Abstract
Knowledge of “actionable” somatic genomic alterations present in each tumor (e.g., point mutations, small insertions/deletions, and copy number alterations that direct therapeutic options) should facilitate individualized approaches to cancer treatment. However, clinical implementation of systematic genomic profiling has rarely been achieved beyond limited numbers of oncogene point mutations. To address this challenge, we utilized a targeted, massively parallel sequencing approach to detect tumor genomic alterations in formalin-fixed, paraffin embedded (FFPE) tumor samples. Nearly 400-fold mean sequence coverage was achieved, and single nucleotide sequence variants, small insertions/deletions, and chromosomal copy number alterations were detected simultaneously with high accuracy compared to other methods in clinical use. Putatively actionable genomic alterations, including those that predict sensitivity or resistance to established and experimental therapies, were detected in each tumor sample tested. Thus, targeted deep sequencing of clinical tumor material may enable mutation-driven clinical trials and, ultimately, ”personalized” cancer treatment.
INTRODUCTION
The maturation of cancer genome characterization efforts has fueled the notion that many treatment decisions might ultimately be guided by the genetic makeup of individual tumors (1). Moreover, the rapid proliferation of targeted agents in development has called specific attention to the importance of molecular profiling approaches that pinpoint in situ those tumors most likely to respond. Knowledge of such alterations in the clinical and translational arenas—including mutations, somatic copy number alterations, and polymorphisms affecting drug metabolism—should ultimately facilitate individualized approaches to cancer treatment. However, systematic genetic profiling of cancers remains underdeveloped in the clinical setting. Since many targeted agents in development are designed to intercept proteins and/or pathways commonly perturbed by tumor genetic changes, an urgent need exists to implement robust approaches that determine the “actionable” genetic profiles of individual tumors. If widely obtained, such information might better identify those patients most likely to respond to existing and emerging anticancer regimens.
We and others have developed tumor mutation profiling platforms that use mass-spectrometric genotyping (2, 3) or allele-specific PCR based technologies (4). Each of these approaches interrogates known oncogene or tumor suppressor gene mutations present in DNA obtained from either frozen or formalin-fixed, paraffin embedded (FFPE) tumor tissue. However, genotyping-based platforms have certain limitations that may preclude their applicability as definitive cancer diagnostics modalities. These include the finite number of pre-specified point mutations that can be assayed (designated a priori from a restricted subset of known cancer genes), difficulties in detecting small insertions or deletions (“indels”), insensitivity to most tumor suppressor gene mutations (which may occur anywhere within the gene), inability to detect gene amplifications or deletions, and decreased sensitivity in tumor samples with high stromal admixture. At the present time, no systematic mechanism exists whereby clinical tumor specimens might be interrogated in situ for a fully comprehensive panel of “actionable” cancer gene alterations.
The advent of massively parallel sequencing is transforming the cancer genomics landscape by enabling comprehensive cancer genome characterization at an unprecedented scope (1, 5, 6). Concomitantly, hybrid selection based methods that enrich for coding sequences prior to sequencing (“exon capture”) (7, 8) are routinely being implemented in discovery-oriented settings (5). Here, we describe an adaptation of exon capture and massively parallel sequencing for robust detection of somatic genomic alterations in FFPE samples. The approach leverages a targeted exon capture technique to enrich for a cancer-relevant genomic territory consisting of 137 genes (~400,000 coding bases), thereby allowing multiple barcoded samples to be pooled into a single sequencing reaction while preserving deep (e.g., >300-400-fold) sequencing coverage of targeted regions. This approach simultaneously identifies mutations and chromosomal copy number alterations in clinical tumor material, and may inform a comprehensive means to achieve DNA-based patient stratification in the clinical and translational oncology arena.
RESULTS
We generated a list of 137 “druggable” or potentially “actionable” genes known to undergo somatic genomic alterations in cancer (Supplementary Table 1). These include targets of existing and novel therapeutics, prognostic markers, and other oncogenes and tumor suppressors that are frequently mutated in cancer. In addition, we included 79 pharmacogenomic polymorphisms in 34 genes that may predict heightened sensitivity/resistance or toxicity to conventional cancer therapies (Supplementary Table 2). Altogether, these genes are comprised of 2372 exons encoding 433,159 bases. We then designed and synthesized 7,021 unique biotinylated RNA baits corresponding to these genomic regions.
We leveraged a solution-based exon capture/massively parallel sequencing approach in which a pool of long oligonucleotides complimentary to these exons of interest were used to reduce the complexity of tumor genomic DNA for clinically-oriented sequencing. Here, a 6-nucleotide DNA barcode was appended to the ends of DNA fragments during library construction, thus allowing multiple samples to be pooled prior to hybrid selection in order to expand the scope of genomic profiling (9). The approach is illustrated schematically in Supplementary Figure 1.
Capture Performance and Reproducibility
We first optimized the approach using genomic DNA from normal samples and tumor cell lines known to harbor mutations and/or chromosomal copy number alterations affecting multiple cancer genes represented in our hybrid capture baits. Ten cancer cell lines with well characterized, mutually exclusive cancer gene mutations were chosen (Supplementary Table 3) as well as control diploid genomic DNA. Equimolar amounts of the resulting sequencing libraries were pooled together with an additional library from the HT-29 cell line, which was added at a 50% molar ratio compared to the other libraries. This pool of 12 libraries was subjected to a single hybrid selection reaction and sequenced in a single Illumina lane using 100-bp paired end reads.
The 11 equimolar DNAs were evenly represented, with 12-17 million purity-filtered (PF) reads generated per sample (average of ~14.6 million PF reads; Supplementary Table 4); whereas the sample present at 50% concentration (HT-29, index 2) had ~7.8 million PF reads, as expected. The percent of bases mapping “on-target” averaged 60% (range 56% to 64%) across all samples in the pool, yielding a mean target coverage 527x (range 441x to 593x) for the 11 equimolar samples. More than 95% of target exons exhibited >30x coverage after sequencing (sufficient to call “high-confidence” variants in a sample with 70-80% tumor purity), while only 1% had no coverage (Supplementary Figure 2A-B). In general, poorly captured exons had greater than 70% GC content, though GC content did not account for all of the poorly captured targets (Supplementary Figure 2C). The capture performance for a particular target exon was highly reproducible from sample to sample (Supplementary Figure 2D-F).
Detection of Single Nucleotide Variants, Insertions/Deletions, and Copy Number Alterations
In total, 102 single nucleotide variants (SNVs) and 6 indels (excluding known germline polymorphisms) were detected in coding sequences across the 10 cell lines, including all 21 SNVs and 3 out of 4 indels reported for these lines in the Catalogue of Somatic Mutations in Cancer database (COSMIC) (10) (Supplementary Table 5). The single indel that was not initially identified—a 9 bp deletion in PIK3CA in the NCI-H69 cell line—was readily detected by manual inspection of the raw sequencing data. Therefore, all previously reported point mutations and indels for this small collection were detectable by this approach. (A complete listing of all alterations identified in these cell lines can be found in the Supplementary Appendix.)
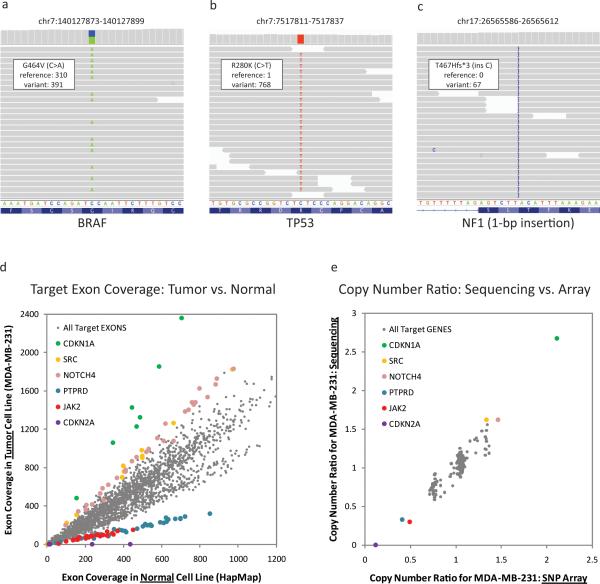
In the absence of paired normal samples, the majority of variants detected are germline alterations. Nonetheless, previously unreported variants were still informative in several instances. For example, 12 single nucleotide variants were detected in the breast cancer cell line MD-MBA-231, including all 4 alterations in the COSMIC database (BRAF, TP53, KRAS, and NF2) (10) (Figures 1A-B, Supplementary Table 6). One of the additional alterations was a 1-bp frameshift insertion involving the NF1 tumor suppressor predicted to generate a truncated protein product (Figure 1C). This NF1 insertion likely represents a bona fide cancer-associated mutation. The MDA-MB-231 cell line has previously been shown to lack both an NF1 mRNA isoform and the neurofibromin protein (the product of NF1); thus, these findings may provide a genetic basis for neurofibromin loss in this setting (11).
Figure 1. Genomic alterations in breast cancer cell line MDA-MB-231.
(A-C) Representative genome images from the Integrated Genome Viewer (IGV) for several alterations found in the breast cancer cell line MD-MBA-231. The number of reads for the reference allele and the variant allele are shown for each alteration. (A) BRAF oncogene point mutation. (B) Point mutation in the TP53 tumor suppressor gene. (C) A 1-bp insertion in tumor suppressor NF1. (D) Sequence coverage for each target exon in breast cancer cell line MD-MBA-231 as compared to a normal diploid sample. Targets from several genes with copy number gains and losses are highlighted. (E) Comparison of gene-level copy number alterations as detected by exon capture and copy number data previously obtained using a high-density SNParray (Affymetrix SNP 6.0 platform). Several genes with copy number gains and losses are highlighted. Copy number data are highly correlated, with a correlation coefficient of 0.94.
Although detection of point mutations and indels by targeted, massively parallel sequencing has become increasingly common, the simultaneous detection of chromosomal copy number alterations by this approach is less well established, particularly in the clinical arena. To determine copy number alterations, the accumulated sequence coverage for each exon in the tumor sample was compared to the coverage obtained for the same exon in the diploid normal control (after normalization for global differences in “on-target” sequence coverage). When tumor and normal reads are displayed as a scatter plot (normal = X axis and tumor = y axis), exons with a neutral copy number across the two samples should be distributed along a diagonal with a slope of 1. Amplified exons present in the tumor should have a greater number of relative reads and therefore fall above the diagonal, while deleted exons should have fewer reads and fall below the diagonal.
Guided by this framework, we determined relative copy number ratios for all targeted exons across the cell line collection. An example for the MDA-MB-231 breast cancer cell line (compared to a normal diploid sample) is shown in Figure 1D. In total, 8 genes with amplifications (defined as mean sequence coverage >3-fold higher than the reference normal) and another 8 with deletions (mean sequence coverage >3-fold lower than the reference normal) were seen across the cell lines. Comparison of overall copy number values derived by sequencing to those obtained from high-density SNP array data (Affymetrix SNP 6.0 platform) demonstrated a robust correlation at the gene level, with correlation coefficients ranging from 0.89 to 0.98 (Supplementary Table 7). As an example, the correlation for the MDA-MB-231 cell line (r2 = 0.94) is shown in Figure 1E.
Profiling of Archival Tumor Samples by Massively Parallel Sequencing
Having established a robust approach for high-throughput exon capture and massively parallel sequencing of 137 cancer genes, we next sought to determine whether this approach might prove useful in the clinical setting. As a proof-of-principle, we characterized a pilot collection of 10 formalin-fixed, paraffin-embedded (FFPE) tumor samples from patients with breast or colon cancer. As was the case with the cell line experiment above, each of the 12 barcoded samples was evenly represented, with a mean coverage of 391x (Table 1). There was greater variation in the tumor samples compared to the cell lines, with coverage ranging from 116x to 537x. This variance may reflect differences in quality of FFPE-derived input DNA. For 11 samples, 94% of exons targeted had >30x coverage after sequencing and 1% had no coverage. In one sample (FFPE sample #9, Table 1), 86% of exons showed >30x coverage and 2% had zero coverage—this sample also had the lowest mean coverage of the group (116X). The tumor purity for eight samples was greater than 50%, whereas two samples had tumor purities of ≤ 20% (FFPE 2 and FFPE 3) (Table 1).
Table 1.
Summary of sequencing results for FFPE samples
| SAMPLE | TUMOR TYPE | TUMOR PURITY | PF READS | PERCENT OF TOTAL PF READS IN POOL | PERCENT SELECTED BASES | MEAN TARGET COVERAGE | PERCENT OF TARGET BASES WITH AT LEAST 30X COVERAGE |
|---|---|---|---|---|---|---|---|
| HAPMAP | N/A | N/A | 9,655,996 | 7% | 46% | 394 | 96% |
| FFPE 1 | Colon | 60% | 11,161,868 | 8% | 46% | 457 | 96% |
| FFPE 2 | Colon | 10% | 8,841,660 | 7% | 48% | 353 | 94% |
| FFPE 3 | Colon | 20% | 13,047,230 | 10% | 44% | 498 | 96% |
| FFPE 4 | Colon | 60% | 10,144,562 | 8% | 38% | 300 | 95% |
| FFPE 5 | Breast | 80% | 16,450,558 | 12% | 36% | 472 | 95% |
| FFPE 6 | Breast | 70% | 15,188,624 | 11% | 42% | 532 | 96% |
| FFPE 7 | Colon | 50% | 8,480,282 | 6% | 39% | 250 | 94% |
| FFPE 8 | Breast | 80% | 15,758,604 | 12% | 41% | 537 | 96% |
| FFPE 9 | Colon | 60% | 3,640,236 | 3% | 42% | 116 | 86% |
| FFPE 10 | Colon | 50% | 14,429,284 | 11% | 36% | 410 | 96% |
| HT-29 (cell line) | Colon | N/A | 7,519,880 | 6% | 53% | 369 | 96% |
Barcoded and pooled genomic DNA from FFPE tumor samples was subjected to exon capture and sequenced in a single 100 bp paired-end Illumina HiSeq2000 lane. Purity filtered (PF) sequence reads for each sample are shown; the percent of total PF reads shows the relative representation of each sample within the pool. “Percent selected bases” indicates bases that mapped within 250 bp of a target exon, including both on- and near-target sequence. “Mean target coverage” represents the average number of unique reads in which each base was sequenced.
In total, 155 sequence variants and 14 indels were detected across the samples. In addition, 2 gene amplifications (>3-fold increase in mean sequence read counts compared to a reference normal sample) and 2 gene-level deletions (3-fold decrease in mean sequence read counts) were seen. (Summary information for all 10 samples is shown in Supplementary Table 8; a complete listing of all alterations can be found in the Supplementary Appendix.)
Detection of Clinically Actionable Genomic Alterations in FFPE Tumor Samples
Next, we developed an initial framework to segregate genetic alterations based on their predicted clinical utility. Toward this end, we designated three categories of alterations. One category, termed “Actionable in Principle”, includes variants that predict tumor sensitivity or resistance to FDA-approved (Tier 1) or experimental therapies (Tier 2). Another category contains prognostic or diagnostic variants. The remaining alterations are termed “Variants of Unclear Significance”, which may include biologically important mutations without known therapeutic implications as well as uncharacterized mutations in genes with presumed clinical relevance.
We detected biologically or clinically meaningful alterations in all 10 FFPE samples, including the two samples that contained only 10-20% tumor cells. These include known somatic mutations in KRAS, BRAF, PIK3CA, and CTNNB1; nonsense mutations in tumor suppressors APC, MSH2, SMAD2, TSC1, and TP53; and a 2-bp deletion in BRCA1. In particular, 12 of the 155 SNVs and 1 of the 14 indels were deemed plausibly actionable (“Actionable in Principle” or “Prognostic/Diagnostic”; Table 2). KRAS mutations in colon cancer predict resistance to cetuximab (12, 13), and exemplify Tier 1 actionable alterations. In addition, mutations in PIK3CA have been shown in some studies to promote resistance to cetuximab in colon cancer (13-16) and trastuzumab in breast cancer (17, 18), and therefore may conceivably represent Tier 1 alterations (although this has not been shown definitively). Multiple Tier 2 actionable alterations (targeted by drugs currently in clinical development) were also seen, including mutations in PIK3CA (PI3K pathway inhibitors (19)), KRAS (MEK inhibitors (20)), TSC1 (TOR inhibitors (21)), BRAF (MAPK pathway inhibitors (22, 23)) and BRCA1 (PARP inhibitors (24)). Other noteworthy alterations included a nonsense mutation in MSH2, which is diagnostic for hereditary non-polyposis coli (HNPCC) and is a prognostic marker in colon cancer; and a nonsense mutation in SMAD2, which has been suggested to be associated with advanced disease and decreased survival in colon cancer (25).
Table 2.
Actionable or prognostic genomic alterations in 10 FFPE tumor samples
| SAMPLE | TUMOR TYPE | MEAN TARGET COVERAGE | Actionable in Principle | Prognostic / Diagnostic | |
|---|---|---|---|---|---|
| Tier 1 | Tier 2 | ||||
| FFPE 1 | Colon | 457 | KRAS (Q61H)b | KRAS (Q61H)a | |
| FFPE 2 | Colon | 353 | KRAS (G13C)a | KRAS (G13C)a | |
| FFPE 3 | Colon | 498 |
KRAS (G13C)a PIK3CA (H1047R)b |
KRAS (G13C)a PIK3CA (H1047R)a |
MSH2 (R680*)a |
| FFPE 4 | Colon | 300 |
BRAF (D594G)d TSC1 (E258*)c |
||
| FFPE 5 | Breast | 472 |
CCND1 ampb FGFR1 ampd |
CCND1 ampd FGFR1 ampa |
CCND1 ampb |
| FFPE 6 | Breast | 532 | BRCA1 (2-bp del)a | BRCA1 (2-bp del)a | |
| FFPE 7 | Colon | 250 |
KRAS (G13D)a PIK3CA (E545K)b |
KRAS (G13D)a PIK3CA (E545K)a |
|
| FFPE 8 | Breast | 537 | PIK3CA (H1047R)b | PIK3CA (H1047R)a | |
| FFPE 9 | Colon | 116 | SMAD2 (S306*)b | ||
| FFPE 10 | Colon | 410 | KRAS (Q61H)b | KRAS (Q61H)a | |
clinically validated and approved alteration (for Tier 1 or Prognostic/Diagnostic) or specifically targeted alteration (for Tier 2) [shown in bold]
limited clinical evidence
clinical evidence in a different tumor type only
pre-clinical evidence only
Abbreviations: amp = amplification, del = deletion
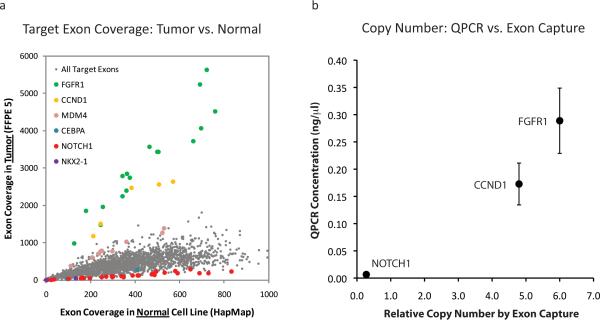
Plausibly actionable amplifications of both FGFR1 and CCND1 were observed in a breast tumor sample (Figure 2A). In preclinical studies, FGFR1 amplification was shown to predict resistance to hormonal therapy in breast cancer (26), and thus may be considered a candidate Tier 1 copy number event for this FDA-approved indication. Clinical trials are currently underway to test FGFR inhibitors against tumors with amplified or overexpressed FGFR1, making FGFR1 amplification a Tier 2 actionable variant as well. Amplification of CCND1 (which encodes the Cyclin D1 cell cycle regulator) has also been suggested to predict resistance to hormonal therapy (27, 28). Moreover, this alteration may predict sensitivity to cyclin-dependent kinase inhibitors (Tier 2 actionable event) (29), as well as overall disease prognosis in breast cancer (Prognostic alteration) (27, 28, 30). Lower level copy number alterations (between 2- and 3-fold relative changes) were observed in several known or putative cancer genes, including CDK8, GNAS, MYC, and SRC. Although these events are most likely to reflect aneuploidy, some may represent higher level copy number alterations in samples with low tumor purity.
Figure 2. Copy number alterations in an archival breast cancer sample.
(A) Sequence coverage is shown for each target in the tumor sample as compared to a normal diploid sample. Exon targets from several genes with copy number gains and losses are highlighted. (B) Copy number correlation between exon capture and QPCR in sample FFPE 5. Quantitative PCR of FGFR1, CCND1, and NOTCH1 using 3 independent sets of primers was performed and average values for each gene were compared to exon capture copy number.
Examination of 79 pharmacogenomic loci facilitated inspection of plausibly actionable polymorphisms (Supplementary Table 9). The ERCC2-K751QC allele, associated with increased risk of FOLFOX-induced grade 3 or 4 hematologic toxicity (31), was present in 2 samples (FFPE 2 and FFPE 9). The UGT1A1-G3156A allele was found to be heterozygous in 5 samples but homozygous in none of them. This allele is associated with irinotecan-related neutropenia when present as a homozygous event (32).
To validate these findings, a representative subset of alterations (31 nonsynonymous variants and 2 indels; samples 4-7) were independently queried by mass spectrometric genotyping (2, 3). All 31 SNVs and 2 indels tested were confirmed, demonstrating 100% specificity of the targeted exon capture approach in the small subset examined. Copy number alterations involving 3 genes that were amplified or deleted in sample FFPE 5 (FGFR1, CCND1, NOTCH1) were also tested by quantitative PCR (QPCR) using 3 independent primer pairs for each gene. As shown in Figure 2B, the QPCR results were highly correlated to the copy number ratios detected by targeted exon capture/sequencing in FFPE 5 (r2 = 0.94). The correlation coefficient for these same genes in sample FFPE 9—which has a 2.3-fold amplification of FGFR1 but no copy number changes in CCND1 or NOTCH1—was r2 = 0.99 (Supplementary Figure 3).
Comparison to an Existing Mutation Profiling platform
We next wished to compare the sensitivity and specificity of targeted hybrid capture/sequencing to an existing mass spectrometric genotyping based platform, since this type of approach is currently being used in several clinical and translational oncology settings (2, 33-35). We thus performed OncoMap, a mass-spectrometric genotyping technology that interrogates more than 400 known mutations in 33 cancer genes. Of the 155 SNVs seen by hybrid capture/sequencing of the FFPE samples described above, 13 were also interrogated by assays present in OncoMap (Table 3). However, OncoMap only detected 10 of these 13 mutations. To determine the basis for this discrepancy, we assayed all 13 mutations by an orthogonal genotyping approach that uses distinct reagent chemistry (hME genotyping; see Methods). All 13 mutations were confirmed by this orthogonal genotyping method, suggesting that the 3 mutations not detected by OncoMap were false negatives by mass spectrometric genotyping (Table 3, shown in bold). All mutations seen by OncoMap were also detected by targeted exon capture.
Table 3.
Comparison of OncoMap and targeted exon capture profiling in FFPE samples.
| FFPE SAMPLE | SAMPLE TYPE | GENE | MUTATION | SEEN VIA ONCOMAP | SEEN VIA EXON CAPTURE | VALIDATED BY hME |
|---|---|---|---|---|---|---|
| FFPE 1 | Colon | KRAS | Q61H | YES | YES | YES |
| FFPE 2 | Colon | KRAS | G13C | YES | YES | YES |
| FFPE 3 | Colon | KRAS | G13D | YES | YES | YES |
| FFPE 3 | Colon | PIK3CA | H1047R | YES | YES | YES |
| FFPE 4 | Colon | BRAF | D594G | NO | YES | YES |
| FFPE 4 | Colon | APC | Q1367* | NO | YES | YES |
| FFPE 4 | Colon | APC | Q1378* | YES | YES | YES |
| FFPE 6 | Breast | TP53 | R248Q | YES | YES | YES |
| FFPE 7 | Colon | KRAS | G13D | YES | YES | YES |
| FFPE 7 | Colon | PIK3CA | E545K | YES | YES | YES |
| FFPE 7 | Colon | CTNNB1 | S45F | NO | YES | YES |
| FFPE 8 | Breast | PIK3CA | H1047R | YES | YES | YES |
| FFPE 10 | Colon | KRAS | Q61H | YES | YES | YES |
DISCUSSION
We have developed a targeted, massively parallel sequencing platform to detect actionable genomic alterations in clinical tumor samples. In this initial proof-of-concept effort, we sequenced 137 cancer genes from 10 pooled FFPE tumor DNA samples (plus 2 control samples) and achieved 391x mean coverage per sample within a single paired-end sequencing lane. This depth of coverage afforded robust, simultaneous detection of base mutations, indels, amplifications and deletions. Thus, targeted massively parallel sequencing provides a unifying approach for detection of multiple categories of actionable genetic alterations.
In our pilot study, all of the tumor samples profiled contained biologically or clinically meaningful genomic alterations, including several that might predict sensitivity or resistance to targeted agents or provide useful prognostic information. In particular, 15 alterations (at least one per sample) were plausibly actionable, and might thus be predicted to impact clinical decision-making or clinical trial enrollment if identified as part of an experimental therapeutics or phase I trial program. Several actionable somatic alterations (KRAS, PIK3CA, and MSH2) were detected in samples with tumor purity as low as 10-20%, highlighting the utility of this approach in “real world” clinical tumor samples.
Comparison with OncoMap, a mass spectrometric genotyping platform in current translational use, confirmed robust performance of targeted massively parallel sequencing, even when applied to FFPE tumor specimens. In our prior study, the sensitivity and specificity of OncoMap in FFPE tissue was 89.3% and 99.4%, respectively, based on a focused comparison to massively parallel sequencing of KRAS (codon 12) in 93 FFPE samples. In the current study, OncoMap detected 10 of 13 mutations (79% sensitivity) that were seen by sequencing at multiple loci (including KRAS). The OncoMap approach involves iPLEX genotyping of >500 mutations followed by hME validation of all candidates (see Methods)—the iPLEX method allows increased multiplexing, but in our hands has proved somewhat less sensitive than hME genotyping. The fact that all 13 mutations were subsequently confirmed by hMe chemistry suggests that massively parallel sequencing to several hundred fold mean coverage affords enhanced sensitivity compared to mass spectrometric genotyping. Moreover, most alterations found by sequencing are not assayed by genotyping or allele-specific PCR based mutation profiling platforms. Thus, the sequencing-based approach may uncover more actionable options for patients than allele-specific approaches.
Hybrid selection approaches have been widely used to promote gene discovery by reducing genome complexity prior to sequencing (5). In this study, we adapted this technique to capture a highly restricted genomic territory comprised of 137 known cancer genes and 400,000 coding bases. This afforded an expanded depth of coverage (to >400-fold) while also enabling multiple barcoded samples to be pooled within a single sequencing lane, thereby increasing throughput and lowering costs. We previously employed a similar approach to characterize a frozen tumor sample from a patient with metastatic melanoma who developed resistance to the RAF-inhibitor vemurafenib, and identified an activating mutation in MEK1 that caused resistance to RAF- and MEK-inhibition (36). Here, we have adapted the approach to capture and sequence multiple barcoded samples and to identify distinct categories of genomic alterations simultaneously.
An advantage of solution-phase hybrid capture is that redesign and synthesis of long oligonucleotides for bait generation is a straightforward process that may be performed iteratively until an optimal set of baits has been developed. Thus, prioritized genomic regions can be readily amended as new knowledge of cancer gene mutations becomes available. Furthermore, DNA barcoding and pooling decreases the sequencing cost per sample in a manner proportional to the number of pooled samples present within a sequencing lane. Achieving deep sequencing coverage increases the sensitivity of mutation detection—particularly in the setting of high stromal admixture, which can pervade clinical tumor tissue. As such, this study extends earlier barcoding and hybrid capture/sequencing efforts (36-49) by identifying multiple types of actionable somatic alterations in archival (i.e., FFPE) tumor specimens. Since the majority of clinical samples are stored as FFPE material, this approach may prove suitable for many translational and clinical applications.
At the same time, variations in FFPE sample quality may adversely affect library construction, hybrid selection, or sequencing. Potential solutions include the incorporation of additional pre-processing steps to enrich for high-quality FFPE DNA, pooling of fewer samples prior to hybrid selection, and/or increasing the overall depth of sequencing if the starting library complexity is sufficiently high (50). The use of orthogonal technologies such as direct genotyping, quantitative PCR, or FISH to validate actionable alterations may prove useful in the short term, as these techniques are widely employed in existing clinical laboratories. However, if the superior sensitivity and specificity is confirmed in independent clinical studies, massively parallel sequencing may become increasingly used in diagnostic or CLIA laboratory settings.
Several additional areas for technical and analytical optimization remain. Although we generally achieved robust sequence coverage of targeted regions, genomic territory with very high or very low GC content presents certain challenges. Options to improve coverage of these regions include redesign or inclusion of additional baits targeting regions that are difficult to capture. On the analytical side, detection of longer indels (such as the 9bp PIK3CA deletion in the NCI-H69 cell line) remains difficult with current algorithms. Since actionable indels occur in multiple genes including EGFR, ERBB2, and KIT, supplemental assays may be needed to ensure sensitive indel detection. Moreover, exon-directed capture approaches do not detect clinically relevant gene rearrangements such as those involving ALK, ABL, and PDGFR. One potential strategy to detect known rearrangements would involve design of baits tiled across common translocation breakpoints. Furthermore, whereas both amplifications and deletions could be detected in cell line DNA, such events were only observed in a single FFPE sample, which had 80% tumor purity. Detection of copy number aberrations by targeted sequencing may be more problematic in samples with significant stromal contamination. Future analytical methods that incorporate allelic information to infer tumor purity may enhance detection of copy gains and losses in samples with variable tumor purity.
Emerging frameworks for clinical interpretation of genome sequencing data typically categorize alterations based on “actionability” or prognostic utility. Potentially actionable alterations may be further subdivided depending on the level of evidence about a particular alteration, ranging from those with established therapies to others with sound pre-clinical evidence. Plausibly actionable alterations may also include those for which the predictive implications within a particular cancer type are not known (e.g. BRAF mutations in lung cancer), or for which there is no established clinical proof of concept (e.g. RET mutations in lung cancer), even though a particular therapy against the target (sorafenib) may be commercially available. This category may also includes mutations in tumor suppressor genes (e.g., PTEN) hypothesized to predict vulnerability to targeted agents (e.g., PI3-kinase inhibitors).
More than 160 variants of unclear significance were identified in our sample set. Undoubtedly, many such variants represent uncharacterized germline polymorphisms. Differentiating somatic from germline alterations is readily accomplished by including matched normal samples (36), although paired normal material is not always available in research settings. Even amongst alterations that are clearly somatic, additional approaches to interpret their potential significance and communicate the results to clinicians and patients will be needed. Development of a rigorous formalism for clinical interpretation of complex genomic data will likely become an active research area, with the goal of enabling optimal, genomics-driven decision making for therapy or clinical trial enrollment.
Potential applications for targeted hybrid capture/massively parallel sequencing in translational and clinical oncology research include both retrospective and prospective profiling of tumor cohorts. Here, the goal may be to identify predictive and prognostic genes or validate pharmacogenomic polymorphisms. Ultimately, similar approaches may be used for prospective genomic profiling of cancer patients to guide clinical decision making. Toward this end, the potential turnaround time for the current approach is ~2 weeks. Emerging sequencing instruments promise vast reductions in turnaround time. Cost, a significant consideration in clinical sequencing, can also be reduced dramatically by sample pooling. Indeed, it is likely that a combination of multiplexing together with falling sequencing costs may ultimately eliminate cost as a limiting barrier to sequencing data generation.
In conclusion, the results described herein suggest that targeted, massively parallel sequencing offers a promising method to detect actionable genetic alterations across a large panel of cancer genes in the clinical diagnostic arena. If widely deployed, such implementation may open new opportunities to link cancer genomics with molecular features, clinical outcomes, and treatment response in a manner that empowers multiple directions in molecular cancer epidemiology. In addition, this approach may ultimately impact clinical practice by offering a categorical means to identify genetic changes affecting genes and pathways targeted by existing and emerging drugs, thereby speeding the advent of personalized cancer medicine.
METHODS
High-throughput, targeted deep sequencing: overview
Massively parallel sequencing libraries (Illumina) that contain barcoded universal primers (9) were generated using genomic DNA from FFPE tumor material. After pre-amplification and DNA quantification, equimolar pools were generated consisting of 12 barcoded tumor DNAs. These DNA pools were subjected to solution-phase hybrid capture with biotinylated RNA baits targeting all exons from 137 “actionable” cancer genes. Each hybrid capture reaction was sequenced in a single paired-end lane of an Illumina flow cell. Subsequently, the sequencing data was deconvoluted to match all high-quality barcoded reads with the corresponding tumor samples, and genomic alterations (single nucleotide sequence variants, small insertions/deletions, and DNA copy number alterations) were identified. The approach is illustrated schematically in Supplementary Figure 1.
Tumor Tissue and Cell Line DNA
Discarded and de-identified tumor specimens were obtained from the Cooperative Human Tissue Network (CHTN). Institutional review board (IRB) exemption was obtained for all samples from the Dana-Farber/Partners Cancer Care Office for the Protection of Research Subjects (Protocol 10-380). Genomic DNA was extracted from tumor tissue using methods previously described (2). Cell line genomic DNA was purchased directly from the American Type Culture Collection (ATCC). Authentication of cell line genomic DNA was performed by ATCC using short tandem repeat (STR) profiling, which employs multiplex PCR to simultaneously amplify the amelogenin gene and eight of the most informative polymorphic markers in the human genome. Control genomic DNA was from the HapMap consortium, purchased from the Coriell Institute for Medical Research.
Barcoded Genomic DNA Library Construction
Genomic DNA was quantified using Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, California). 1 μg of genomic DNA from each sample was sheared by sonication with the following conditions: Duty Cycle 10%, Intensity 5, Cycles per Burst 200, and 135 seconds (Covaris S2 instrument). Paired-end adapters for massively parallel sequencing (Illumina) were added as previously described (51), with the following modifications to the paired end library preparation step (Basic Protocol 2). First, the multiplex adapter provided with the Multiplex Paired-End Library Sample Preparation Kit (Illumina) was used instead of the standard paired-end adapter. Second, PCR enrichment was conducted in 150 μl total volume with 3 primers from the Multiplexing Sample Preparation Oligonucleotide Kit (Illumina). Each PCR enrichment reaction contained 75 μl Phusion polymerase (Finnzymes), 3 μl Multiplexing PE Primer 1.0 (25 μM), 3 μl Multiplexing PE Primer 2.0 (0.5 μM), 3 μl of an Index primer (25 μM), 36 μl paired-end library, and 30 μl nuclease-free water. Samples were denatured for 5 min at 95°C; 18 cycles of 10 sec at 95°C, 30 sec at 65°C, and 30 sec at 72°C; and final 5 min at 72°C before cooling to 4°C. PCR primers were removed by using 1.8× volume of Agencourt AMPure PCR Purification kit (Agencourt Bioscience Corporation).
Selection of Targeted Genes
We identified 137 genes that are biologically or clinically relevant in cancer, including targets of new and existing therapies, genes that predict sensitivity or resistance to therapies, genes that are prognostic markers, and oncogenes and tumor suppressors that are known to undergo recurrent somatic genomic alterations in cancer (Supplementary Table 1). These genes were identified by mining existing databases including the Catalogue of Somatic Mutations in Cancer (COSMIC) (10) and The Cancer Genome Atlas (52). In addition, we identified 79 pharmacogenomic polymorphisms described in the literature, which might predict sensitivity or resistance to conventional cancer therapies (Supplementary Table 2).
Biotinylated RNA Baits
The Agilent SureSelect E-array program was used to design 7,021 unique RNA baits corresponding to the coding sequence of the 137 genes described above, as well as to the 79 pharmacogenomic polymorphisms and to 24 SNPs for fingerprinting. Target loci were covered with a tiling density of 2x. Baits were replicated 8 times on the 55,000 bait library array. The sequences of all 7,021 baits are listed in the Supplementary Appendix. Biotinylated RNA baits were synthesized by Agilent for the SureSelect Target Enrichment system.
Pooling and Hybrid Capture
DNA libraries were pooled by mixing 300 ng of each library in a single 1.5 mL polypropylene sample tube, lyophilizing using a speedvac evaporator, and resuspending in 4 μl of nuclease-free water. This entire amount (3600 ng DNA in 4 μl) was used for hybrid selection. Solution-phase hybrid capture was performed as previously described (51) with three modifications to the hybrid selection step (Basic Protocol 3). First, instead of 1.5 μl of Blocking Oligo 2.0, 0.125 μL of each of 12 additional 200 μM blocking oligonucleotides with sequences complementary to the barcodes were added to the hybridization reaction (see Supplementary Methods for sequences). Second, the biotinylated oligonucleotide baits were diluted 1:8 with nuclease-free water from a concentration of 100 ng/μl to 12.5 ng/μl immediately prior to hybridization and 5 μl of this solution was added to the hybridization reaction. The final volume of the hybridization reaction was 19 μl, consisting of the following components: 4 μl pooled DNA libraries, 2.5 μl 1.0 mg/ml human Cot-1 DNA, 2.5 μl 10.0 mg/ml salmon sperm DNA, 1.5 μl 200 μM blocking oligo 1.0, 1.5 μl total of the twelve 200 μM blocking oligonucleotides, 5.0 μl 12.5 ng/μl biotinylated oligonucleotide baits, 1.0 μl 20 U/μl Superase-In RNAse inhibitor, and 1 μl nuclease-free water. Third, during PCR enrichment of the captured DNA (“the catch”), PCR was performed with primers P5 (5’-AAT GAT ACG GCG ACC ACC GA-3’) and P7 (5’-CAA GCA GAA GAC GGC ATA CGA-3’), both at 100 μM, instead of PCR primers PE1.0 and PE2.0. PCR conditions remained as described. All custom primers were obtained from Integrated DNA Technologies (IDT).
Sequencing and Analysis
We sequenced 100 bases from both ends of library DNA fragments using an Illumina HiSeq 2000 instrument. The sequence reads were aligned to human reference genome hg18 with the Burrows-Wheeler Alignment tool (BWA) (53), using the following parameters: –q 5 –l 32 –k 2 –o 1. Artifactual duplicate read pairs were removed using Picard tools (picard.sourceforge.net). An average of 450 megabases of aligned sequence was generated for each library.
Single nucleotide variants (SNVs) and small insertions/deletions were identified using algorithms from the Genome Analysis Toolkit developed at the Broad Institute (54). A local multiple sequence alignment was performed on intervals suspected to harbor indels in order to derive the most probable underlying genomic structure of the query sample. SNVs were called separately on each sample using UnifiedGenotyper and annotated using GenomicAnnotator. Variants were discarded if they were present in dbSNP and not in the COSMIC database (10), they exhibited an unfavorable strand balance score (> -20), or they were detected in the HapMap normal control. Novel recurrent SNVs were manually reviewed to eliminate additional systematic artifacts. Indels were called using IndelGenotyperV2 and were retained if they occurred in protein-coding exons and on both DNA strands, in <2% of reads in the HapMap normal control, and were absent from dbSNP.
To calculate relative copy number levels of the 137 target gene loci, we computed the mean sequence coverage for each gene across all protein-coding exons using the DepthOfCoverage tool in the Genome Analysis Toolkit. All bases in reads with mapping quality <5 were ignored, as were any additional bases with base quality <5. Gene-level coverage in each tumor was normalized by the gene-level coverage for an indexed HapMap diploid cell line included in the same pooled hybrid selection experiment (after adjusting for differences in the overall amount of aligned sequence per sample). Sequence-derived estimates of copy number were then compared to SNP array-derived estimates of copy number for the cancer cell lines.
Mass Spectrometric Genotyping
Mass spectrometric genotyping was performed with the OncoMap 3.0 platform as previously described (2, 3), using iPLEX chemistry for initial mutation profiling and validation by multi-base hME™ extension chemistry. Genomic DNA from all tumor samples was quantified using Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, California).
To validate alterations detected by massively parallel sequencing that were not included in the OncoMap assay collection, base substitutions and indels were queried using multi-base hME™ extension chemistry with plexing of ≤6 assays per pool. Conditions for hME validation were implemented as described previously (2, 3). Primers and probes used for hME validation were designed using the Sequenom MassARRAY Assay Design 3.0 software, applying default multi-base extension (MBE) parameters but with the following modifications: maximum multiplex level input equal to 6; maximum pass iteration base adjusted to 200.
Microarray Analysis of Chromosomal Copy Number
Chromosomal copy number information was obtained from the Broad-Novartis Cancer Cell Line Encyclopedia project, which has high-density SNP array data from the Affymetrix SNP 6.0 platform for all cancer cell lines profiled in this study (55).
Quantitative PCR Analysis of Chromosomal Copy Number
Quantitative PCR (QPCR) was performed using the SYBR® Green PCR Master Mix kit (Applied Biosystems) according to the manufacturer's instructions. To determine the chromosomal copy number of each gene, three sets of gene-specific primers were designed to interrogate the genetic locus. Primers recognizing LINE sequences were used for reference amplification/normalization as described previously (56). Primer sequences are provided in Supplementary Methods. Male genomic DNA (Promega) was included as a standard, and HapMap DNA (Coriell) was used as a normal diploid control. QPCR reactions were performed in triplicate for each sample using an ABI 7300 instrument, in 25 μl reactions containing 0.5 ng genomic DNA and forward and reverse primers each at a concentration of 600 nM.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Despite the rapid proliferation of targeted therapeutic agents, systematic methods to profile clinically relevant tumor genomic alterations remain underdeveloped. Here we describe a sequencing-based approach to identifying genomic alterations in FFPE tumor samples. These studies affirm the feasibility and clinical utility of targeted sequencing in the oncology arena and provide a foundation for genomics-based stratification of cancer patients.
Acknowledgments
Financial Support:
This work was supported by the NIH Director's New Innovator Award DP2OD002750 (L.A.G.), the National Cancer Institute R33CA126674 (L.A.G.), the National Cancer Institute U24CA143867 (M.M.), the Snyder Medical Foundation (W.C.H.), and the Starr Cancer Consortium (M.F.B., L.A.G.).
Footnotes
Disclosures: Consultant/Advisory Role: Foundation Medicine (N.W., M.F.B., M.J.D., M.M., L.A.G.), Novartis (W.C.H., M.M., L.A.G.), Daiichi Sankyo (L.A.G.). Ownership Interest: Foundation Medicine (N.W., M.M., L.A.G.). Research Support: Novartis (W.C.H., M.M., L.A.G.). Patents: Laboratory Corporation of America (M.M.). Honoraria: Illumina (M.F.B.).
REFERENCES
- 1.Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol. 2010;28(35):5219–28. doi: 10.1200/JCO.2009.27.4944. PMCID: 3020694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. PMCID: 2774511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 4.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–58. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 6.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–24. doi: 10.1038/nature07943. PMCID: 2821689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–9. doi: 10.1038/nbt.1523. PMCID: 2663421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc. 2009;4(6):960–74. doi: 10.1038/nprot.2009.68. PMCID: 2990409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5(10):887–93. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. PMCID: 3013785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogata H, Sato H, Takatsuka J, De Luca LM. Human breast cancer MDA-MB-231 cells fail to express the neurofibromin protein, lack its type I mRNA isoform and show accumulation of PMAPK and activated Ras. Cancer Lett. 2001;172(2):159–64. doi: 10.1016/s0304-3835(01)00648-6. [DOI] [PubMed] [Google Scholar]
- 12.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27(12):2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 13.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28(7):1254–61. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 14.Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One. 2009;4(10):e7287. doi: 10.1371/journal.pone.0007287. PMCID: 2750753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–7. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 16.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 17.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Zhang Q, Zhang J, Sun S, Guo H, Jia Z, et al. PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer. 2011;11(1):248. doi: 10.1186/1471-2407-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28(6):1075–83. doi: 10.1200/JCO.2009.25.3641. PMCID: 2834432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffy A, Kummar S. Targeting mitogen-activated protein kinase kinase (MEK) in solid tumors. Target Oncol. 2009;4(4):267–73. doi: 10.1007/s11523-009-0125-x. [DOI] [PubMed] [Google Scholar]
- 21.Wagner AJ, Malinowska-Kolodziej I, Morgan JA, Qin W, Fletcher CD, Vena N, et al. Clinical activity of mTOR inhibition with sirolimus in malignant perivascular epithelioid cell tumors: targeting the pathogenic activation of mTORC1 in tumors. J Clin Oncol. 2010;28(5):835–40. doi: 10.1200/JCO.2009.25.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical Characteristics of Patients With Lung Adenocarcinomas Harboring BRAF Mutations. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smalley KS, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letrero R, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28(1):85–94. doi: 10.1038/onc.2008.362. PMCID: 2898184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 25.Xie W, Rimm DL, Lin Y, Shih WJ, Reiss M. Loss of Smad signaling in human colorectal cancer is associated with advanced disease and poor prognosis. Cancer J. 2003;9(4):302–12. doi: 10.1097/00130404-200307000-00013. [DOI] [PubMed] [Google Scholar]
- 26.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70(5):2085–94. doi: 10.1158/0008-5472.CAN-09-3746. PMCID: 2832818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bostner J, Ahnstrom Waltersson M, Fornander T, Skoog L, Nordenskjold B, Stal O. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene. 2007;26(49):6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- 28.Lundgren K, Holm K, Nordenskjold B, Borg A, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10(5):R81. doi: 10.1186/bcr2150. PMCID: 2614516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. PMCID: 2790859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy PG, Pratt N, Purdie CA, Baker L, Ashfield A, Quinlan P, et al. High CCND1 amplification identifies a group of poor prognosis women with estrogen receptor positive breast cancer. Int J Cancer. 2010;127(2):355–60. doi: 10.1002/ijc.25034. [DOI] [PubMed] [Google Scholar]
- 31.Boige V, Mendiboure J, Pignon JP, Loriot MA, Castaing M, Barrois M, et al. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000-05. J Clin Oncol. 2010;28(15):2556–64. doi: 10.1200/JCO.2009.25.2106. [DOI] [PubMed] [Google Scholar]
- 32.Lara PN, Jr., Natale R, Crowley J, Lenz HJ, Redman MW, Carleton JE, et al. Phase III trial of irinotecan/cisplatin compared with etoposide/cisplatin in extensive-stage small-cell lung cancer: clinical and pharmacogenomic results from SWOG S0124. J Clin Oncol. 2009;27(15):2530–5. doi: 10.1200/JCO.2008.20.1061. PMCID: 2684855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13(1):64–73. doi: 10.1016/j.jmoldx.2010.11.005. PMCID: 3070595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer. 2011;73(1):96–102. doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beadling C, Heinrich MC, Warrick A, Forbes EM, Nelson D, Justusson E, et al. Multiplex Mutation Screening by Mass Spectrometry: Evaluation of 820 Cases from a Personalized Cancer Medicine Registry. J Mol Diagn. 2011 doi: 10.1016/j.jmoldx.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting Therapeutic Resistance to RAF Inhibition in Melanoma by Tumor Genomic Profiling. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3(65):65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berg JS, Evans JP, Leigh MW, Omran H, Bizon C, Mane K, et al. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: Implications for application to clinical testing. Genet Med. 2011;13(3):218–29. doi: 10.1097/GIM.0b013e318203cff2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chmielecki J, Peifer M, Jia P, Socci ND, Hutchinson K, Viale A, et al. Targeted next-generation sequencing of DNA regions proximal to a conserved GXGXXG signaling motif enables systematic discovery of tyrosine kinase fusions in cancer. Nucleic Acids Res. 2010;38(20):6985–96. doi: 10.1093/nar/gkq579. PMCID: 2978357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossmann V, Kohlmann A, Klein HU, Schindela S, Schnittger S, Dicker F, et al. Targeted next-generation sequencing detects point mutations, insertions, deletions and balanced chromosomal rearrangements as well as identifies novel leukemia-specific fusion genes in a single procedure. Leukemia. 2011;25(4):671–80. doi: 10.1038/leu.2010.309. [DOI] [PubMed] [Google Scholar]
- 41.Johansson H, Isaksson M, Sorqvist EF, Roos F, Stenberg J, Sjoblom T, et al. Targeted resequencing of candidate genes using selector probes. Nucleic Acids Res. 2011;39(2):e8. doi: 10.1093/nar/gkq1005. PMCID: 3025563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiialainen A, Karlberg O, Ahlford A, Sigurdsson S, Lindblad-Toh K, Syvanen AC. Performance of microarray and liquid based capture methods for target enrichment for massively parallel sequencing and SNP discovery. PLoS One. 2011;6(2):e16486. doi: 10.1371/journal.pone.0016486. PMCID: 3036585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao GJ, Lun FM, Zheng YW, Chan KC, Leung TY, Lau TK, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem. 2011;57(1):92–101. doi: 10.1373/clinchem.2010.154336. [DOI] [PubMed] [Google Scholar]
- 44.Meder B, Haas J, Keller A, Heid C, Just S, Borries A, et al. Targeted Next-Generation Sequencing for the Molecular Genetic Diagnostics of Cardiomyopathies. Circ Cardiovasc Genet. 2011 doi: 10.1161/CIRCGENETICS.110.958322. [DOI] [PubMed] [Google Scholar]
- 45.Nijman IJ, Mokry M, van Boxtel R, Toonen P, de Bruijn E, Cuppen E. Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat Methods. 2010;7(11):913–5. doi: 10.1038/nmeth.1516. [DOI] [PubMed] [Google Scholar]
- 46.Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, 2nd, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–9. doi: 10.1073/pnas.1012989107. PMCID: 3000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summerer D, Schracke N, Wu H, Cheng Y, Bau S, Stahler CF, et al. Targeted high throughput sequencing of a cancer-related exome subset by specific sequence capture with a fully automated microarray platform. Genomics. 2010;95(4):241–6. doi: 10.1016/j.ygeno.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20(10):1420–31. doi: 10.1101/gr.106716.110. PMCID: 2945191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zang ZJ, Ong CK, Cutcutache I, Yu W, Zhang SL, Huang D, et al. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer Res. 2011;71(1):29–39. doi: 10.1158/0008-5472.CAN-10-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12(1):R1. doi: 10.1186/gb-2011-12-1-r1. PMCID: 3091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blumenstiel B, Cibulskis K, Fisher S, DeFelice M, Barry A, Fennell T, et al. Targeted exon sequencing by in-solution hybrid selection. Curr Protoc Hum Genet. 2010 doi: 10.1002/0471142905.hg1804s66. Chapter 18:Unit 18 4. [DOI] [PubMed] [Google Scholar]
- 52.The Cancer Genome Atlas. Available from: cancergenome.nih.gov [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. PMCID: 2705234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. PMCID: 2928508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broad-Novartis Cancer Cell Line Encylopedia. Available from: www.broadinstitute.org/ccle.
- 56.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.