Abstract
The prion-forming domain of the fungal prion protein HET-s, HET-s(218-289), is known from solid-state NMR studies to have a β-solenoidal structure; the β-solenoid has the cross-β structure characteristic of all amyloids, but is inherently more complex than the generic stacked β-sheets found in studies of small synthetic peptides. At low pH HET-s(218-289) has also been reported to form an alternative structure, which has not been characterized. We have confirmed by x-ray fiber diffraction that HET-s(218-289) adopts a β-solenoidal structure at neutral pH, and shown that at low pH, it forms either a β-solenoid or a stacked β-sheet structure, depending on the integrity of the protein and the conditions of fibrillization. The low pH stacked-sheet structure is usually formed only by proteolyzed HET-s(218-289), but intact HET-s(218-289) can form stacked sheets when seeded with proteolyzed stacked-sheet HET-s(218-289). The polymorphism of HET-s parallels the structural differences between the infectious brain-derived and the much less infectious recombinant mammalian prion protein PrP. Taken together, these observations suggest that the functional or pathological forms of amyloid proteins are more complex than the simple generic stacked-sheet amyloids commonly formed by short peptides.
Introduction
Mammalian prions were first recognized as the infectious agents in diseases such as scrapie in sheep, bovine spongiform encephalopathy (mad cow disease), and Creutzfeldt-Jakob disease, and consist only of a conformational isoform of the mammalian prion protein PrP (1). Prions derived from other proteins have now been shown to act as non-Mendelian genetic elements in organisms such as yeast and fungi, where they exhibit phenotypes that may be beneficial to survival (2). Prions often aggregate into long unbranched amyloid fibrils that exhibit well-defined cross-β structure: β-strands running approximately perpendicular to the fibril axis, assembled into β-sheets running parallel to the fibril axis (3). It is becoming apparent that the mechanisms of prion structure propagation may be applicable to most biologically important amyloids, including those associated with Alzheimer's, Parkinson's, and many other diseases (4).
HET-s is a functional prion found in the filamentous fungus Podospora anserina and used in a self/nonself recognition event leading to programmed cell death, termed a heterokaryon incompatibility reaction (5). The proteinase K resistant core, residues 218–289, is both necessary and sufficient for infectivity (6,7). Recombinantly expressed HET-s(218-289) fibrillized in vitro at physiological pH can yield fibrils that are infectious in vivo. By varying the fibrillization conditions, primarily by lowering pH, noninfectious amyloid fibril species can also be obtained (8). These species exhibit different fibril morphologies, with the noninfectious fibrils displaying greater structural heterogeneity (8,9). Infectious and noninfectious HET-s(218-289) fibrils have been shown by solid-state NMR (ssNMR) to have different protein folds (10,11); an atomic model, a two-layered β-solenoid with extensions and loops, has been generated for the infectious form (10,12). Low-resolution structures of one single- and two three-protofilament forms of low pH HET-s(218-289) have been determined by cryo-electron microscopy (cryo-EM) (13). These structures were consistent with the β-solenoid fold determined by ssNMR.
We show here by x-ray fiber diffraction that HET-s(218-289) fibrils can adopt two very different amyloid structures. Although fibrils formed at physiological pH have the β-solenoid structure found by ssNMR, fibrils formed at low pH may be either β-solenoids or simple stacked β-sheets. The stacked β-sheet structure, in its simplest or generic form, consists of identical but more or less independent β-sheets stacked together, with the β-strands approximately perpendicular to the fibril axis, and the sheets parallel to the axis (14). It was first described without molecular details by Astbury in 1935 (15), and subsequently characterized in molecular detail using fiber diffraction data from a number of amyloids including cross-β silk (14,16–19). It has been found repeatedly in amyloids formed by short synthetic peptides with amino acid sequences taken from natural amyloidogenic proteins, and more recently similar structures have been observed crystallographically for a large number of such peptides (20,21). We show that the HET-s stacked sheets are formed as a result of proteolytic degradation, although the degraded form can seed solutions of the undegraded protein to form stacked β-sheets.
Methods
Expression and purification of HET-s(218-289)
HET-s(218-289) protein was expressed as a C-terminally tagged 6-histidine construct in BL-21 Escherichia coli cells using ZYM-5052 autoinduction media (22). HET-s(218-289) inclusion bodies were purified from cells using lysozyme digestion, sonication, and centrifugation, and solubilized under denaturing conditions (50 mM Tris-HCl pH 8, 6 M Gdn-HCl, 5 mM imidazole, 1 mM β-mercaptoethanol). Solubilized inclusion body solution was clarified by ultracentrifugation and the HET-s(218-289) protein was purified from the supernatant by affinity chromatography on Ni-NTA resin (Qiagen, Hilden, Germany) while maintaining denaturing conditions. Protein was eluted by switching to low pH (50 mM citric acid pH 2.0, 50 mM imidazole, 6 M Gdn-HCl). Buffer was exchanged for nondenaturing buffer using Sephadex G-25 superfine (GE Lifesciences, Uppsala, Sweden).
Fibrillization of HET-s(218-289)
High pH HET-s(218-289) was prepared from monomeric HET-s(218-289) by exchanging denaturing buffer for 150 mM acetic acid pH 2.5, titrating to pH 7.5 with 3 M Tris, and allowing to fibrillize overnight at room temperature (10). For low pH fibrils, HET-s(218-289) was exchanged into 40 mM boric acid, 10 mM citric acid, 7 mM NaCl buffer at pH 2 (8). Other low pH fibrillization buffers were made by titrating 50 mM citric acid or acetic acid to pH 2, 2.25, 2.5, 2.75, or 3 with 1 M HCl or NaOH. Fibrils were allowed to form at room temperature. Fibrillization was judged by solution turbidity and presence of fibrils in negative stain EM.
Negative stain electron microscopy
Fibril suspensions were diluted to ∼1 mg/ml concentration with water and adsorbed into carbon-coated copper grids, rinsed with water, and stained with 1% uranyl acetate. Micrographs were recorded using a Phillips CM12 transmission electron microscope operating at 80 KeV with a side-mounted 2 k × 2 k charge-coupled device camera.
X-ray fiber diffraction
Fiber diffraction specimens were prepared by suspending 12 μl of concentrated fibril suspension (∼20 mg/ml) between two wax-tipped glass capillaries and allowed to dry under high humidity conditions (97% or nominal 100% relative humidity) (23). Fibril solutions were prepared by ultracentrifugation and resuspension into solutions that provided optimal orientation (water adjusted to pH 4.0 with HCl for high pH fibrils, 2.5 mM pH 4.0 acetate, or 2.5 mM acetic acid/HCl pH 2.5 for low pH and seeded low pH fibrils). High humidity was maintained throughout data collection. X-ray fiber diffraction data were collected at the Biological Small-Angle X-ray Scattering beamline 4-2 at the Stanford Synchrotron Radiation Lightsource; some preliminary data were collected at the BioCAT and BioCARS beamlines of the Advanced Photon Source synchrotron, Argonne National Laboratory. Diffraction patterns were processed using the program WCEN (24).
SDS-PAGE
Fibril suspensions were mixed 2:1 with 6× Laemmli buffer for a final 2× sodium dodecyl sulfate (SDS) concentration in silanized microcentrifuge tubes and placed into a boiling water bath for 5 min. Samples were then run on NuPAGE Novex 10% Bis-Tris gels using NuPAGE MES buffer (Invitrogen, Carlsbad, CA) and stained with SimplyBlue SafeStain (Invitrogen). Gels were imaged with a Bio-Rad ChemiDoc Imaging System (Bio-Rad, Hercules, CA). For seeded low pH samples, SDS-polyacrylamide gel electrophoresis (PAGE) was run on fiber diffraction specimens from which data had already been collected, resuspended in 10 μl of water, and prepared as previously mentioned.
Mass spectrometry
Bands were excised from SDS-PAGE gels and subjected to in-gel tryptic digests. Digested peptide solutions were then analyzed using tandem mass spectrometry (MS/MS) on an Applied Biosystems 4700 Proteomics Analyzer.
Thioflavin T fluorescence
Thioflavin T (ThT) measurements were performed on solutions containing 10 mM HET-s(218-289) fibrils and 25 mM ThT in Tris-buffered saline pH 7.5 at 25°C. Measurements were performed using a Horiba Jobin Yvon Fluoromax-3 with an excitation wavelength of 450 nm and emission range from 470 to 570 nm.
Results
Fiber diffraction
HET-s(218-289) fibrillized at pH 7.5 exhibited the cable-like morphologies considered to be the hallmark of infectious HET-s(218-289) fibrils (8) (Fig. 1 A). Intensities in x-ray fiber diffraction patterns from these fibrils (see Fig. 1 E) are on layer lines with a repeat of 9.5 Å, corresponding to the two-rung structure of the β-solenoid. The second layer line includes a very strong sharp meridional intensity at d = 4.75 Å, the signature of cross-β structure, corresponding to the spacing between β-strands. Intensity maxima away from the meridian do not all lie exactly on the layer lines, because of the helical twist of the fibrils (25). The equator consists of a series of diffraction maxima including strong intensities at 17 Å and 11 Å. Such series are typical of the patterns expected from fibrils whose structures approximate to that of a solid cylinder, for example β-solenoids (26,27). The pattern in Fig. 1 A thus confirms the β-solenoidal structure found by ssNMR for HET-s(218-219) fibrillized under these conditions.
Figure 1.
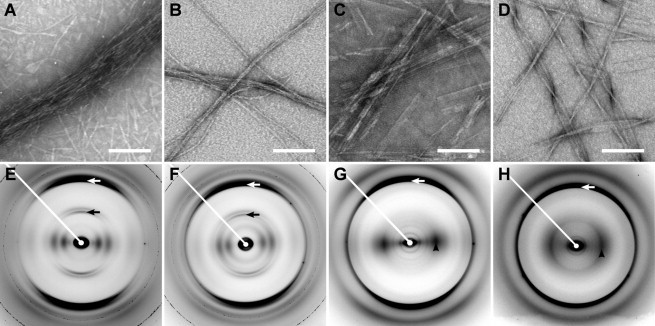
Negative stain electron micrographs (top row), and x-ray fiber diffraction patterns (bottom row) of HET-s(218-289). (A and E) High pH fibrillization; (B and F) low pH fibrillization; (C and G) low pH fibrillization resulting in degradation; (D and H) low pH fibrillization, seeded by degraded low pH fibrils. Meridians in the diffraction patterns (axes parallel to the fiber axis) run approximately vertically, whereas equators (axes orthogonal to the meridian) are approximately horizontal. White arrows: 4.7 Å meridional reflections. Black arrows: 9.4 Å meridional reflections. Black arrowheads: ∼10 Å equatorial intensities. Scale bars = 100 nm.
Diffraction patterns from HET-s(218-289) fibrillized at low pH fell into two very different classes, with little variation within each class. Patterns in the first class (see Fig. 1 F) were remarkably similar to those of the pH 7.5 fibrils (see Fig. 1 E), although the fibrils exhibited a slightly twisted heterogeneous morphology (Fig. 1 B) different from the cable-like bundles of the pH 7.5 HET-s(218-289) fibrils (Fig. 1 A). These patterns were obtained from fibrils formed at pH 2, 2.5, 2.75, and 3, but most commonly above pH 2.5. In some cases fibrillization took only a few days, whereas in others many weeks were required. The example in Fig. 1, B and F, is from a monomer solution allowed to fibrillize at pH 2.0 in a boric acid/citric acid buffer (the buffer used in (8)) at room temperature for about 3 months. The similarities between these diffraction patterns and those from the pH 7.5 fibrils imply a high degree of structural similarity at the levels of both protein fold and fibril symmetry, although they do not necessarily mean that the two structures are identical in detail.
In contrast to this structural similarity, the appearance of the second class of patterns implies major structural differences between these fibrils and the pH 7.5 fibrils. These patterns were obtained from fibrils formed at pH 2, 2.25, and 2.5, sometimes over very long periods but again, sometimes after only a few days. Although fibrillization in this pH range usually led to the second type of pattern, some samples under apparently identical conditions yielded the first type of pattern. The example in Fig. 1, C and G, is from fibrils formed in aliquots from the same purification used for Fig. 1, B and F, and fibrillized under the same conditions (pH 2.0, boric acid/citric acid buffer at room temperature). They were more rigid and sheet-like (Fig. 1 C) than the twisted fibrils in Fig. 1 B. In the diffraction patterns in this second class (Fig. 1 G), the 4.75 Å meridional reflection still showed the presence of cross-β structure, however there was no longer a 9.5 Å layer line, indicating that the two-rung structure was not present. The equatorial reflections were also dramatically different from those of the pH 7.5 fibrils and the fibrils giving rise to the first class of low pH patterns, being dominated by a single strong 11 Å reflection.
The combination of a 4.75 Å meridional reflection and a strong equatorial intensity maximum close to 10 Å, first observed by Astbury (15), has long been recognized as the distinctive signature of stacked β-sheet structure (14–21,28). The interstrand distance (along the fibril) of 4.75 Å gives rise to the meridional reflection, whereas the intersheet distance (across the fibril; ∼10 Å depending on the side chains in the interface between adjacent sheets) gives rise to the equatorial diffraction. The complete lack of a 9.5 Å meridional reflection even in patterns from the exceptionally high-quality beamline (29) used indicates that the β-sheets are parallel, not antiparallel. The strong isolated equatorial 11 Å intensity indicates that the sheets are stacked and not solenoidal; the distinction has been noted for the yeast prion protein Sup35 NM (23,28), and the differences are illustrated in the extensive simulations described in the online Supporting Information of 27; in no case did a solenoidal structure give rise to a pattern resembling the stacked-sheet pattern. The differences between the two types of patterns are illustrated by calculated HET-s diffraction patterns in Fig. S1 in the Supporting Material.
HET-s(218-289) has been shown (8) to form noninfectious amyloid fibrils at pH 2, under the solution conditions used to form both types of fibril seen here. The EM observations (Fig. 1, B and C), and diffraction patterns (Fig. 1, F and G), do not indicate which of these two forms corresponds to the fibrils described in 8, but the experiments with ThT described below indicate that those fibrils were the stacked-sheet fibrils.
Degradation in low pH fibrils
SDS-PAGE analysis of HET-s(218-289) solutions and redissolved fiber diffraction samples (Fig. 2) showed that although newly purified high pH HET-s(218-289) fibrils were composed of homogeneous full-length peptide (Fig. 2 A), all of the low pH fibrils that exhibited stacked β-sheet-like diffraction were extensively proteolyzed, forming several well-defined molecular species (Fig. 2 B). Tryptic digests were performed on the degraded protein bands and MS/MS was used to analyze the composition of the resulting peptides (Fig. 2 D). There was no coverage between residues Asn-266 and Arg-238 because of the large number of tryptic proteolytic sites in this area. Nevertheless, the presence of the first peptide and otherwise complete coverage to the 6×His-tag from band 1 showed that this was the complete recombinant protein. Band 2 did not contain the first tryptic peptide, indicating N-terminal proteolysis into the uncovered region, most probably including strand β1-a. Band 3 contained a unique tryptic peptide showing proteolysis up to Gly-242 and the loss of the first rung of the β-solenoid core. Band 4 contained the first tryptic peptide but was missing the last three tryptic peptides, indicating proteolysis from the C-terminus, including the loss of the second rung of the β-solenoid core. With these significant losses of secondary structure, it is not surprising that these molecular species do not form solenoids, but instead polymerize into simple stacked β-sheets.
Figure 2.
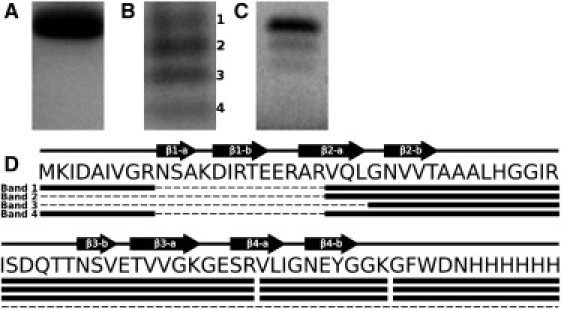
Examples of SDS-PAGE and MS/MS analysis of HET-s(218-289) fibril preparations. (A) SDS-PAGE after high pH fibrillization. (B) SDS-PAGE after low pH fibrillization over 12 months. Numbers indicate the bands analyzed by MS/MS. (C) SDS-PAGE after seeded low pH fibrillization. (D) Secondary structure from ssNMR (10), amino acid sequence, and peptides found by MS/MS analysis. Black arrows: β-strands. Solid lines connecting arrows: loops or unordered regions. Heavy solid lines below the sequence correspond to tryptic peptides from SDS-PAGE bands in (B).
Seeding undegraded HET-s propagates the degraded structure
Fibrils were also made at pH 2.0 by seeding freshly prepared HET-s(218-289) solutions with degraded fibrils that had been formed at low pH. Seeded fibrils formed orders of magnitude more rapidly than unseeded fibrils under comparable conditions, requiring times from minutes to 2 or 3 days, in contrast to unseeded fibrils, which required from a few days to several months to form. The seeded fibrils exhibited heterogeneous, flexible morphology (Fig. 1 D) distinct from that of proteolyzed fibrils (Fig. 1 C), and stacked β-sheet diffraction (Fig. 1 H), but showed little proteolysis in SDS-PAGE (Fig. 2 C). Densitometry of the seeded fibril gel shown in Fig. 2 C, which was from a redissolved fiber used for diffraction, indicated that intact protein made up ∼80% of the diffracting sample, but the diffraction pattern showed no evidence of solenoid structure. We conclude that stacked β-sheets formed from the degraded protein were able to act as templates for undegraded protein, even though we did not at any time observe spontaneous formation of stacked β-sheets by undegraded protein.
ThT fluorescence
ThT fluorescence induced by binding to the fibrils (Fig. 3) confirmed the structural differences observed by fiber diffraction. The β-solenoid structure of the high pH fibrils induced minimal ThT fluorescence, whereas samples composed primarily of stacked β-sheet fibrils (degraded low pH fibrils and seeded freshly made low pH fibrils) showed significant fluorescence (Fig. 3). Seeded low pH fibrils with minimal degradation exhibited ThT fluorescence even stronger than that of degraded fibrils. Some short incubation time low pH HET-s(218-289) fibrils also showed significant amounts of fluorescence, although their diffraction patterns were characteristic of β-solenoid structure. We attribute this fluorescence to the presence of a low proportion of degraded fibrils with the stacked β-sheet conformation, enough to bind a significant fraction of the ThT and induce fluorescence, but not enough to greatly affect the diffraction pattern. The noninfectious low pH fibrils described in 8 also exhibited strong ThT fluorescence, suggesting that those fibrils may have been degraded and contained significant amounts of stacked β-sheet structures.
Figure 3.
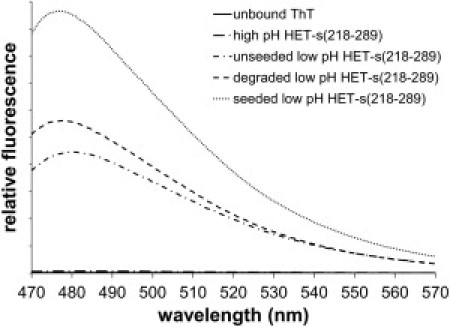
Induced ThT fluorescence on binding to several forms of HET-s(218-289) amyloid. Units of relative fluorescence intensity are arbitrary. The sample for the unbound ThT curve (almost coincident with the axis) contained no protein.
Discussion
Amyloids were for many years believed to have a common stacked β-sheet structure representing a generic low-energy protein fold (14,15,20,21). This commonality, with variations in detail, does appear to hold for short synthetic amyloidogenic peptides, which have been the subject of most detailed amyloid structural studies, but as HET-s illustrates particularly well, biological amyloids may have more complex structures. These complex structures retain the fundamental cross-β motif (β-strands perpendicular to the fibril axis) (30), but often do not retain the simple regular intersheet stacking that gives rise to equatorial diffraction at ∼10 Å. Examples include the HET-s β-solenoid structure (10,12), PrP, for which EM, modeling, and fiber diffraction have also suggested a β-solenoid structure (27,31), and even the Alzheimer's-associated Aβ peptide and the very similar diabetes-associated islet amyloid polypeptide, which form β-structures assembled into hinged sheets similar to but more complex than the simple generic stacked sheets of the synthetic peptide models (32,33).
The two forms of HET-s amyloid are strikingly parallel to two forms of PrP amyloid; highly infectious brain-derived PrP fibrils most probably have a β-solenoid structure, and certainly do not appear to be made up of stacked β-sheets, whereas recombinant amyloid PrP fibrils exhibit a very clear stacked-sheet fiber diffraction pattern. The diffraction patterns of the two forms of PrP (27) bear a considerable resemblance to the two types of HET-s(218-289) diffraction pattern (Fig. 1). Brain-derived PrP fibrils are highly infectious, whereas recombinant PrP amyloid, although infectious, is much less so (34). The infectivity of the recombinant amyloid could be due to a very small fraction of β-solenoid structure, too little to detect by fiber diffraction, or it could be due to an inherently low level of infectivity in the stacked sheet structure (27). The infectivity of fibrils formed from intact HET-s(218-289) at low pH, either the β-solenoid form or the seeded stacked β-sheet form, has not yet been determined, but previous studies of low pH fibrils that may have been degraded and in the stacked β-sheet form (Results) showed very marginal infectivity (8). Although our experiments do not address infectivity, the seeding experiments do demonstrate templating and point to the possibility of transmissibility, although under far from physiological conditions. Nevertheless, the seeding results do provide an illustration of how prion strains that are structurally very different from each other might arise.
The contrast between the solenoidal structure of intact HET-s(218-289) and the generic stacked-sheet structure formed by degraded HET-s(218-289) fragments highlights the possibility that simple stacked-sheet amyloids, although commonly formed by short synthetic peptides, may be only rarely biologically functional or pathological. Stacked β-sheets are generic low-energy structures that can aggregate into amyloid fibrils (35), but the example of HET-s (and probably PrP) suggests that they are not in themselves sufficient to make a biologically active amyloid, that is, a faithfully templating amyloid that propagates itself under physiological conditions. For the propagation of a distinct structure that confers explicit biological effects, a more complex structure may be needed, although this may still be a relatively simple low-energy structure. The increased complexity need not be great: we might speculate that it could be provided by the parts of the connecting loops or hinges closest to the β-strands, or by side-chain interactions between the strands. The β-solenoid of HET-s and probably PrP, and the hinged β-sheets of Aβ and islet amyloid polypeptide, in all of which there are specific additional interactions and connectivities that distinguish them from simple generic stacked β-sheets, could provide this small but necessary additional level of complexity.
Acknowledgments
We thank the staff of beamlines 4-2 at SSRL, BioCAT and BioCARS at the Advanced Photon Source, Argonne National Laboratory, M. McDonald and W. Bian for discussions and assistance with computations, X. Meng for assistance with expression of HET-s, R. Riek for discussions about HET-s proteolysis, A. Steven for encouragement and valuable insights, and D. Friedman in the Vanderbilt University Mass Spectrometry Research Center for the mass spectrometry data.
This work was supported by U.S. National Institutes of Health (NIH) grants AG002132 and T32-GM008320-21. The Stanford Synchrotron Radiation Laboratory is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource (SSRL) Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH National Center for Research Resources. Use of the Advanced Photon Source was supported by the U.S. Department of Energy. BioCAT and BioCARS are NIH-supported Research Centers RR-007707 and RR-008630.
Footnotes
Holger Wille's present address is Centre for Prions and Protein Folding Diseases and Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada.
Ulrich Baxa's present address is Electron Microscopy Laboratory, SAIC-Frederick, Inc., National Cancer Institute at Frederick, Frederick, MD.
Supporting Material
References
- 1.Prusiner S.B. Prions. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott, Williams, and Wilkins; Philadelphia, PA: 2007. pp. 3059–3091. [Google Scholar]
- 2.Wickner R.B., Taylor K.L., Roberts B.T. Prions in Saccharomyces and Podospora spp.: protein-based inheritance. Microbiol. Mol. Biol. Rev. 1999;63:844–861. doi: 10.1128/mmbr.63.4.844-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 4.Frost B., Diamond M.I. Prion-like mechanisms in neurodegenerative diseases. Nat. Rev. Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saupe S.J. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin. Cell Dev. Biol. 2011;22:460–468. doi: 10.1016/j.semcdb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Maddelein M.L., Dos Reis S., Saupe S.J. Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl. Acad. Sci. USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balguerie A., Dos Reis S., Saupe S.J. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabaté R., Baxa U., Saupe S.J. Prion and non-prion amyloids of the HET-s prion forming domain. J. Mol. Biol. 2007;370:768–783. doi: 10.1016/j.jmb.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Sen A., Baxa U., Steven A.C. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J. Biol. Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- 10.Wasmer C., Lange A., Meier B.H. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 11.Wasmer C., Soragni A., Meier B.H. Infectious and noninfectious amyloids of the HET-s(218-289) prion have different NMR spectra. Angew. Chem. Int. Ed. Engl. 2008;47:5839–5841. doi: 10.1002/anie.200704896. [DOI] [PubMed] [Google Scholar]
- 12.Van Melckebeke H., Wasmer C., Meier B.H. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno N., Baxa U., Steven A.C. Structural dependence of HET-s amyloid fibril infectivity assessed by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA. 2011;108:3252–3257. doi: 10.1073/pnas.1011342108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunde M., Serpell L.C., Blake C.C. Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 15.Astbury W.T., Dickinson S., Bailey K. The x-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 1935;29:2351–2360. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eanes E.D., Glenner G.G. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 17.Geddes A.J., Parker K.D., Beighton E. “Cross-beta” conformation in proteins. J. Mol. Biol. 1968;32:343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 18.Bonar L., Cohen A.S., Skinner M.M. Characterization of the amyloid fibril as a cross-beta protein. Proc. Soc. Exp. Biol. Med. 1969;131:1373–1375. doi: 10.3181/00379727-131-34110. [DOI] [PubMed] [Google Scholar]
- 19.Jahn T.R., Makin O.S., Serpell L.C. The common architecture of cross-beta amyloid. J. Mol. Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 20.Nelson R., Sawaya M.R., Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawaya M.R., Sambashivan S., Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 22.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 23.McDonald M., Kendall A., Stubbs G. Enclosed chambers for humidity control and sample containment in fiber diffraction. J. Appl. Cryst. 2008;41:206–209. [Google Scholar]
- 24.Bian W., Wang H., Stubbs G. WCEN: a computer program for initial processing of fiber diffraction patterns. J. Appl. Cryst. 2006;39:752–756. [Google Scholar]
- 25.Stubbs G., Makowski L. Coordinated use of isomorphous replacement and layer-line splitting in the phasing of fiber diffraction data. Acta Crystallogr. A. 1982;38:417–425. [Google Scholar]
- 26.Franklin R.E., Holmes K.C. Tobacco mosaic virus: application of the method of isomorphous replacement to the determination of helical parameters and radial density distribution. Acta Crystallogr. 1958;11:213–220. [Google Scholar]
- 27.Wille H., Bian W., Stubbs G. Natural and synthetic prion structure from x-ray fiber diffraction. Proc. Natl. Acad. Sci. USA. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishimoto A., Hasegawa K., Yoshida M. β-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem. Biophys. Res. Commun. 2004;315:739–745. doi: 10.1016/j.bbrc.2004.01.117. [DOI] [PubMed] [Google Scholar]
- 29.Smolsky I.L., Liu P., Tsuruta H. Biological small-angle x-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J. Appl. Cryst. 2007;40:s453–s458. [Google Scholar]
- 30.Rudall, K. M. 1946. The structure of epidermal protein. The Society of Dyers and Colourists: Symposium on Fibrous Proteins 15–23. Chorley and Pickersgill, Leeds.
- 31.Govaerts C., Wille H., Cohen F.E. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci. USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paravastu A.K., Leapman R.D., Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luca S., Yau W.M., Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legname G., Baskakov I.V., Prusiner S.B. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 35.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 36.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 37.The PyMOL Molecular Graphics System, version 1.2r1. Schrödinger, LLC., Portland, OR.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


