviroc (VCV) involves changes solely in the gp41 fusion peptide (FP). Here, we show that the G516V change is critical to VCV resistance in PBMC and TZM-bl cells, although it must be accompanied by either M518V or F519I to have a substantial impact. Modeling VCV inhibition data from the two cell types indicated that G516V allows both double mutants to use VCV-CCR5 complexes for entry. The model further identified F519I as an independent determinant of preference for the unoccupied, high-VCV affinity form of CCR5. From inhibitor-free reversion cultures, we also identified a substitution in the inner domain of gp120, T244A, which appears to counter the resistance phenotype created by the FP substitutions. Examining the interplay of these changes will enhance our understanding of Env complex interactions that influence both HIV-1 entry and resistance to CCR5 inhibitors.
Keywords: Entry inhibitors, CCR5 coreceptor, vicriviroc, escape mutants, gp41, gp120 inner domain
Introduction
Maraviroc (MVC, Selzentry; Pfizer, Inc.) is the first, and so far only, small molecule CCR5 inhibitor to be approved for treating HIV-1 infection (MacArthur and Novak, 2008). Vicriviroc (VCV, Schering-Plough Research Institute; now Merck Research Laboratories) is an investigational compound whose development was abandoned because it did not meet primary efficacy endpoints in late stage clinical trials. The small molecule CCR5 inhibitors all impede the coreceptor engagement step of the HIV-1 entry process via a noncompetitive or allosteric mechanism. To do so, they bind in a hydrophobic pocket within the CCR5 transmembrane helices, stabilizing the receptor in a conformation that is not recognized efficiently by the HIV-1 envelope (Env) glycoprotein, gp120 [reviewed in (Kuhmann and Hartley, 2008)].
The genetic plasticity of HIV-1 allows it to become resistant to antiretroviral drugs in vitro and in vivo, the CCR5 inhibitors being no exception. Escape mutants arise when the virus is cultured for prolonged periods in the presence of these compounds. The genetic and phenotypic changes that take place in vitro are similar to those seen in resistant viruses that emerge in patients [reviewed in (Moore and Kuritzkes, 2009)]. Virologic failure in vivo often involves the expansion of pre-existing, CXCR4-using viruses that are naturally insensitive to CCR5 inhibitors (Westby et al., 2006). However, resistant variants can also emerge de novo. Sometimes the escape pathway involves coreceptor switching, the virus now entering cells via CXCR4, but more usually the resistant variant acquires the ability to use the drug-bound conformation of CCR5 (Moore and Kuritzkes, 2009).
The determinants of resistance generally map to the V3 region of gp120 (Baba et al., 2007; Berro et al., 2012; Kuhmann et al., 2004; Laakso et al., 2007; Ogert et al., 2010; Ogert et al., 2008; Tsibris et al., 2008; Westby et al., 2007). This region plays a role in CCR5 binding but can tolerate a considerable range of sequences without loss of function (Hartley et al., 2005; Kuhmann and Hartley, 2008). Four V3 substitutions (K305R, H308P, A316V and G321E) were, for example, responsible for the resistant phenotype of isolate CC101.19 when the CCR5-using virus CC1/85 was cultured with the small molecule CCR5 inhibitor AD101 (Kuhmann et al., 2004; Trkola et al., 2002). A much rarer, indeed to date unique, route to resistance was undertaken by viruses derived from the same CC1/85 lineage in an AD101/VCV selection culture (Marozsan et al., 2005). The D101.12 escape mutants contained at least one substitution in V3 (H308P) as well as either of the following combinations of substitutions within and immediately downstream of the gp41 fusion peptide (FP): G514V+V535M or M518V+F519L+V535M (Anastassopoulou et al., 2011). We have also described a VCV escape pathway for viruses from the D1/85.16 lineage that involved no V3 changes; instead, resistance mapped to three conservative substitutions in the FP: G516V, M518V and F519I (Anastassopoulou et al., 2009). The phenylalanine (F) residue at position 519 can change either to isoleucine (I) or to leucine (L) depending on the resistance pathway (Anastassopoulou et al., 2011, 2009). Here, we have studied the pathway including the F519I change.
Unlike a clone derived from CC101.19 (V3 mutations), the D1/85.16 cl.23 virus (FP mutations) did not have an increased dependency on the CCR5 N-terminus, and its CCR5 binding site was not obviously more exposed (Berro et al., 2009). The FP-mutant was, however, atypically sensitive to a CCR5 monoclonal antibody (MAb) that stains discrete cell surface clusters of CCR5 that might correspond to lipid rafts (Berro et al., 2011). Nevertheless, the precise mechanism by which the FP changes confer resistance remains to be determined. To better understand the latter escape mechanism, and also Env structure-function relationships that influence HIV-1 entry and its inhibition, we have now studied the effects of the three FP sequence changes, individually and in combination.
Here, we report that the G516V change is critical to VCV resistance, although it must be accompanied by either M518V or F519I to have a substantial impact. Thus, two different double mutants are partially resistant, but all three FP changes are required for complete resistance. During reversion cultures without VCV, the only time the complete cluster of three FP changes persisted was within the genetic context of a gp120 derived from the reference resistant clone of this lineage, D1/85.16 cl.23. However, the FP changes were then accompanied by a substitution in the inner domain of gp120, T244A, that is part of a β-sandwich structure involved in gp41 interactions (Finzi et al., 2010; Pancera et al., 2010; Yang et al., 2003). The emergence of the T244A change counters the VCV-resistance phenotype created by the FP substitutions. These findings add to the complexity of inter-subunit interactions within the Env complex and may eventually help our understanding of how FP mutations can confer CCR5 inhibitor resistance.
Results
Two pair-wise combinations of FP changes confer partial resistance to VCV in PBMC
We have shown that the VCV-resistant phenotypes of the HIV-1 R5 primary isolate D1/85.16, and viruses derived from it, were attributable to three conservative sequence changes (G516V, M518V and F519I) in the gp41 FP (Anastassopoulou et al., 2009). To further study the effects of these substitutions on VCV sensitivity, we performed site-directed mutagenesis to introduce each change individually, and in all possible pair-wise combinations, into the reference parental, VCV-sensitive clone CC1/85 cl.6 (= S). From here on, we refer to the G516V, M518V and F519I changes as a, b and c, respectively (see Table 1 for virus nomenclature). The mutated coding sequences were reconstituted into replication-competent viruses, most of which replicated comparably to their parent. Thus, titers in peripheral blood mononuclear cells (PBMC) on day 7 post-infection were generally in the range 104–105 TCID50/mL (3–5 ng/mL of p24 antigen). However, although the double mutant clone S+a,c was replication-competent, its titers were consistently several-fold lower (103–104 TCID50/mL; ~3 ng/mL of p24).
Table 1.
Nomenclature and properties of viruses used in this study
| Virus | Type | Env origin | Engineered FP mutations (*) |
|---|---|---|---|
| S | Parental, VCV-sensitive clone | CC1/85 cl.6 | N/A |
| S+a | Single mutant parental clone | CC1/85 cl.6 | G516V |
| S+b | Single mutant parental clone | CC1/85 cl.6 | M518V |
| S+c | Single mutant parental clone | CC1/85 cl.6 | F519I |
| S+a,b (.r#) | Double mutant parental clone (Clone reversion#) | CC1/85 cl.6 | G516V, M518V |
| S+b,c | Double mutant parental clone | CC1/85 cl.6 | M518V, F519I |
| S+a,c (.r#) | Double mutant parental clone(Clone reversion#) | CC1/85 cl.6 | G516V, F519I |
| S+a,b,c (.r#) | Triple mutant parental clone (Clone reversion#) | CC1/85 cl.6 | G516V, M518V, F519I |
| R (.r#) | VCV-resistant clone (Clone reversion#) | D1/85.16 cl.23 | N/A |
N/A, not applicable.
Clones designated as R.r, S+a,b.r, S+a,c.r, and S+a,b,c.r followed by a number were derived from the PBMC cultures in which the reference VCV-resistant clone R, the double mutants S+a,b and S+a,c and the engineered resistant triple mutant S+a,b,c, correspondingly, were cultured for a prolonged period in the absence of VCV. The designation.
r# indicates the number of weekly passages the virus was in a reversion culture, without an inhibitor present.
Amino acid numbering is based on HXB2 Env.
All the single and double mutants were tested for VCV sensitivity in a PBMC infection-inhibition assay, alongside the reference VCV-sensitive and –resistant clones S and D1/85.16 cl.23 (= R), and also the engineered, VCV-resistant triple mutant S+a,b,c [previously referred to as S+3FP, (Anastassopoulou et al., 2009)] (Fig.1). For each virus, we determined the half-maximum inhibitory concentration (IC50) and the maximum percent inhibition (MPI) value (Table 2). The dose-response curves of CCR5 inhibitor-resistant viruses are characterized by the “plateau effect,” i.e., the inability of an inhibitor to reduce infectivity below a certain threshold value, the MPI. This parameter reflects the relative ability of a virus to use the inhibitor-bound and inhibitor-free forms of CCR5 for entry; the lower the MPI, the more efficiently a resistant virus uses the inhibitor-CCR5 complex (Pugach et al., 2007; Westby et al., 2007).
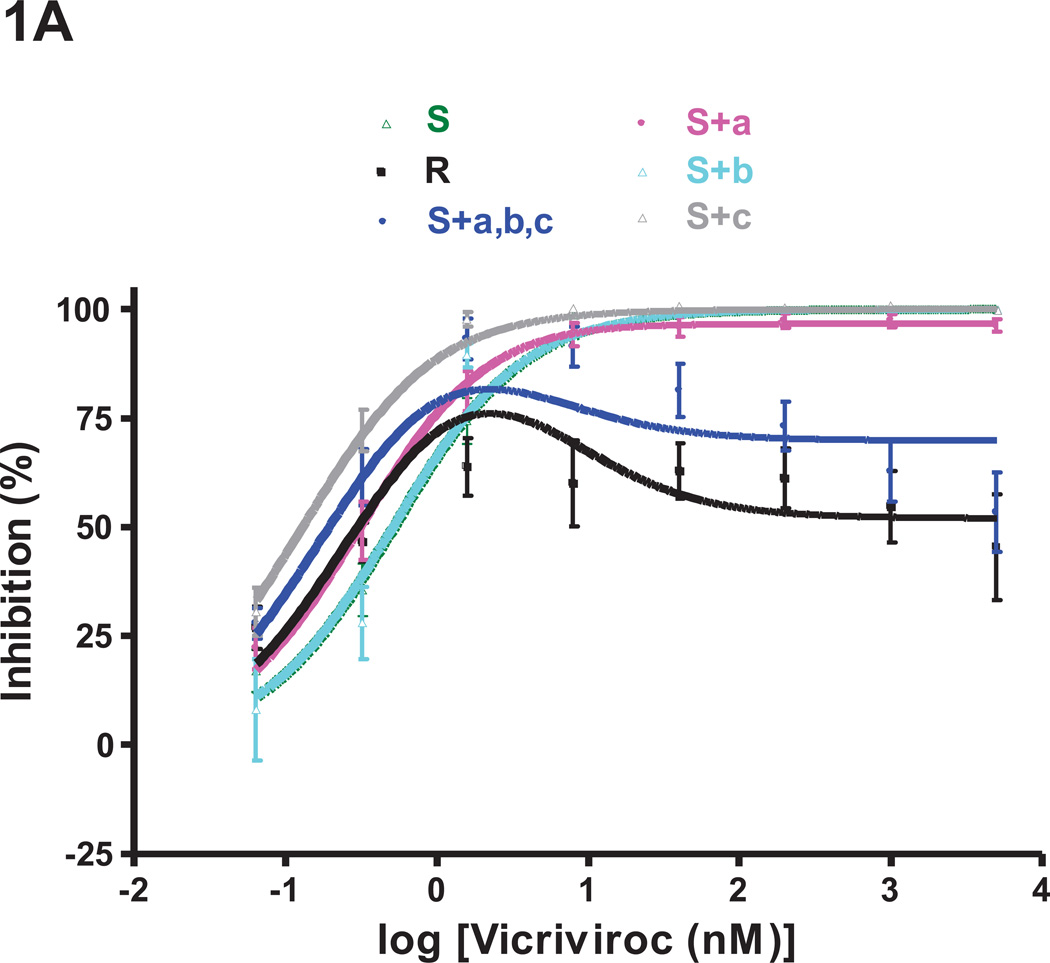
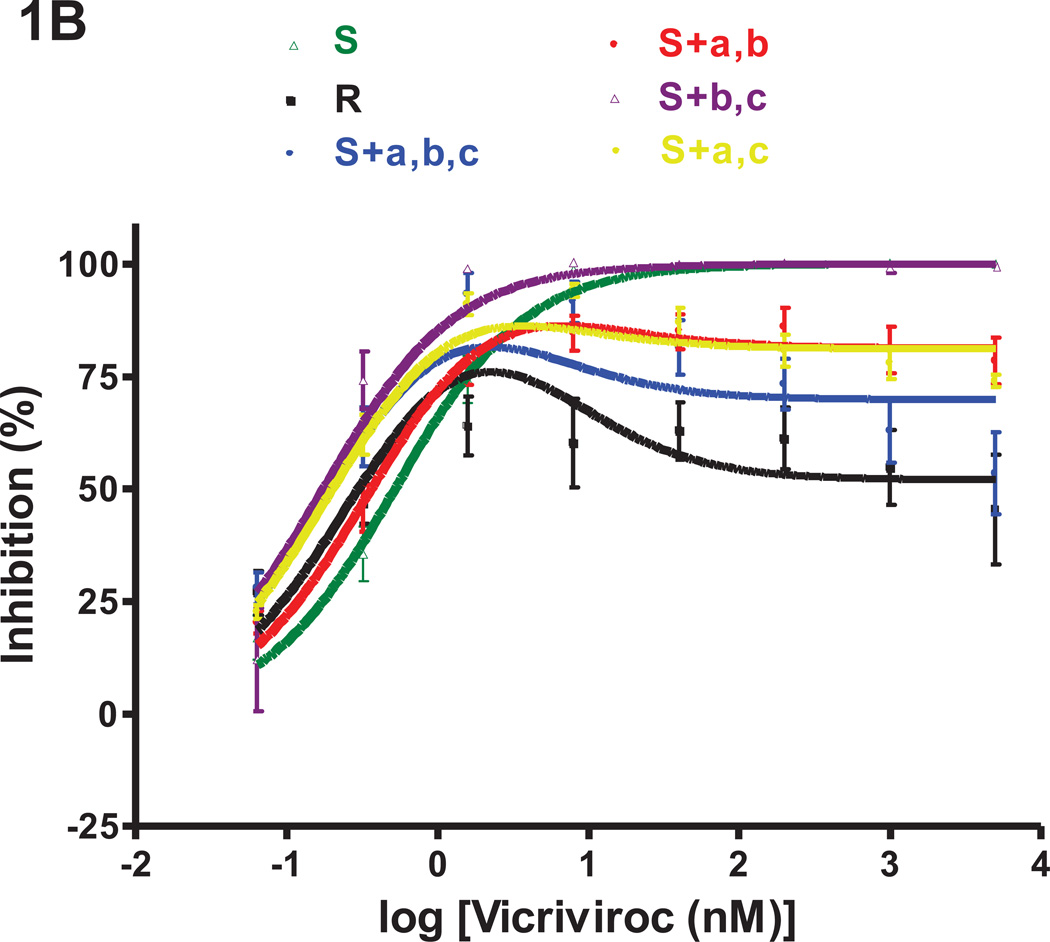
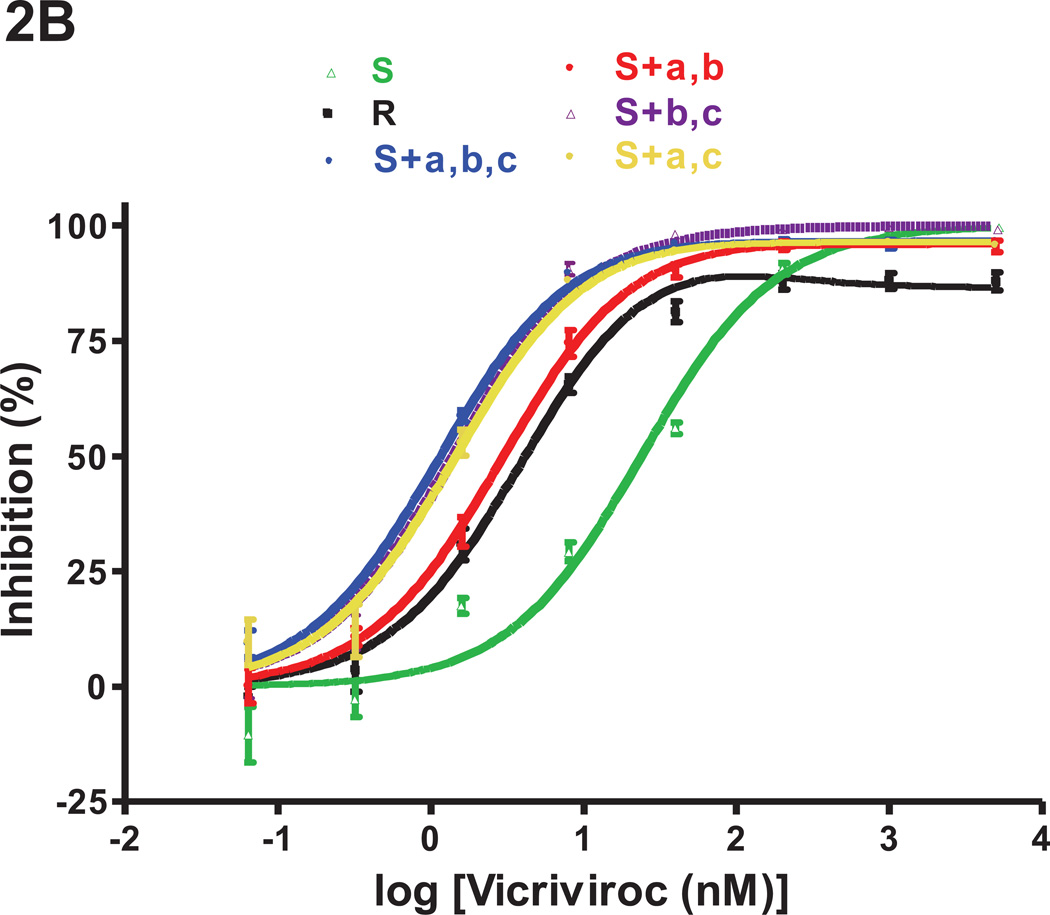
Fig.1. Effects of the three FP mutations individually and in combination on VCV resistance on primary cells.
The engineered FP mutant viruses were tested for VCV sensitivity in a PBMC-based, multi-cycle replication assay. For comparison, the parental, VCV-sensitive clone S, the VCV-resistant clone R and the engineered resistant triple mutant S+a,b,c [previously referred to as S+3FP, (Anastassopoulou et al., 2009)] were also tested in the same experiments. Each experiment was performed on a pool of PBMC from two individuals. For clarity, the dose-response curves are shown in separate panels for (A) single and (B) double FP mutants. The results shown are the average of 7–10 independent experiments each performed using triplicate wells, with the error bars indicating the SEM.
Table 2.
VCV inhibition of replication of FP mutants in PBMC and TZM-bl cellsa
| PBMC | TZM-bl | ||||
|---|---|---|---|---|---|
| Virus | MPI (%) | IC50 | MPI (%) | IC50 | VCV phenotype in PBMCb |
| S | 100 | 0.51 | 99 | 23 | Sensitive |
| S+a | 97 | 0.31 | 99 | 4.6 | Weakly resistant |
| S+b | 100 | 0.51 | 100 | 10 | Sensitive |
| S+c | 100 | 0.13 | 100 | 1.4 | Sensitive |
| S+a,b | 86 | 0.36 | 96 | 3.0 | Intermediately resistant |
| S+b,c | 100 | 0.17 | 100 | 1.4 | Sensitive |
| S+a,c | 86 | 0.20 | 96 | 1.4 | Intermediately resistant |
| S+a,b,c | 82 | 0.19 | 97 | 1.1 | Fully resistant |
| R | 76 | 0.30 | 87 | 4.1 | Fully resistant |
MPI, maximum percent inhibition.
The changes marked as a, b and c are G516V, M518V and F519I, respectively.
The model function for inhibition, Q=(1−(1−(C/(Kdi+C))+w *(C/(22*Kdi+C))))*100%, was fitted to the average PBMC and TZM-bl cell data (see Figs. 1 and 2), to determine MPI and IC50 values (see Materials and Methods and Results; R2 values are given in Table 3). The IC50 is defined as the VCV concentration (C) that yields 50% inhibition (Q). The IC50 therefore applies only when MPI > 50%; accordingly, it can be substantially larger than the VCV concentration that would inhibit infection to an extent equal to 50% of the MPI value. MPI is defined as the upper plateau for sigmoid curves, or as the maximum inhibition value for peaked curves.
Viruses with MPI values for VCV inhibition in PBMC in the range 90–98% are arbitrarily defined as weakly resistant, 85–90% as having intermediate resistance, and <85% fully resistant. The fully resistant viruses, according to this definition, are highlighted in bold.
Individually, none of the FP changes had a marked impact on VCV sensitivity (Fig.1A). The inhibition curve for the S+b mutant (M518V substitution) completely coincided with that of the sensitive virus S. The curves for mutants S+a and S+c were similar in shape to that for S, and not to the unusual, biphasic inhibition curves seen with the resistant viruses R and S+a,b,c (i.e., S+3FP). We have described and modeled the latter type of curve previously, noting the substantial influence of PBMC donors on the complex curve-shapes that can arise with VCV-resistant viruses (Anastassopoulou et al., 2011, 2009). In the present study, both R and S+a,b,c were substantially inhibited (~50–80%) by low VCV concentrations of ~0.3–2 nM (Fig.1). The MPI values for R and S+a,b,c were 76% and 82%, respectively, at a VCV concentration of ~1.6 nM, while the corresponding plateau values at higher concentrations (>40 nM) were 52% and 70% (Fig.1, Table 2).
When the PBMC inhibition curves for the single FP mutants were analyzed in more detail, it was apparent that the G516V change does confer very low resistance to the S+a mutant, as manifested by the marginal (~3%) level of replication (MPI = 97%) that can be detected at high VCV concentrations (up to 5 µM). The G516V change also causes a leftward shift of the dose-response curve and, thus, a slight reduction in IC50 compared to the sensitive, virus S (0.31 nM vs. 0.51 nM, for S+a and S, respectively; Fig.1A, Table 2). The F519I substitution causes a more pronounced leftward shift and a further reduction in the IC50 value (0.13 nM vs. 0.51 nM, for S+c and S, respectively), but without affecting the MPI value (= 100%) (Fig.1A, Table 2). The S+c mutant is clearly a VCV-sensitive virus, indeed one that is hypersensitive to low VCV concentrations (~0.3–2 nM) compared to S+a, S+b and S (Fig.1A).
Although all three FP changes are required for complete VCV-resistance, two double mutants, S+a,b and S+a,c (changes G516V+M518V and G516V+F519I, respectively), were partially resistant (Fig.1B). Thus, these viruses both had MPI values of 86 and plateau values (see Methods) of ~81 at VCV concentrations above ~40 nM (Fig.1B, Table 2). At low concentrations (~0.3–2 nM), the third substitution, F519I, confers an increase in VCV sensitivity; thus under these conditions, S+a,c is more sensitive than S+a,b (IC50 0.20 vs. 0.36 nM, respectively). The S+b,c virus is VCV-sensitive but its IC50 was reduced to 0.17 nM, presumably due to the impact of the F519I change.
Overall, the G516V change appears to play the most important role in VCV-resistance; when it is accompanied by either of the M518V or F519I substitutions, the resulting virus is significantly, but incompletely, resistant. All three FP changes are required for complete resistance.
FP changes that confer partial VCV-resistance have cell type-dependent effects
To further study the phenotype conferred by FP changes, we tested the replication-competent single and double mutants for VCV resistance in TZM-bl cells, again in comparison to the reference parental and VCV-resistant clones S and R, and the engineered resistant triple mutant S+a,b,c (Fig.2). The target cell is a known influence on how resistance to small molecule CCR5 inhibitors is manifested, particularly MPI values (Anastassopoulou et al., 2011, 2009; Ogert et al., 2008; Pugach et al., 2007; Westby et al., 2007). Furthermore, the differences between the MPI values of VCV-resistant and - sensitive viruses are generally much smaller in cell lines engineered to express CCR5, such as TZM-bl, than in PBMC (Anastassopoulou et al., 2011, 2009). The VCV-resistant viruses carrying FP changes also have paradoxically lower IC50 values than wild-type viruses, particularly in TZM-bl cells. Hence although by one measure (MPI) the FP-mutant viruses display a modest level of resistance in TZM-bl cells, by another (IC50) they appear to be hypersensitive (Anastassopoulou et al., 2011, 2009). Our present results are consistent with these earlier findings in this regard.
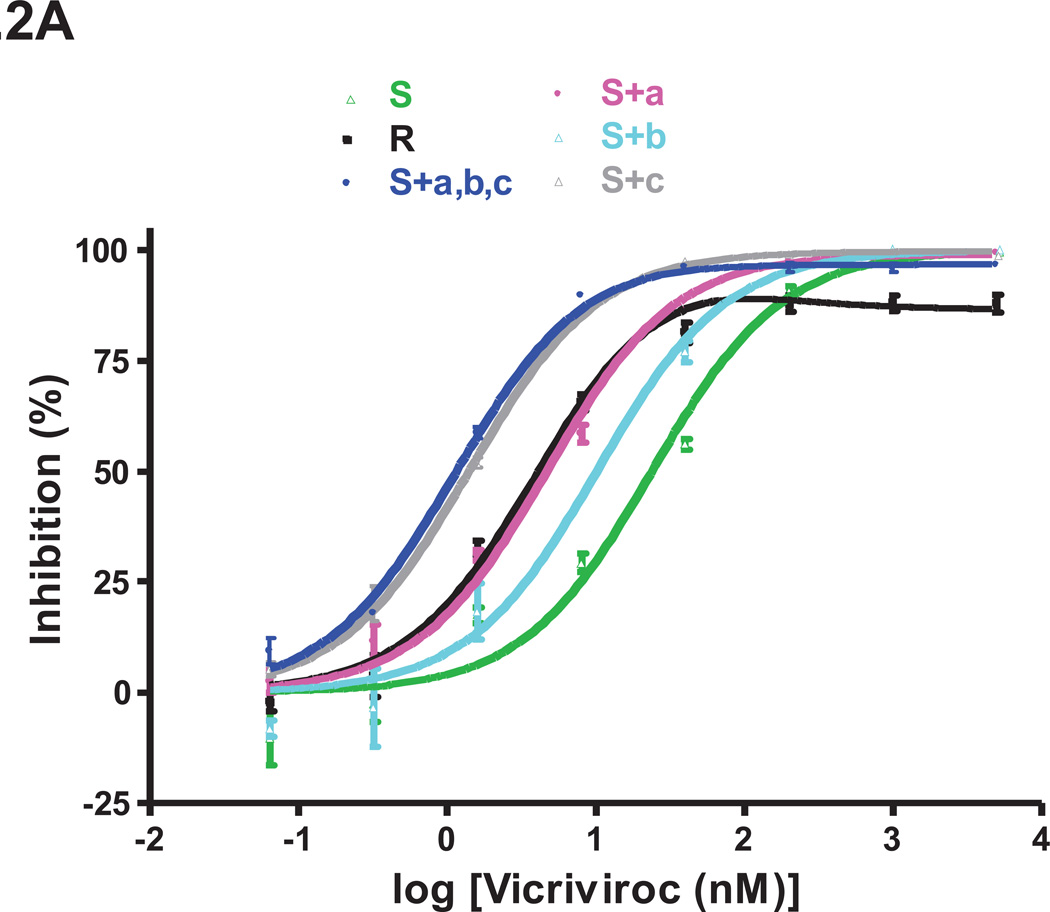
Fig.2. Determinants of VCV resistance in TZM-bl cells.
The same clonal viruses studied in PBMC were assessed for VCV sensitivity in a TZM-bl cell-based, single-cycle infection assay. For clarity, the dose-response curves are shown in separate panels for (A) single and (B) double FP mutants. The results shown are the average of 3 independent experiments, with the error bars indicating the SEM.
In the TZM-bl cell assay, the VCV sensitivity profiles of the single FP mutants were similar to those seen in PBMC, with a few exceptions. As before, the G516V change in the S+a mutant caused a leftward shift of the inhibition curve and a consequent IC50 reduction compared to S (4.6 nM vs. 23 nM; Fig.2, Table 2). The very low level, but consistently observed, replication of the S+a mutant at very high VCV concentrations in PBMC (MPI = 97%) was not seen with TZM-bl cells (MPI = 99%; Table 2). The S+b mutant was clearly VCV-sensitive in TZM-bl cells, albeit with a reduced IC50 compared to S (10 nM vs. 23 nM; Fig.2, Table 2). Among the three FP changes, F519I caused the biggest leftward shift of the dose-response curve, and thus the greatest reduction in IC50 (1.4 nM for S+c vs. 23 nM for S; Fig.2, Table 2). Nonetheless, S+c was completely sensitive to VCV in TZM-bl cells, just as it was in PBMC (MPI = 100% in both cell types; Figs.1, 2 and Table 2).
The VCV sensitivity profiles for the double FP mutants were similar in TZM-bl cells and PBMC. Thus, the MPI values of 96% in the TZM-bl cells were consistently below 100% for both S+a,b and S+a,c, indicating that a very low level of replication could still occur at high inhibitor concentrations. The paradoxical reductions in IC50 values in TZM-bl cells were again seen for both double mutants (3.0 nM and 1.4 nM for S+a,b and S+a,c vs. 23 nM for S; Fig.2 and Table 2). Similar phenotypes were also observed when the corresponding Env-pseudotyped viruses were tested in U87.CD4.CCR5 cells (data not shown). The F519I change caused the most pronounced leftward shift in the dose-response curves in both PBMC and TZM-bl cells, and consequently the greatest IC50 reductions (Figs.1,2). Thus, as in PBMC, S+b,c was a fully sensitive virus (MPI = 100%) in TZM-bl cells, with a reduced IC50 compared to S (1.4 nM vs. 23 nM) that reflected the influence of the F519I change (Figs.1,2 and Table 2).
Modeling VCV resistance of FP mutants
We applied a previously described model (Anastassopoulou et al., 2009) to VCV inhibition data derived from both PBMC and TZM-bl cells. The model is a quantitative formulation of the hypothesis that two distinct forms of CCR5 with high or low affinity for VCV exist in varying proportions on the surfaces of different cell types. The inhibition of viral infectivity, Q, is a function of the VCV concentration, C, according to the equation:
Q = (1−(1−(C/(Kdi+C))+w*(C/(Kde+C))))*100%
where Kdi is the postulated dissociation constant for any VCV binding that blocks viral usage of CCR5, and Kde, is the corresponding constant for any binding that allows the coreceptor still to be used. The w term reflects the relative abundance of the CCR5 form that can bind VCV with low affinity, and also the efficacy with which a resistant virus can use that form of CCR5 when it is complexed with VCV. According to the model, a VCV-sensitive virus may be able to use both the high- and low-affinity forms of CCR5 to various extents when they are unoccupied, whereas resistant viruses use preferentially the unoccupied high-affinity receptor and the occupied low-affinity receptor (Anastassopoulou et al., 2009). As previously described, resistant viruses are characterized by w>0 (large values on PBMC, small on TZM-bl cells) and lower Kdi values than apply to sensitive viruses on TZM-bl cells.
To investigate whether these two features of the resistant phenotype could be attributed to different changes in the FP, we modeled VCV inhibition data derived using the nine viruses described above and both PBMC (means of 7–10 replicates) and TZM-bl cells (means of 3 replicates). Nine VCV concentrations were used in the titrations, and the modeling was based on values calculated from 963 individual inhibition data points (a single outlying value was censored from this matrix). Even so, the data were insufficient to determine all three parameters (w, Kdi and Kde) simultaneously with acceptable precision. Therefore, and also to make the w and Kd values comparable across the panel of viruses, we set the ratio of Kde/Kdi to 22, a value between previous estimates and one that was amenable to good fits for all resistant viruses (Anastassopoulou et al., 2011, 2009). All the resulting non-linear regression fits (Prism, Graphpad) yielded R2 values > 0.9, in most cases ~1 (Table 3).
Table 3.
Modeling VCV inhibition of viral usage of two hypothetical forms of CCR5
| Virus: | S | R | S+a | S+b | S+c | S+a,b | S+b,c | S+a,c | S+a,b,c | |
|---|---|---|---|---|---|---|---|---|---|---|
| PBMC | Kdia | 0.52±0.051 | 0.29±0.074 | 0.31±0.021 | 0.51±0.11 | 0.13±0.010 | 0.35±0.034 | 0.17±0.042 | 0.15±0.052 | 0.18±0.066 |
| wb | 0±0.016 | 0.47±0.037 | 0.033±0.011 | 0±0.037 | 0±0.011 | 0.19±0.015 | 0±0.036 | 0.15±0.050 | 0.30±0.050 | |
| R2c | 0.99 | 0.90 | 1.0 | 0.98 | 1.0 | 0.99 | 0.97 | 0.92 | 0.90 | |
| TZM-bl | Kdia | 24±5.1 | 4.0±0.40 | 4.7±0.51 | 10±1.5 | 1.4±0.039 | 3.0±0.13 | 1.4±0.16 | 1.4±0.16 | 1.1±0.091 |
| wb | 0±0.059 | 0.14±0.020 | 0.0096±0.023 | 0±0.035 | 0.0021±0.0051 | 0.041±0.0086 | 0±0.021 | 0.035±0.020 | 0.033±0.014 | |
| R2c | 0.98 | 1.0 | 0.99 | 0.99 | 1.0 | 1.0 | 0.99 | 0.99 | 1.0 | |
Modeled dissociation constants, Kdi, have the dimension of concentration [nM]. The values are not measurements of affinity: That is only one interpretation under the simplest assumptions in the model, as explained (Anastassopoulou et al., 2009).
Values of w ≥ 20% of that for R virus on the respective cells are in italics; values lower than that are given in bold.
R2 describes the goodness of the fit of the model function, Q=(1−(1−(C/(Kdi+C))+w*(C/(22*Kdi+C))))*100%, by non-linear regression.
The modeling results for the reference viruses S and R reproduce what we reported previously (Anastassopoulou et al., 2011, 2009). Thus, w = 0 for S, and w was substantial for R in both cell types but larger for PBMC than TZM-bl cells (Table 3). In comparison, the three single-point mutants and the S+b,c double mutant gave w values of 0 or close to it (only for the S+a single mutant were w values marginally, but consistently, above 0). The S+a,b and S+a,c double mutants and the S+a,b,c triple mutant all reproduced the R phenotype, with w>0 to different extents. The Kdi values were less variable from virus to virus with PBMC than TZM-bl cells. On both cell types, the greatest differences in Kdi compared to the S virus were seen with S+c and all the other mutants containing the F519I change (i.e., mutation-c). In contrast, the Kdi value for the single S+b mutant was the most similar to that for S.
These results indicate that the capacity of VCV-resistant viruses to use VCV-CCR5 complexes is separable genetically from their apparent preference for using unoccupied forms of CCR5 that have higher affinities for VCV, as depicted graphically in Fig.3. Thus, the G516V change (i.e., mutation-a) is a crucial but insufficient contributor to the usage of VCV-CCR5 complexes (Fig.3A); the F519I mutation (i.e., mutation-c) is an independent determinant of preference for the unoccupied high-affinity form of CCR5 (Fig.3B). The M518V (i.e., mutation-b) and F519I changes can both synergize with G516V to confer the ability to use VCV-CCR5 complexes. When M518V is added to the other two changes, it has an incremental effect on the capacity to use VCV-CCR5 complexes on PBMC. We conclude that M518V and F519I complement the G516V substitution in qualitatively distinct ways, but together the three changes largely recreate the VCV-resistance phenotype of the reference virus, R.
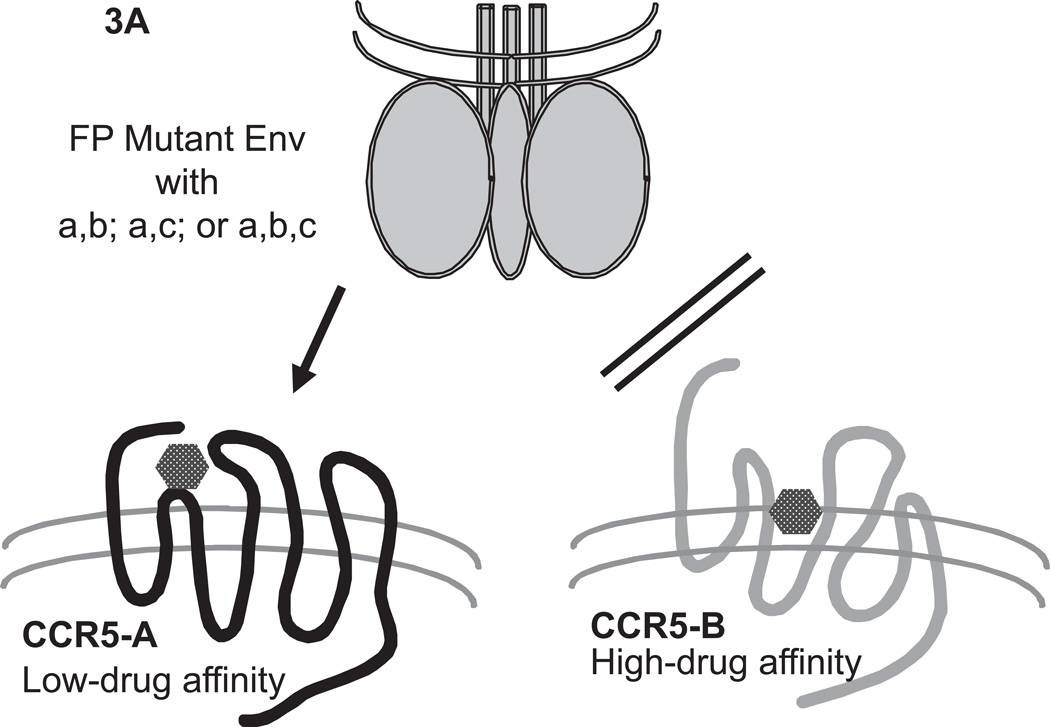
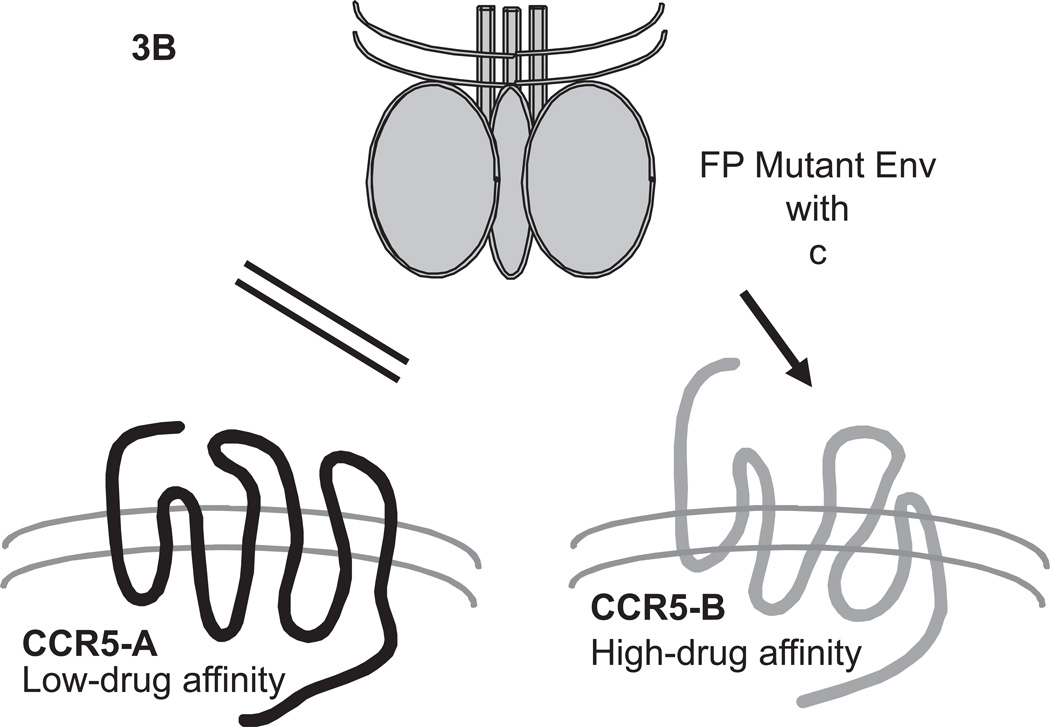
Fig.3. Pictorial model of the usage of different CCR5 conformations by VCV-resistant FP mutants.
CCR5 at the cell surface is postulated to exist in two distinct forms that could be conformers (conformational isomers) or differentially tyrosine-sulfated forms. The two forms of the coreceptor, CCR5-A (black) and CCR5-B (gray), are assumed to bind small molecule CCR5 inhibitors such as VCV with low and high affinity, respectively. According to the model (Anastassopoulou et al., 2009), wild-type virus cannot enter via the VCV-complexes of either CCR5-A or CCR5-B, although it can use both forms of unligated CCR5 for entry, possibly with a preference between them. (A) The VCV-resistant FP mutant virus (trimeric Env sketched in gray on top) has acquired the ability to use specifically the VCV-complex of CCR5-A. The sequence changes that allow for the specific usage of CCR5-A complexed with VCV involve either two double FP combinations (a,b or a,c), or all three FP changes (a,b,c; i.e. G516V, M518V and F519I, respectively). (B) Mutation-c (F519I) confers or enhances the capacity of resistant virus also to enter via the unligated form of CCR5-B. The VCV-resistant FP mutant virus cannot use the unligated form of CCR5-A. This figure is adapted from Fig.S5 of (Anastassopoulou et al., 2009).
Stability of the VCV-resistant phenotypes of FP mutants in the absence of the inhibitor
The genetic routes to VCV resistance may involve solely V3 changes [as in isolate CC101.19, (Kuhmann et al., 2004)]; solely FP changes [as in isolate D1/85.16, (Anastassopoulou et al., 2009)]; or a combination of substitutions in gp41 acting in concert with at least the H308P change in V3 [as in resistant viruses from the D101.12 lineage, (Anastassopoulou et al., 2011)]. But whatever pathway was taken, the resulting isolates all have highly stable VCV-resistance phenotypes in PBMC, with no apparent fitness loss (Anastassopoulou et al., 2011, 2009, 2007). Here, we have assessed the stability of individual clones of the D1/85.16 lineage.
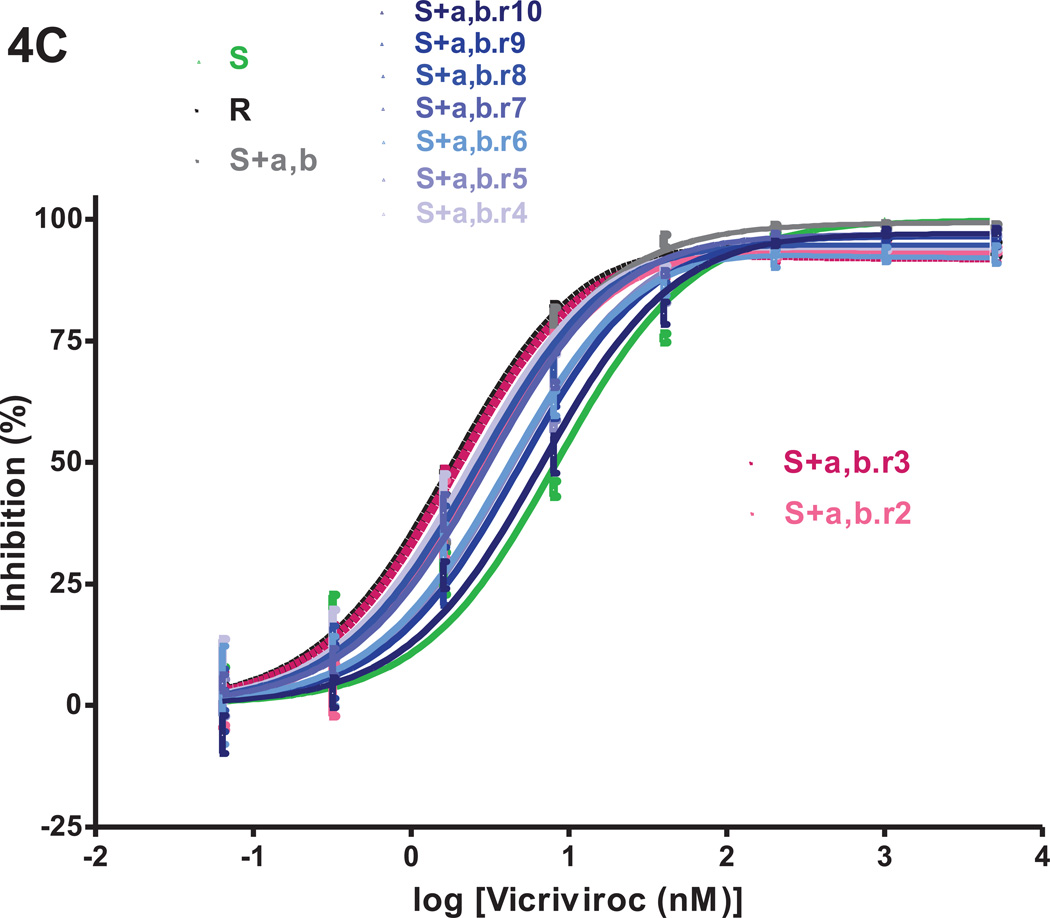
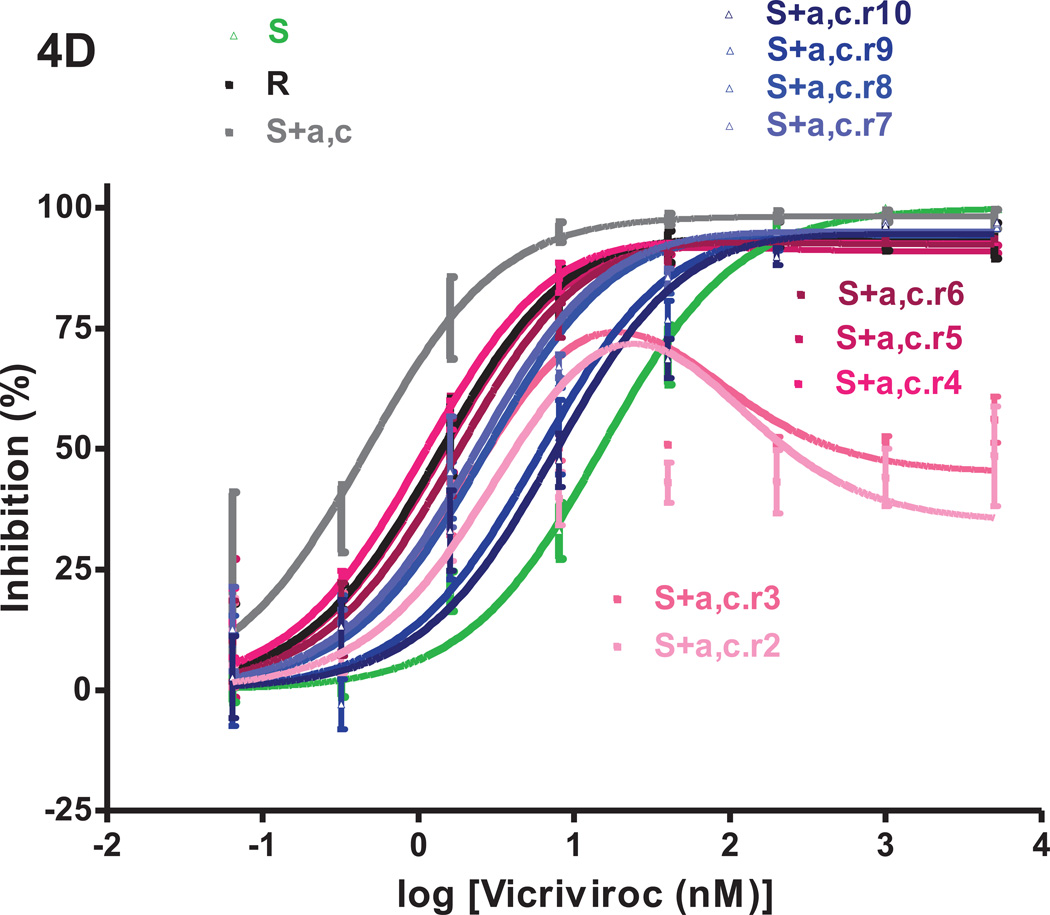
We returned the VCV-resistant reference clone R, the engineered triple mutant S+a,b,c and the two intermediately resistant double mutants S+a,b and S+a,c to culture in PBMC for 10 weekly passages without VCV. The inhibitor sensitivity of replication-competent viruses derived from the four reversion cultures was assessed in TZM-bl cells in comparison to clones S and R and the corresponding input viruses (Fig.4). The model function was fitted to the average inhibition data to determine MPI and IC50 values (Fig.4 and Table 1S). To track genetic changes, we sequenced the env genes from all ten passages of the four reversion cultures (Fig.5). In a few instances, it was not possible to retrieve phenotypic or genotypic data during passage 1, possibly due to a “bottleneck effect” caused by the return to culture of frozen virus stocks. After the first passage, replication became more consistently efficient.
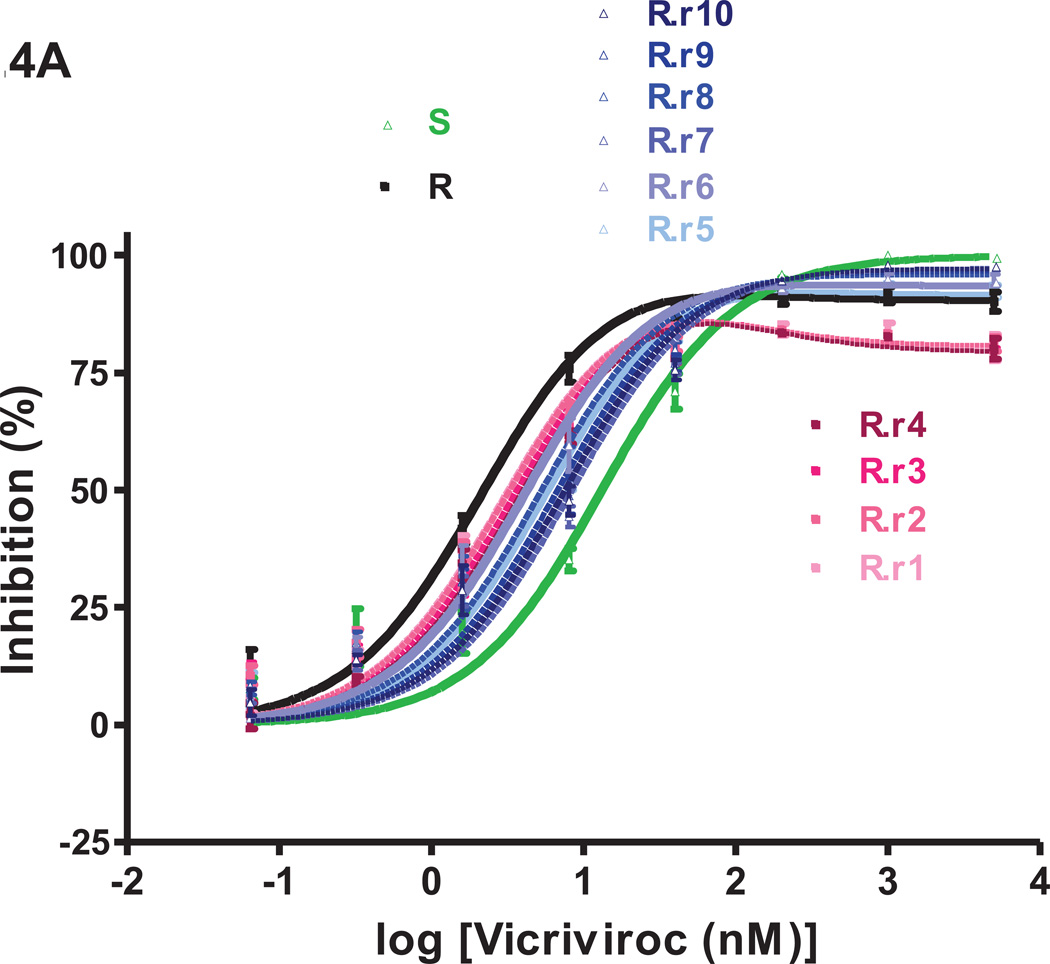
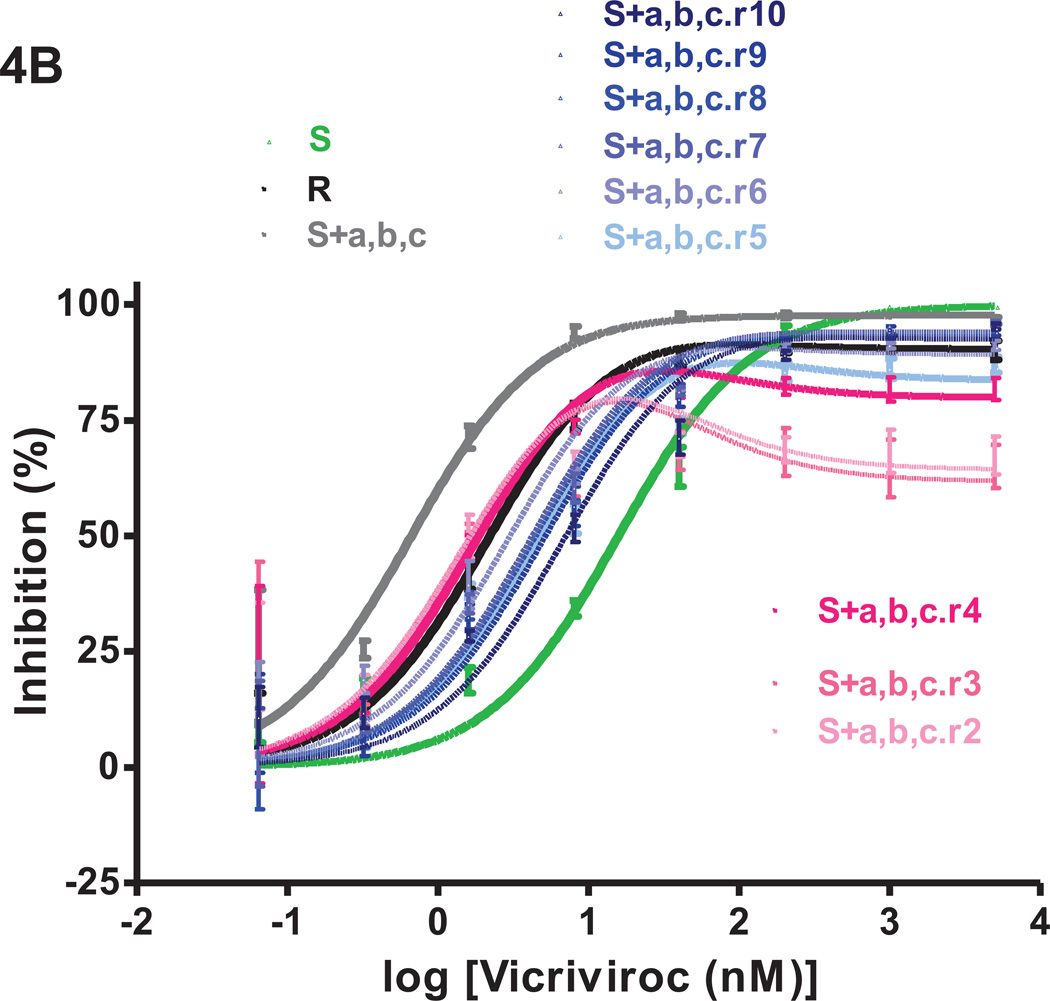
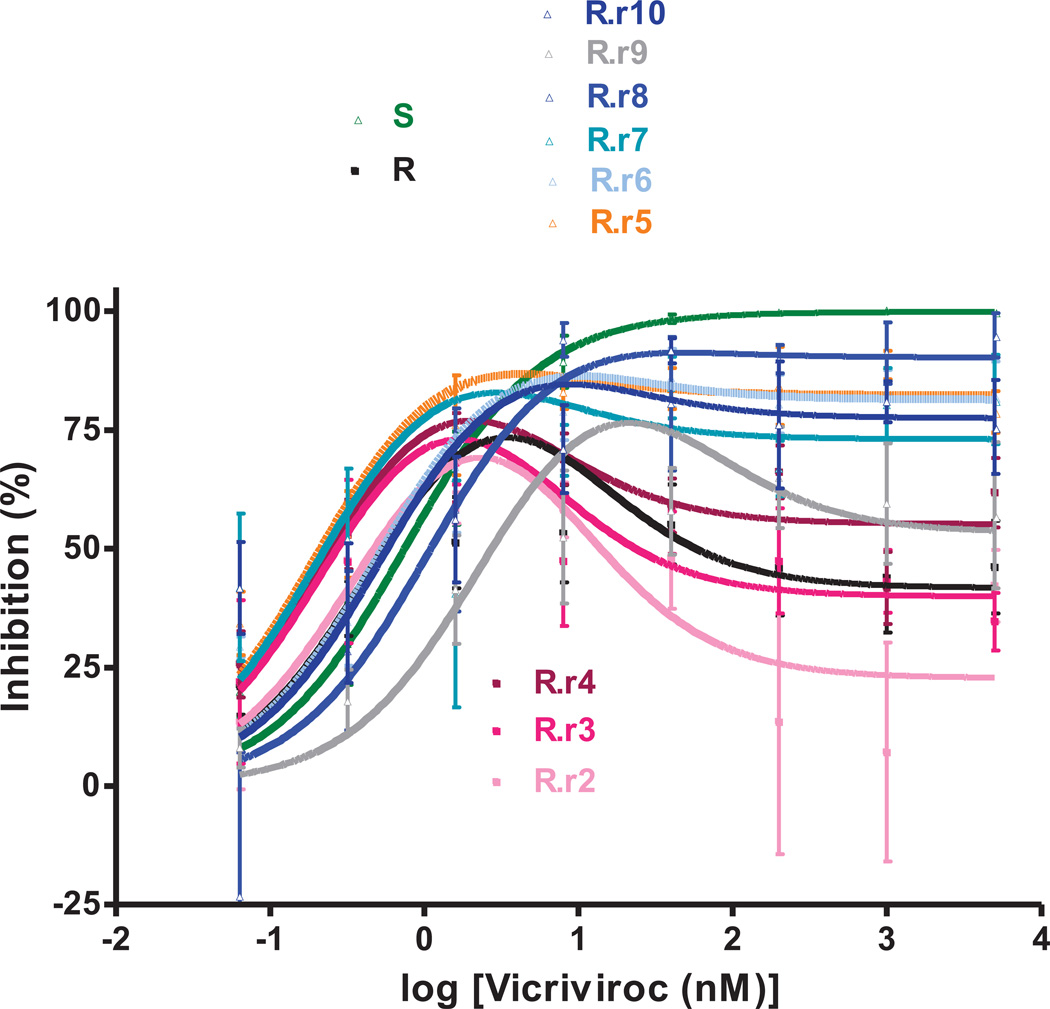
Fig.4. VCV-sensitivity in TZM-bl cells of the VCV-resistant FP mutants after extensive passage in PBMC in the absence of the inhibitor.
Viruses designated (clone name).r1–r10 were derived from the reversion cultures of: (A) the VCV-resistant reference clone R; (B) the engineered VCV-resistant triple mutant S+a,b,c; the two intermediately resistant double FP mutants (C) S+a,b and (D) S+a,c. Each virus was assessed for VCV sensitivity in TZM-bl cells in comparison to clones S and R and the corresponding input resistant virus, with luciferase expression measured after 3 days. The results shown are the average of 2–5 (typically 3) independent experiments, with the error bars indicating the SEM.
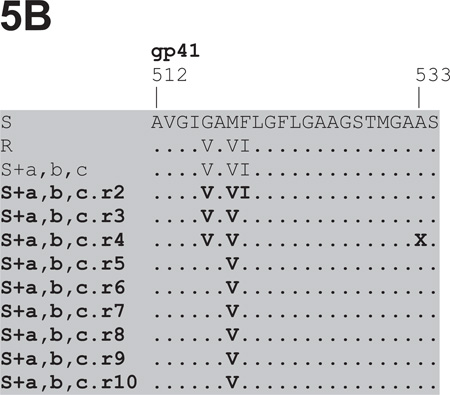
Fig.5. Env sequence alignments of viruses derived from the reversion cultures of VCV-resistant FP mutants.
The first 23 amino acids from the gp41 N-terminus are shown for viruses derived from the 10 weekly VCV-free passages in PBMC of: (A) the reference clone R (D1/85.16 cl.23, GenBank accession no. FJ713453); (B) the triple mutant S+a,b,c; the double FP mutants (C) S+a,b and (D) S+a,c. Additional regions where differences were noted, i.e. either in the C2 of gp120 or further downstream in gp41, are shown for R and S+a,c, respectively. Passage 1 sequence data were unavailable for S+a,b,c and S+a,c. Amino acid sequences are also shown for 10 clones from passage 4, for 12 clones from passage 5 and for 11 clones from passage 9 of the clone R reversion culture. No gp41 sequence data were obtained for the 10 clones from passage 4 and for clone R.r9 cl.39, as indicated by the dashes in the alignment. The bulk sequences of the reversion isolates are highlighted in bold, while those of individual clones are shown in regular fonts. The sequences are recorded relative to that of the reference parental clone S (CC1/85 cl.6, GenBank accession no. AY 357338, top line), with dots indicating amino acid sequence identity. Amino acid numbering is based on HXB2 Env, with the first residue of gp41 at position 512.
The VCV inhibition profiles of the reference clones S and R were similar to those seen before in TZM-bl cells; for R, the MPI and IC50 values were lower than for S (91–94% vs. 100% and 1.4–2.2 nM vs. 8.5–16 nM, respectively; Fig.4, Table 1S). The inhibition curves for viruses from passages 1–4 of the R virus reversion culture overlapped. Compared to R, the MPI values for these viruses were modestly reduced from 90% to 85–86% while the IC50 values gradually increased from 2.2 nM to 4.0 nM (Fig.4A, Table 1S). From passage 5 on, the dose-response curves shifted up- and rightwards, leading to increasing MPI and IC50 values that peaked at 97% and 8.3 nM for virus R.r7. Thus the viruses from the reversion culture of clone R became gradually more similar to the parental virus S, in that they became less able to use the VCV-CCR5 complex for entry.
Genetic sequencing showed that all three FP sequence changes persisted throughout the 10 passages of the R virus reversion culture (Fig.5A). However, by passage 5, an additional Thr-to-Ala substitution had become stabilized at position 244 of the second conserved region (C2) of gp120. This residue is located in strand β7, part of a β-sandwich structure in the inner domain that is involved in CD4-induced conformational changes within gp120 and the interaction with gp41 (Finzi et al., 2010; Helseth et al., 1991; Pancera et al., 2010; Yang et al., 2003). Once it emerged, the T244A substitution persisted together with the 3FP changes. Thus, 2/10 clones from passage 4, 11/12 clones from passage 5 and all 11 clones from passage 9 contained the T244A change (Fig.5A). We describe below studies on the phenotype conferred by the T244A substitution.
Several other amino acid changes also appeared elsewhere in Env as the R virus reversion culture progressed. In particular, three different passage 9 clones contained one of the V3 substitutions T303A, F515L or T317A in addition to the three FP changes (data not shown). The most influential resistance-associated change in the FP, mutation-a, had reverted to the cognate amino acid residue (V516G) in 1/12 passage 5 clones, R.r5 cl.11, but all 11 clones from virus R.r9 retained the three FP substitutions.
Stability of the engineered VCV resistant triple mutant S+a,b,c
On addition to the reversion culture, S+a,b,c again had an MPI value marginally below 100% and a reduced IC50 value of 0.7 nM (compared to 2.2 nM for R and 16 nM for S) (Fig.4B, Table 1S, see also Fig.2, Table 2). Viruses from passages 2 and 3 had much lower MPI values of 79–80% and modestly higher IC50 values of 1.6 nM. The pattern then changed from passage 4 onwards; the later viruses had gradually increasing MPI values that peaked at 94% by passage 7. The IC50 values also tended to rise over time in culture, reaching 7.1 nM by passage 10 (Fig. 4B, Table 1S).
In contrast to the R virus reversion culture, the three FP changes were maintained only during the first two VCV-free passages of S+a,b,c (Fig.5B). Mutation-c was the first to disappear, during the third passage, followed by mutation-a by the fifth passage; mutation-b, however, remained stable during all ten passages. A transient replacement of Ala by Ser at position 533 of gp41 during passage 4 (indicated by X in Fig.5B) coincided temporally with an increase in MPI from 79% to 86% between passages 3 and 4 (Figs. 4B, 5B and Table 1S).
Stability of the double mutants, S+a,b and S+a,c
At the start of the reversion cultures, the double FP mutants S+a,b and S+a,c had MPI values of 99% and 98%, respectively, which were higher than the 96% values seen previously in TZM-bl cells (Figs.4C, 4D, Table 1S, and Fig.2, Table 2). The MPI for clone R was also higher in this experiment than previously (94% compared to 91% in the other two reversion cultures and 87% in previous experiments, Fig.4, Table 1S, and Fig.2, Table 2). The IC50 values for S+a,c and S+a,b (0.5 vs. 2.6 nM, respectively) were comparable to those obtained previously (1.4 vs. 3.0 nM; Fig.4C,4D, Table 1S and Fig.2, Table 2).
Only minor changes in VCV sensitivity occurred during the S+a,b reversion culture. By passage 10, the MPI and IC50 values for the S+a,b.r10 virus were 97% and 6.8 nM, indicating a modest decrease in MPI and a slight (~3-fold) increase in IC50 compared to S+a,b (Fig.4C, Table 1S). As seen in the corresponding experiments with R and S+a,b,c, mutation-b remained stable throughout the S+a,b culture, while mutation-a had disappeared by the fourth passage (Fig.5C). Thus, at the end of this culture, the S+a,b.r10 virus was genetically and phenotypically similar to S+b (see Table 2).
Initial bottleneck events in the S+a,c reversion culture led to a further suppression of the MPI to 72–74% and an increase in the IC50 up to 4.2 nM by passage 3 (Fig.4D, Table 1S). The VCV dose-response curves of the passage 4–6 viruses resembled that for R, with similar MPI (92–93 %) and IC50 values (1.1–1.8 nM), but from passage 7 onwards, the inhibition curves tended to shift only rightwards. The MPI values were maintained at 94–95% but with increasing IC50 values that peaked at 7.6 nM during the tenth passage. Again, the critical resistance mutation, a, reverted rapidly, in this case by passage 3, even more quickly than in the corresponding cultures of S+a,b (passage 4) and S+a,b,c (passage 5) (Fig.5B–D). Mutation-c had disappeared by passage 6, later than in the S+a,b,c culture (passage 3). Two new gp41 mutations V703A and W755R, located in the transmembrane region and the cytoplasmic tail, appeared during passage 4 but they were sustained only for one and two more passages, respectively (Fig.5D). The transient appearance of these mutations coincided with substantial shifts in the inhibition curves, irrespective of whether mutation-c was present in the FP.
In summary, the reversion experiments show that mutation-b, a naturally occurring polymorphism that becomes enriched under VCV or AD101 selection pressure (Anastassopoulou et al., 2009), remained unaltered throughout the inhibitor-free cultures of each relevant input virus. This change, M518V, has little or no effect on VCV sensitivity on its own and appears to be under no pressure to revert. The complete cluster of three FP changes only persisted within the genetic context of a gp120 derived from the resistant clone R. Here, however, they were accompanied by a gp120-C2 substitution, T244A, that appears to counter the VCV-resistance phenotype created by the FP substitutions.
VCV-sensitivity of reversion viruses of clone R in a PBMC assay
We also used the more physiologically relevant, multi-cycle PBMC assay to study replication-competent viruses R.r2-r10 from the R virus reversion culture (Fig.6). Samples of virus R.r1 were unavailable for testing. To determine MPI and IC50 values, the model function was fitted to the average VCV-inhibition data (Table 1S). Viruses R.r2-r10 replicated comparably to R, with PBMC titers of 104–105 TCID50/mL (3–5 ng/mL of p24) on day 7 post-infection.
Fig.6. VCV-sensitivity of viruses from the reversion culture of clone R in a PBMC assay.
Viruses derived from the PBMC reversion culture of clone R were tested for VCV sensitivity in a PBMC-based, multi-cycle replication assay in comparison to the parental, VCV-sensitive clone S and the VCV-resistant reference clone R. Each infection-inhibition experiment was performed on a pool of PBMC from two individuals. The results shown are the average of 2–6 (typically 4) independent experiments, each performed using triplicate wells, with the error bars indicating the SEM. No data were obtained for virus R.r1.
Taking into account PBMC donor-dependent influences on the biphasic VCV-inhibition curves for resistant viruses, clone R behaved comparably to what was seen previously in this assay (compare Fig.6, Table 1S with Fig.1, Table 2). Thus, clone R had lower MPI and IC50 values than S (73% and 0.55 nM vs. 100% and 0.74 nM, respectively). During the reversion culture, the shapes of the VCV-inhibition curves gradually shifted to more closely resemble S than R, although the process appeared to be somewhat more complex in this multi-cycle replication system than in the TZM-bl-based single-cycle assay (Fig.6 cf. Fig.4A). The curves for R.r2–R.r4 were all similar in shape to R with MPI values that ranged from 69–77 %, but those for R.r3 and R.r4 were shifted to the left such that the IC50 values were reduced to 0.25–0.27 nM (Fig.6, Table 1S). From passage 5 onwards, the curves gradually shifted upwards and rightwards, generating MPI and IC50 values that peaked at 91% and 1.1nM for virus R.r8. An exception to this pattern was R.r9, in that it resembled clone R by having a similar MPI value, albeit accompanied by an increased IC50 value (76% and 2.8 nM compared to 73% and 0.55 nM, respectively; Fig.6 and Table 1S).
Effects of the T244A change on resistance of FP mutants
To study the T244A substitution, we introduced it into the VCV-sensitive clone S and the resistant triple mutant S+a,b,c and tested the resulting replication-competent viruses for VCV sensitivity in PBMC and TZM-bl cells. The T244A mutation reduced the PBMC titers of the parental clone S by several-fold (103–104 compared to 104–105 TCID50/mL). The S+a,b,c mutant was little affected by the T244A change, with both viruses replicating in the 103–104 TCID50/mL range.
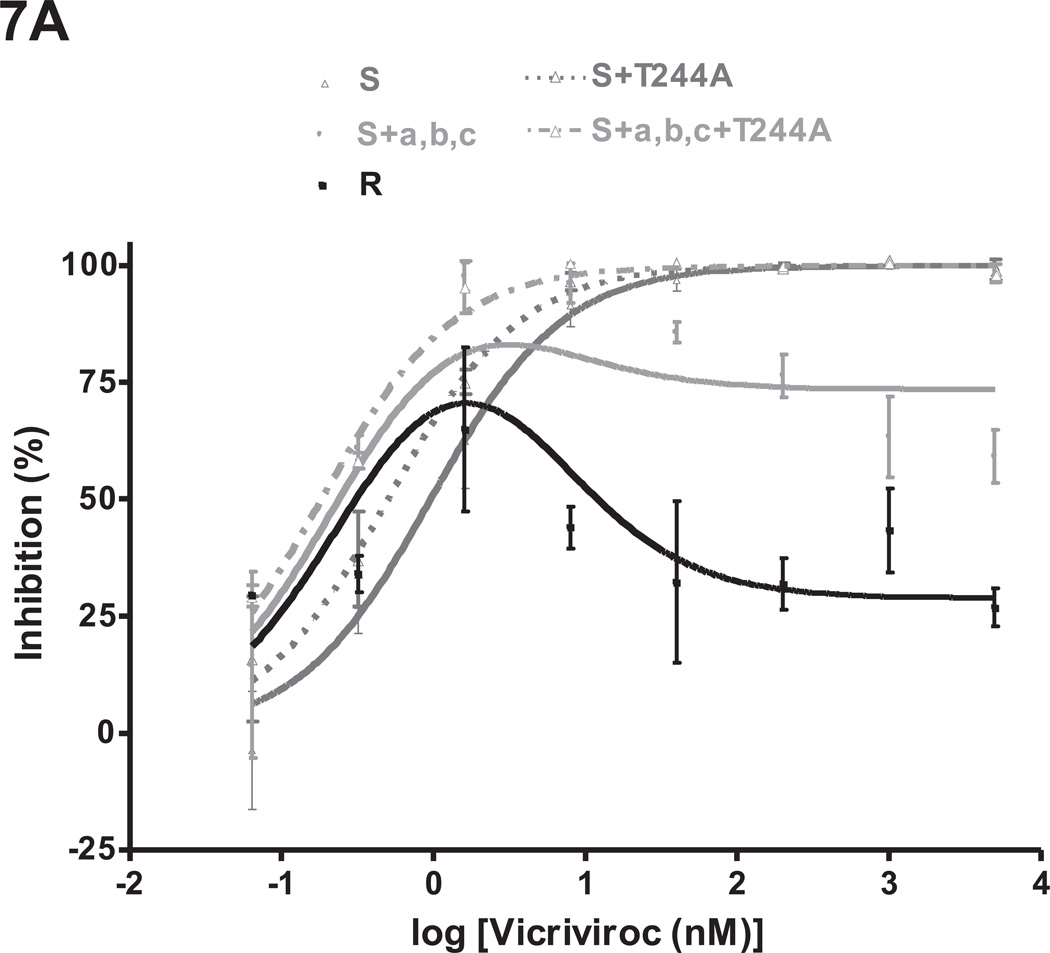
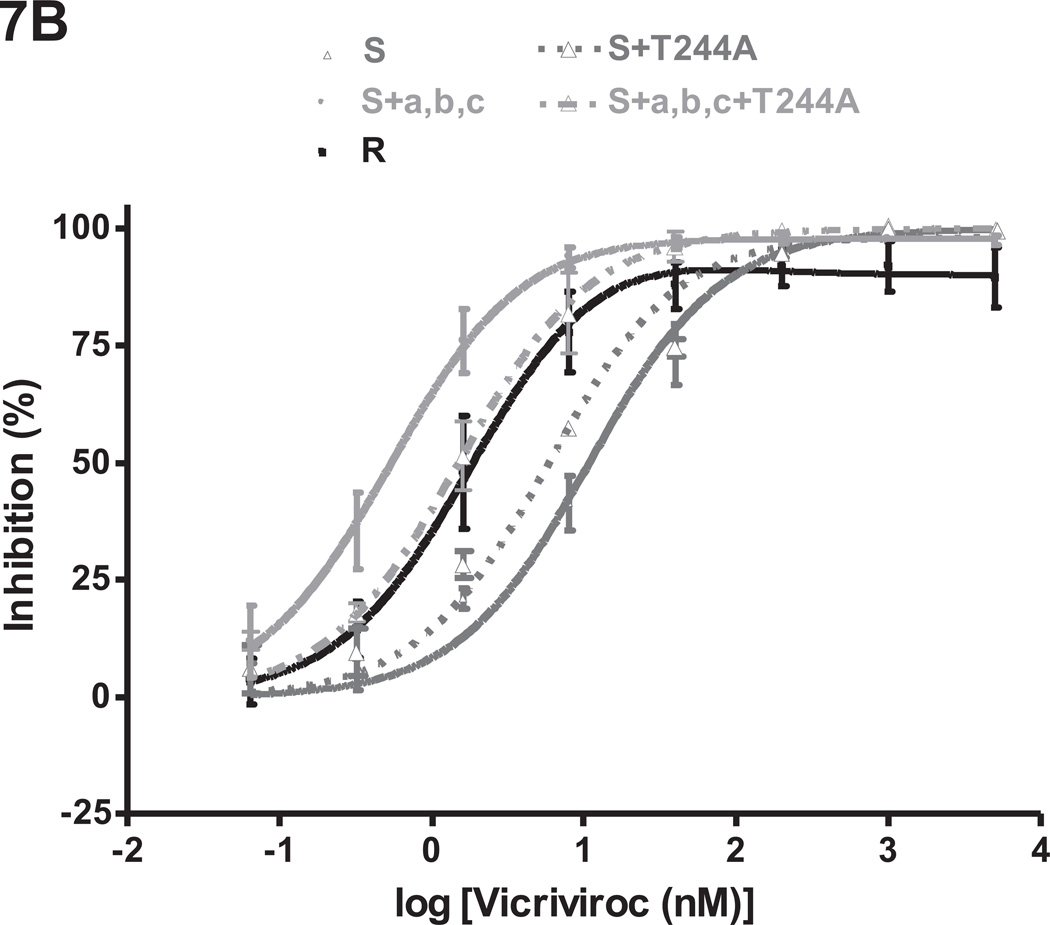
The S+a,b,c+T244A mutant was VCV-sensitive in a PBMC-based multi-cycle assay. Thus, whereas the VCV-inhibition curve for S+a,b,c has a complex shape, the one for S+a,b,c+T244A resembles the parental clone S (Fig.7A). The corresponding MPI values were 83% and 100% (Table 2S). The inhibition curves for S+a,b,c+T244A and S+a,b,c were both still shifted to the left compared to S (IC50 values 0.18 nM and 0.24 nM vs. 0.95 nM), presumably due to the influence of mutation-c.
Fig.7. The T244A change compromises the VCV-resistance phenotype created by the FP mutations.
Replication-competent viruses from the VCV-sensitive clone S and the VCV-resistant triple mutant S+a,b,c were tested for VCV sensitivity in comparison to the corresponding viruses harboring the T244A change in gp120-C2 using (A) PBMC and (B) TZM-bl cells. The VCV-resistant clone R was also tested in the same experiments. Each infection-inhibition experiment on PBMC was performed on a pool of cells from two individuals. The results shown are the average of 2–5 (typically 3) independent experiments in each assay system, each performed using triplicate wells, with the error bars indicating the SEM.
In the TZM-bl cell assay, the inhibition curve for S+a,b,c+T244A was shifted to the right and slightly upward compared to S+a,b,c, yielding increased IC50 and MPI values (1.5 nM vs. 0.54 nM and 100% vs. 98%) (Fig.7B, Table 2S). Similar results were obtained when AD101 was used instead of VCV (data not shown). By itself, the T244A change modestly (~2 fold) reduced the IC50 for VCV when introduced into clone S (0.50 nM vs. 0.95 nM for S+T244A vs. S in PBMC; 5.8 nM vs. 11 nM, respectively, in TZM-bl cells).
Discussion
In vitro resistance to small molecule CCR5 inhibitors, such as MCV and VCV, principally stems from an acquired ability of HIV-1 to utilize the drug-bound form of the coreceptor for entry in addition to free CCR5. The mutations that allow this to happen typically map to the V3 region of gp120 (Moore and Kuritzkes, 2009). However, we have described two alternative routes to resistance. One involved at least an H308P change in V3 and either of the following combinations of gp41 changes: G514V+V535M or M518V+F519L+V535M (Anastassopoulou et al., 2011). In the other, three sequence changes (G516V, M518V and F519I) in the gp41 FP were sufficient for resistance (Anastassopoulou et al., 2009).
We have now studied how the above three FP sequence changes (termed a, b, and c here), individually and in combination, affect VCV sensitivity. Although mutation-a (G516V) confers only a very low level of VCV resistance by itself, an effect evident in PBMC but not TZM-bl cells, it is critical overall. To have a substantial impact, mutation-a must be accompanied by either of the other two FP substitutions. Thus, both the S+a,b and S+a,c double mutants are partially VCV-resistant, but only the S+a,b,c triple mutant is fully resistant. No naturally occurring viruses in the HIV Sequence Database (http://www.hiv.lanl.gov/, updated as of 1/30/2012) contain all three FP changes, although some subtype B and C strains have the a+b (G516V+M518V) combination.
These above two changes are both present in isolate Du151 from a South African who was dually infected with two phylogenetically distinct subtype C strains. During disease progression in this patient, a coreceptor switch to CXCR4 use was associated with specific amino acid changes in the V3 crown, an increased net charge in V3, and an increase in the length of V1 (Coetzer et al., 2007). Together with several other changes scattered throughout Env, the G516V+M518V combination was also present in 8/12 clones at the time of virologic failure (compared to 0/12 at baseline) in a previously treatment-naïve, subtype B-infected patient given the CCR5 inhibitor, aplaviroc, in combination with lopinavir-ritonavir (Kitrinos et al., 2009). This patient underwent a phenotypic switch from R5 to dual/mixed during treatment. The G516V change (mutation-a) by itself has been shown to confer CXCR4 usage on a clonal R5 virus (Huang et al., 2008).
Taken together, these various reports imply that FP changes are associated with, and may facilitate, tropism switches from CCR5 to CXCR4. However, our FP mutants were unable to use CXCR4 in PBMC or TZM-bl cells (Anastassopoulou et al., 2009). The implication is that additional changes are required to drive the virus towards CXCR4, at least in the context of the CC1/85 genetic lineage, which may be predisposed to acquiring resistance to MCV and other CCR5 inhibitors via an inherent, low-level ability to use drug-bound CCR5 for entry (Roche et al., 2011). Further evidence for the relevance of the Env genetic context in CCR5 inhibitor resistance is that introducing the three FP changes into a heterologous virus, JR-FL, did not create a VCV-resistant variant (Anastassopoulou et al., 2009). Nonetheless, the presence of the Val-516 and Val-518 polymorphisms in natural viruses further implies that these residues are compatible with the normal functioning of the FP, despite their rarity compared to Gly-516 and Met-518.
Modeling the VCV inhibition data derived from both PBMC and TZM-bl cells confirmed the crucial role of mutation-a; only the S+a single mutant yielded w-values that were marginally, but consistently, above 0. The model further showed that combining mutation-a with either mutation-b or mutation-c conferred the ability to use VCV-CCR5 complexes for entry. Hence, the S+a,b and S+a,c double mutants and the S+a,b,c triple mutant all reproduced the R phenotype to various extents, with w > 0.
The ability of the escape mutants to use the inhibitor-bound form of CCR5 for entry, in addition to the inhibitor-free form, is consistent with noncompetitive resistance [reviewed in (Kuhmann and Hartley, 2008; Moore and Kuritzkes, 2009)]. Competitive resistance results in a shift in the IC50 of an inhibitor to a higher concentration, with complete inhibition potentially achievable at a high enough inhibitor concentration. In contrast, for a noncompetitive resistance mechanism, the IC50 values are typically equivalent to those for the fully sensitive virus but the extent of inhibition is incomplete. The resulting MPI values vary by cell type for inhibition by VCV and related compounds (Anastassopoulou et al., 2011, 2009; Ogert et al., 2008; Pugach et al., 2007; Westby et al., 2007). The MPI value, at a saturating inhibitor concentration, reflects the efficiency with which the inhibitor-CCR5 complex is used, relative to free CCR5, thereby providing a measure of the degree of resistance of the HIV-1 variant under study (Pugach et al., 2007; Westby et al., 2007).
It is an apparent paradox that many of the FP mutants have reduced IC50 values for VCV, particularly in TZM-bl cells but also on occasions in PBMC. As an explanation, we have proposed that the resistant FP-mutants switch to use unoccupied forms of CCR5 for which VCV has a higher affinity, compared to the forms the wild type virus can use (Anastassopoulou et al., 2009). Here, we show that mutation-c, F519I, is an independent determinant of preference for the unoccupied high-affinity form of CCR5. Thus, the three FP changes combine their qualitatively unique effects to create an unusual VCV-resistance phenotype.
A recent study showed that three VCV-resistant clinical isolates with V3 mutations had delayed entry rates that were restored to pre-therapy levels in the presence of the inhibitor (Putcharoen et al., 2012). The authors suggested that the underlying mechanism of resistance to CCR5 antagonists is the need for HIV-1 to maximize its rate of entry into cells in the context of both its own genetic background and the properties of the host cells (e.g., the quantity and properties of the available CCR5 coreceptors). We have yet to examine the fusion efficiency and entry kinetics of the various FP mutants to further explore this admirable concept.
All the VCV-resistant isolates we studied previously have highly stable phenotypes in PBMC, in that they do not rapidly revert to sensitivity when cultured without the inhibitor (Anastassopoulou et al., 2011, 2009, 2007). Here, we assessed the stability in PBMC of individual FP mutant clones with various degrees of VCV-resistance. Mutation-b (M518V), which has little or no effect on VCV sensitivity by itself, persisted throughout the reversion cultures, implying that there is no pressure for it to revert. Val-518 is a naturally occurring FP polymorphism that becomes enriched during VCV or AD101 selection (Anastassopoulou et al., 2009). A study of viral diversification in newly infected individuals identified FP residue 518 as one that was rapidly evolving (from Met to Val or Ile) under positive selection, although without any indications for the involvement of CTL escape or APOBEC hypermutation (Wood et al., 2009). There were, however, no general indications across multiple patients that Met was present at residue 518 during acute infection, but replaced by Val or Ile in chronic infection.
Viruses from the clone R reversion culture gradually became less able to use the VCV-CCR5 complex for entry from passage 5 onwards. The cluster of three FP changes remained unchanged throughout this culture, but by passage 5, an additional Thr-to-Ala substitution had become stabilized at position 244 in the gp120 C2 region. Once it emerged, the T244A change persisted together with the 3FP changes. Several other amino acid changes also appeared in the reversion culture. In particular, three independent passage 9 clones contained one of the V3 substitutions T303A, F515L, or T317A in addition to the three FP changes (data not shown). However, these V3 changes probably represent the detection of minor populations, as they were not found in the bulk sequence of the passage 9 isolate.
As the genotypic and phenotypic data from the reversion culture suggested the T244A change counteracted the causative effect of the three FP changes on VCV resistance, we introduced it into the resistant triple FP mutant. The resulting S+a,b,c+T244A virus was indeed VCV-sensitive in the PBMC-based, multi-cycle assay. Residue 244 is located in strand β7 of the β-sandwich in the inner domain of gp120, which is thought to be involved in transmitting conformational changes from gp120 to gp41 upon CD4 binding (Finzi et al., 2010; Helseth et al., 1991; Pancera et al., 2010; Yang et al., 2003). The side chain of residue 244 is solvent exposed in the crystal structure of the gp120 core that includes the N- and C-termini, suggesting that it may interact with gp41 directly (Pancera et al., 2010). On its own, but in a different genetic context, the T244A change drastically reduces virus entry (Yang et al., 2003). We noted that it did reduce, albeit only by several-fold, the replication of the reference parental CC1/85 clone (clone S).
A Thr-to-Ser substitution at the same position (T244S) has been reported to be one of four changes that rescue the replication capacity of viruses resistant to virus-inhibitory peptide (VIRIP) (Gonzalez-Ortega et al., 2011), an inhibitor corresponding to the C-proximal region of α1-antitrypsin (Munch et al., 2007). Viruses resistant to VIRIP and its derivative VIR-353 contain a combination of three mutations in gp120 (A433T/V489I) and gp41 (V570I), but no changes in or near the FP (Gonzalez et al., 2011). Furthermore, sequence variations in the FP were not responsible for the different virological responses to treatment with another VIRIP analog, VIR-576 (Forssmann et al., 2010). Nonetheless, the involvement of position 244 in both VIRIP and VCV resistance/reversion is worth noting from the perspective of structure/function relationships in Env that influence both HIV-1 entry and resistance development.
Materials and Methods
Reagents
The small molecule CCR5 inhibitors VCV (SCH-D, SCH-417690) and AD101 (SCH-350581) were provided by Dr. Julie Strizki (Schering-Plough Research Institute; now Merck Research Laboratories).
Viruses
The clonal viruses listed in Table 1 were designed to investigate individually and in pair-wise combinations the effects of three FP changes (G516V, M518V, F519I) on the VCV-resistant phenotype of the D1/85.16 lineage (Anastassopoulou et al., 2009). Infectious, replication-competent stocks were prepared by transient transfection of 293T cells with pNL4-3/env proviral plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Env-pseudotyped viruses were prepared by co-transfecting 293T cells with pCI-env and pNLluc-AM plasmids (3:1 ratio), again using Lipofectamine 2000. All viruses were used at similar pre-titrated doses (100 50% tissue culture infective doses (TCID50) per well of a 96-well plate), as described (Anastassopoulou et al., 2009; Kuhmann et al., 2004).
Construction of chimeric NL4-3/env proviruses and pCI-based env-expressing plasmids
NL4-3/env proviruses were constructed as previously described (Anastassopoulou et al., 2009; Kuhmann et al., 2004). Site-directed mutagenesis was performed with QuickChange II (Stratagene), using pBluescript KS(+) plasmids containing EcoRI/XhoI fragments that were then subcloned into pNL4-3. Mutagenesis was similarly performed on a pCI-based plasmid expressing the env gene of the reference VCV-sensitive clone CC1/85 cl.6 (= S, GenBank accession no. AY357338). The construction of the pNLluc-AM and pCI-env plasmids for expression of S has been described elsewhere (Pugach et al., 2007). Env genes were sequenced as described (Marozsan et al., 2005; Trkola et al., 2002) and aligned with MacVector 10.5.1. Env sequences derived from the reversion cultures of the VCV-resistant clone R (D1/85.16 cl.23, GenBank accession no. FJ713453), the triple mutant S+a,b,c and the double mutants S+a,b and S+a,c have been submitted to GenBank (accession nos. JQ707955-JQ708027).
Infection-inhibition assays
HIV-1 sensitivity to VCV was assessed using primary cells (PBMC) or engineered cell lines (TZM-bl and U87.CD4.CCR5), as previously described (Anastassopoulou et al., 2011, 2009; Ketas et al., 2007; Kuhmann et al., 2004; Marozsan et al., 2005). The PBMC assay endpoint was p24 production after 7 days of infection. With TZM-bl and U87.CD4.CCR5 cells, luciferase expression in relative light units (RLU) was measured after 2 and 3 days, respectively. The sensitivity of VCV escape mutants to inhibitors of post-entry stages of the replication cycle has been evaluated previously; D101.12 and D1/85.16 and their parental isolates had comparable sensitivities to the nucleoside reverse transcriptase inhibitor zidovudine (AZT), the non-nucleoside reverse transcriptase inhibitor nevirapine, the protease inhibitor indinavir and the integrase inhibitor 118-D-24 (Marozsan et al., 2005).
The model function for inhibition, Q=(1−(1−((C/(Kdi+C))+w*(C/(22*Kdi+C))))*100% (Anastassopoulou et al., 2011, 2009; Ketas et al., 2012; Klasse, 2007), was fitted to the average PBMC and TZM-bl data by nonlinear regression (Prism, Graphpad). The model function was also fitted by nonlinear regression (Prism, Graphpad) to determine MPI and IC50 (as defined in Tables 2, 1S and 2S) as well as the parameters Kdi and w (Table 3). The MPI is defined as the upper plateau for sigmoid curves, or as the maximum inhibition value for peaked curves, while the IC50 is defined as the VCV concentration (C) that yields 50% inhibition (Q). To generate the VCV-inhibition curves shown in Figs.1, 2, 4, 6 and 7, we used a form of the equation with 10-logarithms of inhibitor concentrations as x values:
Q = (1−(1−(10logC/(Kdi+10logC))+w*(10logC/(22*Kdi+10logC))))*100%, where logC = x.
The G516V change is critical to VCV resistance in PBMC and TZM-bl cells
To have a substantial impact, G516V must be accompanied by either M518V or F519I
G516V allows both double FP mutants to use VCV-CCR5 complexes for entry
F519I specifies a preferred use of the unoccupied, high-VCV affinity form of CCR5
T244A appears to counter the VCV-resistance phenotype created by the FP changes
Supplementary Material
Acknowledgements
We thank Antonia Thomas and Samson Jacob for technical support, and Julie Strizki for supplying CCR5 inhibitors. This work was supported by NIH grant R01 AI41420.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastassopoulou CG, Ketas TJ, Depetris RS, Thomas AM, Klasse PJ, Moore JP. Resistance of a human immunodeficiency virus type 1 isolate to a small molecule CCR5 inhibitor can involve sequence changes in both gp120 and gp41. Virology. 2011;413(1):47–59. doi: 10.1016/j.virol.2010.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassopoulou CG, Ketas TJ, Klasse PJ, Moore JP. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 2009;106(13):5318–5323. doi: 10.1073/pnas.0811713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassopoulou CG, Marozsan AJ, Matet A, Snyder AD, Arts EJ, Kuhmann SE, Moore JP. Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss. PLoS Pathog. 2007;3(6):e79. doi: 10.1371/journal.ppat.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Miyake H, Wang X, Okamoto M, Takashima K. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob. Agents Chemother. 2007;51(2):707–715. doi: 10.1128/AAC.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro R, Klasse PJ, Lascano D, Flegler A, Nagashima KA, Sanders RW, Sakmar TP, Hope TJ, Moore JP. Multiple CCR5 conformations on the cell surface are used differentially by human immunodeficiency viruses resistant or sensitive to CCR5 inhibitors. J Virol. 2011;85(16):8227–8240. doi: 10.1128/JVI.00767-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro R, Klasse PJ, Moore JP, Sanders RW. V3 determinants of HIV-1 escape from the CCR5 inhibitors Maraviroc and Vicriviroc. Virology. 2012 doi: 10.1016/j.virol.2012.02.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro R, Sanders RW, Lu M, Klasse PJ, Moore JP. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 2009;5(8) doi: 10.1371/journal.ppat.1000548. e1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzer M, Cilliers T, Papathanasopoulos M, Ramjee G, Karim SA, Williamson C, Morris L. Longitudinal analysis of HIV type 1 subtype C envelope sequences from South Africa. AIDS Res Hum Retroviruses. 2007;23(2):316–321. doi: 10.1089/aid.2006.0207. [DOI] [PubMed] [Google Scholar]
- Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell. 2010;37(5):656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forssmann WG, The YH, Stoll M, Adermann K, Albrecht U, Barlos K, Busmann A, Canales-Mayordomo A, Gimenez-Gallego G, Hirsch J, Jimenez-Barbero J, Meyer-Olson D, Munch J, Perez-Castells J, Standker L, Kirchhoff F, Schmidt RE. Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide. Sci Transl Med. 2010;2(63) doi: 10.1126/scitranslmed.3001697. 63re63. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Ballana E, Clotet B, Este JA. Development of resistance to VIR-353 with cross-resistance to the natural HIV-1 entry virus inhibitory peptide (VIRIP) AIDS. 2011;25(13):1557–1583. doi: 10.1097/QAD.0b013e328348a733. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ortega E, Ballana E, Badia R, Clotet B, Este JA. Compensatory mutations rescue the virus replicative capacity of VIRIP-resistant HIV-1. Antiviral Res. 2011;92(3):479–483. doi: 10.1016/j.antiviral.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res. Hum. Retroviruses. 2005;21(2):171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65(4):2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Toma J, Fransen S, Stawiski E, Reeves JD, Whitcomb JM, Parkin N, Petropoulos CJ. Coreceptor tropism can be influenced by amino acid substitutions in the gp41 transmembrane subunit of human immunodeficiency virus type 1 envelope protein. J Virol. 2008;82(11):5584–5593. doi: 10.1128/JVI.02676-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketas TJ, Holuigue S, Matthews K, Moore JP, Klasse PJ. Env-glycoprotein heterogeneity as a source of apparent synergy and enhanced cooperativity in inhibition of HIV-1 infection by neutralizing antibodies and entry inhibitors. Virology. 2012;422(1):22–36. doi: 10.1016/j.virol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketas TJ, Kuhmann SE, Palmer A, Zurita J, He W, Ahuja SK, Klasse PJ, Moore JP. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364(2):281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitrinos KM, Amrine-Madsen H, Irlbeck DM, Word JM, Demarest JF. Virologic failure in therapy-naive subjects on aplaviroc plus lopinavir-ritonavir: detection of aplaviroc resistance requires clonal analysis of envelope. Antimicrob Agents Chemother. 2009;53(3):1124–1131. doi: 10.1128/AAC.01057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369(2):245–262. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Hartley O. Targeting chemokine receptors in HIV: a status report. Annu. Rev. Pharmacol. Toxicol. 2008;48:425–461. doi: 10.1146/annurev.pharmtox.48.113006.094847. [DOI] [PubMed] [Google Scholar]
- Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 2004;78(6):2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso MM, Lee FH, Haggarty B, Agrawal C, Nolan KM, Biscone M, Romano J, Jordan AP, Leslie GJ, Meissner EG, Su L, Hoxie JA, Doms RW. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 2007;3(8):e117. doi: 10.1371/journal.ppat.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RD, Novak RM. Reviews of anti-infective agents: maraviroc: the first of a new class of antiretroviral agents. Clin Infect Dis. 2008;47(2):236–241. doi: 10.1086/589289. [DOI] [PubMed] [Google Scholar]
- Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338(1):182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Moore JP, Kuritzkes DR. A piece de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr Opin HIV AIDS. 2009;4(2):118–124. doi: 10.1097/COH.0b013e3283223d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J, Standker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann WG, Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129(2):263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- Ogert RA, Hou Y, Ba L, Wojcik L, Qiu P, Murgolo N, Duca J, Dunkle LM, Ralston R, Howe JA. Clinical resistance to vicriviroc through adaptive V3 loop mutations in HIV-1 subtype D gp120 that alter interactions with the N-terminus and ECL2 of CCR5. Virology. 2010;400(1):145–155. doi: 10.1016/j.virol.2010.01.037. [DOI] [PubMed] [Google Scholar]
- Ogert RA, Wojcik L, Buontempo C, Ba L, Buontempo P, Ralston R, Strizki J, Howe JA. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology. 2008;373(2):387–399. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107(3):1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361(1):212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putcharoen O, Lee SH, Henrich TJ, Hu Z, Vanichanan J, Coakley E, Greaves W, Gulick RM, Kuritzkes DR, Tsibris AM. HIV-1 clinical isolates resistant to CCR5 antagonists exhibit delayed entry kinetics that are corrected in the presence of drug. J Virol. 2012;86(2):1119–1128. doi: 10.1128/JVI.06421-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche M, Jakobsen MR, Ellett A, Salimiseyedabad H, Jubb B, Westby M, Lee B, Lewin SR, Churchill MJ, Gorry PR. HIV-1 predisposed to acquiring resistance to maraviroc (MVC) and other CCR5 antagonists in vitro has an inherent, low-level ability to utilize MVC-bound CCR5 for entry. Retrovirology. 2011;8:89. doi: 10.1186/1742-4690-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, Palani A, Shapiro S, Clader JW, McCombie S, Reyes GR, Baroudy BM, Moore JP. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. U.S.A. 2002;99(1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibris AM, Sagar M, Gulick RM, Su Z, Hughes M, Greaves W, Subramanian M, Flexner C, Giguel F, Leopold KE, Coakley E, Kuritzkes DR. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 2008;82(16):8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, Jenkins TM, Perros M, van der Ryst E. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80(10):4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 2007;81(5):2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000414. e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology. 2003;313(1):117–125. doi: 10.1016/s0042-6822(03)00273-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.