Abstract
Background
Common bean (Phaseolus vulgaris L.) is one of the most important legumes in the world. Several diseases severely reduce bean production and quality; therefore, it is very important to better understand disease resistance in common bean in order to prevent these losses. More than 70 resistance (R) genes which confer resistance against various pathogens have been cloned from diverse plant species. Most R genes share highly conserved domains which facilitates the identification of new candidate R genes from the same species or other species. The goals of this study were to isolate expressed R gene-like sequences (RGLs) from 454-derived transcriptomic sequences and expressed sequence tags (ESTs) of common bean, and to develop RGL-tagged molecular markers.
Results
A data-mining approach was used to identify tentative P. vulgaris R gene-like sequences from approximately 1.69 million 454-derived sequences and 116,716 ESTs deposited in GenBank. A total of 365 non-redundant sequences were identified and named as common bean (P. vulgaris = Pv) resistance gene-like sequences (PvRGLs). Among the identified PvRGLs, about 60% (218 PvRGLs) were from 454-derived sequences. Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis confirmed that PvRGLs were actually expressed in the leaves of common bean. Upon comparison to P. vulgaris genomic sequences, 105 (28.77%) of the 365 tentative PvRGLs could be integrated into the existing common bean physical map. Based on the syntenic blocks between common bean and soybean, 237 (64.93%) PvRGLs were anchored on the P. vulgaris genetic map and will need to be mapped to determine order. In addition, 11 sequence-tagged-site (STS) and 19 cleaved amplified polymorphic sequence (CAPS) molecular markers were developed for 25 unique PvRGLs.
Conclusions
In total, 365 PvRGLs were successfully identified from 454-derived transcriptomic sequences and ESTs available in GenBank and about 65% of PvRGLs were integrated into the common bean genetic map. A total of 30 RGL-tagged markers were developed for 25 unique PvRGLs, including 11 STS and 19 CAPS markers. The expressed PvRGLs identified in this study provide a large sequence resource for development of RGL-tagged markers that could be used further for genetic mapping of disease resistant candidate genes and quantitative trait locus/loci (QTLs). This work also represents an additional method for identifying expressed RGLs from next generation sequencing data.
Keywords: Common bean (Phaseolus vulgaris L.), 454 pyrosequencing, ESTs, Resistance gene-like sequences, Molecular markers
Background
Plant resistance genes (R genes) play an important role in direct or indirect recognition of proteins encoded by specific avirulence (Avr) genes of pathogens [1]. In recent years, a large number of R genes which confer resistance to diverse pathogens have been cloned from different plant species by using either map-based cloning or transposon tagging approaches [2-5]. Based on the amino acid sequence comparison among cloned R genes, several highly conserved domains, such as NBS, LRR, protein kinase (PK), transmembrane domain (TM), and TIR were identified as the majority of known R genes [6-9]. These distinct conserved domains provide scientists a convenient way to identify and then clone additional R genes and RGLs or resistance gene analogs (RGAs).
The largest class consists of R genes which contain NBS and LRR domains (NBS-LRR). This class can be further divided into two sub-classes, TIR-NBS-LRR and non-TIR-NBS-LRR, based on the presence or absence of a TIR domain at the N-terminus [7,10]. The second class of R genes is comprised of those with LRR and PK domains (LRR-PK), such as Fls2 in Arabidopsis and Xa21 in rice [11,12]. The third class contains those with a large extracellular LRR domain, for example, Cf-4 in tomato and Vf1 in apple [13,14]. The fourth class contains those only with the PK domain, such as Pto in tomato and Pbs1 in Arabidopsis [15,16]. And finally, the fifth class includes all other R genes that confer resistance through different mechanisms, for example, Hm1 in maize and mlo in barley [3,17].
Based on conserved domains of known R genes, PCR amplification has been successfully used to isolate RGLs or RGAs from Arabidopsis [18,19], soybean [20-22], rice [23], maize [24,25], wheat [26], and many other plant species [27-30]. For example, Bertioli et al. isolated 78 RGAs from cultivated and wild peanuts by using degenerate primers designed on NBS conserved domains [31]. Leal-Bertioli et al. identified candidate genome regions for disease resistances by using a combination of NBS profiling, sequence characterized amplified region (SCAR) markers, and Southern blot by probes with NBS encoding regions [32]. In common bean, eight classes of disease resistance related sequences and 15 RGAs have been isolated based on the conserved NBS and TIR domains [33,34]. An RGA, SB1, was found to cosegregate with the Crg gene which is known to be responsible for resistance to the rust pathogen Uromyces appendiculatus [35].
Recently, data-mining approaches have been employed to identify RGLs and RGAs from plant EST databases. In maize, 199 RGLs have been identified using data-mining, modified amplified fragment length polymorphism (AFLP), and rapid amplification of cDNA ends (RACE) methods [25]. In sugarcane, Rossi et al. identified 88 R-gene-like ESTs via the data-mining method [36]. In wheat, Dilbirligi and Gill found 220 expressed R-gene candidates using modified RNA fingerprinting and data-mining approaches [37]. In Medicago, Zhu et al. have revealed 179 unique NBS-LRR RGLs by using combination methods of database queries, hybridization with RGLs from related species, and PCR [38]. With the availability of several complete and nearly complete plant genome sequences, R gene analyses at the whole genome level have been carried out in Arabidopsis [10], rice [39], Medicago [40], and Lotus [41]. As a result, 149 NBS-LRR-encoding genes and an additional 58 related genes have been identified in the Arabidopsis genome [10]. Also 489 predicted NBS-LRR genes were identified in the rice genome, all of which belong to the non-TIR-NBS-LRR class [39]. From the Medicago truncatula draft genome (Mt1.0), Ameline-Torregrosa et al. have identified 333 non-redundant NBS-LRRs and predicted 400 to 500 NBS-LRRs in the full genome [40]. In the Lotus genome, 158 NBS-encoding sequences have been isolated and nearly 40% of them are pseudogenes encoding incomplete protein sequences [41]. With advances in sequencing technologies over the past few years, next-generation sequencing technologies have become widely available and have dramatically accelerated biological research [42], making identification of R gene homologs and members of other related gene families by data-mining methods much more efficient.
Common bean (Phaseolus vulgaris L.) is one of the most important legume crops in the world. Due to its high protein and low fat content, it has become a principal source of dietary protein and fiber in many developing countries, especially those in Eastern and Southern Africa and Latin America [43]. Diseases such as anthracnose, angular leaf spot (ALS), rust, common bacterial blight (CBB), and Bean golden yellow mosaic virus (BGYMV) highly reduce bean production and quality. For example, CBB is a serious seed-borne disease that can cause over 40% yield loss in both temperate and tropical bean production regions [44]. Rust can cause 18% to 100% yield loss of susceptible bean genotypes under favorable environmental conditions [45]. Therefore, it is very important to better understand disease resistance in common bean.
The objective of this study was to identify expressed RGLs from common bean and to develop RGL-tagged molecular markers for genetic mapping of candidate genes and QTLs for disease resistance.
Results
Identification of RGLs from 454 sequences and ESTs in GenBank
More than 1.69 million sequences with 349,977,079 base pairs (SRA028837) were generated by 454 sequencing from an adapter-ligated normalized cDNA collection from four different plant organs (leaves, flowers, roots, and pods) of common bean. After clustering and assembling using the Newbler software from Roche (Branford, CT), these sequences were assembled into 39,572 contigs and 19,723 singletons with sequence size more than 100 base pairs [46]. Moreover, 116,716 common bean ESTs are available in GenBank (verified on July 7, 2011). Therefore, a total of 176,011 sequences were used for identification of RGLs.
Full length protein sequences of the 50 known R genes (Table 1) were used to perform tBLASTn searches against 454-derived sequences and common bean ESTs in GenBank. A total of 259 contigs and nine singletons from 454-derived sequences and 629 common bean ESTs were identified with scores more than or equal to 100 and E values less than or equal to 1e-10. After clustering, 365 non-redundant sequences were developed and considered as tentative P. vulgaris RGLs, named as PvRGLs (See Additional file 1: sequences of 365 tentative PvRGLs). The average size of PvRGLs was about 1,110 base pairs, although some fragments were 2,500 base pairs or longer. Among these 365 tentative PvRGLs, 166 (45.48%) PvRGLs were from 454-derived sequences, 147 (40.27%) were from common bean ESTs, and 52 (14.25%) PvRGLs were present in both of the groups (See Additional file 2: Characterization of 365 tentative PvRGLs).
Table 1.
Fifty known R genes from plants used in this study [2]
| Plant | R genes | Protein ID | Structure | Plant | R genes | Protein ID | Structure |
|---|---|---|---|---|---|---|---|
| Apple | Vf1 | CAC40825 | LRR | Potato | R1 | AAU95638 | NBS-LRR |
| Fls2 | BAB11088 | LRR-PK | Rgc1 | AAF76163 | NBS-LRR | ||
| Pbs1 | ABR46085 | PK | Rx | CAB50786 | NBS-LRR | ||
| Arabidopsis | Rpm1 | CAA61131 | NBS-LRR | Rice | Pib | BAA76282 | NBS-LRR |
| Rpp1 | AAC72977 | NBS-LRR | Pi-ta | BAF91352 | NBS-LRR | ||
| Rpp4 | AAM18462 | NBS-LRR | Rpr1 | BAA75812 | NBS-LRR | ||
| Rpp5 | AAF08790 | NBS-LRR | Xa1 | BAA25068 | NBS-LRR | ||
| Rpp8 | AAC78631 | NBS-LRR | Xa21 | AAC80225 | LRR-PK | ||
| Rpp13 | AAF42832 | NBS-LRR | Xa26 | ABD36512 | LRR-PK | ||
| Rps2 | AAM90883 | NBS-LRR | Sugarbeet | Hs1 | AAW03319 | LRR-TM | |
| Rps4 | CAB50708 | NBS-LRR | Tobacco | N | AAA50763 | NBS-LRR | |
| Rps5 | AAC26126 | NBS-LRR | Cf-2 | AAC15779 | LRR-TM | ||
| Rpw8 | ACJ05900 | TM | Cf-4 | CAA05268 | LRR-TM | ||
| Ssi4 | AAN86124 | NBS-LRR | Cf-5 | AAC78591 | LRR-TM | ||
| Rcy1 | AAM13905 | NBS-LRR | Cf-9 | CAA05274 | LRR-TM | ||
| Barley | Rpg1 | ABK51312 | PK | Tomato | Hero | CAD29728 | NBS-LRR |
| Mla1 | AAG37354 | NBS-LRR | I2 | AAD27815 | NBS-LRR | ||
| Mla6 | CAC29242 | NBS-LRR | Mi-1 | AAC67238 | NBS-LRR | ||
| Mlo | CAB06083 | TM | Prf | AAC49408 | NBS-LRR | ||
| Flax | L6 | AAA91022 | NBS-LRR | Pto | AAB47421 | PK | |
| M | AAB47618 | NBS-LRR | Sw-5 | ACQ42910 | NBS-LRR | ||
| P2 | AAK28806 | NBS-LRR | Cre3 | CAD12795 | NBS-LRR | ||
| Maize | Hm1 | AAC04333 | Toxin reductase | Wheat | VRGL 1 | AAF19148 | NBS-LRR |
| Rp1-d | AAD47197 | NBS-LRR | Yr10 | AAG42168 | NBS-LRR | ||
| Lettuce | Dm3 | AAD03156 | NBS-LRR | Pepper | Bs2 | AAF09256 | NBS-LRR |
In addition, all tentative PvRGLs were used to identify their putative functions by BLASTX searches against the GenBank databases. Among the 365 tentative PvRGLs, 29 belonged to the NBS-LRR class, 96 belonged to the LRR, LRR-TM, and LRR-PK classes, 229 belonged to the PK class, six were similar to R gene reated sequences with putative TM domains, and five showed high similarity to R genes with Toxin reductase domains (See Additional file 2: Characterization of 365 tentative PvRGLs).
Validation of expression of selected RGLs
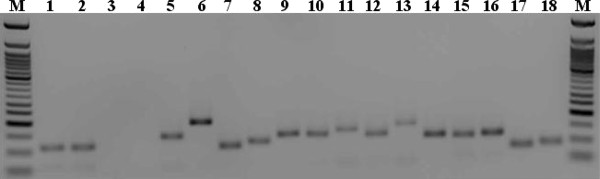
In order to determine whether the identified PvRGLs from common bean were expressed, cDNA was prepared from two-week old leaves of the genotype Sierra grown in the greenhouse. RT-PCR was then used to amplify with primers specific to each of the 27 selected PvRGLs. Twenty four of the 27 primer pairs produced a single product and we showed amplification of 15 of these primer pairs (Figure 1). Furthermore, to ensure that the amplification products actually amplified from expressed mRNA and not from contaminating DNA, we used genomic DNA, cDNA, cDNA control (no reverse transcriptase added to reverse transcription reaction), and water as templates to perform RT-PCR and the results showed that only DNA and cDNA have specific products, while cDNA control and water have no products (Figure 1). Based on these results, we concluded that there was no DNA contamination in the cDNA and our PvRGLs were indeed expressed.
Figure 1.
Validation of fifteen selected tentative PvRGLs by RT-PCR. Confirmation of absence of DNA contamination is shown in lanes 1-4 where RT-PCR amplification was carried out with primers designed from contig11286 in lanes with genomic DNA, leaf cDNA, leaf cDNA control (no reverse transcriptase added to reverse transcription reaction), and water as template to check DNA contamination. In lanes 5-18, RT-PCR products derived by amplification from an additional 14 common bean RGLs using leaf cDNA as a template are shown. M: 50 bp ladder.
Mapping identified PvRGLs to the P. vulgaris genome
Mapping of the identified PvRGLs is important for isolation of candidates of a particular resistance gene or quantitative trait locus (QTL). Recently, a Finger_Printed Contigs (FPC) physical map has been established for common bean http://phaseolus.genomics.purdue.edu/ with about 9X coverage of the genome [47]. The common bean FPCs makes it possible to integrate some of the PvRGLs into the physical map by comparison to BAC-end sequences [45]. BLASTn search was used to determine those PvRGLs matching to a BAC-end sequence with the criteria of scores more than or equal to 100 and E values less than or equal to 1e-50. As a result, 105 (28.77%) of the 365 tentative PvRGLs matched to BAC-end sequences with minimum criteria and could be integrated into the physical map (see Additional file 3: list of 105 tentative PvRGLs having matches to a BAC-end sequences).
Interestingly, there were five cases of two PvRGLs and three cases of three PvRGLs located to one physical contig: for example, PvRGL093 and PvRGL233 were located in the P. vulgaris physical map FPC_Contig13 while PvRGL266, PvRGL275 and PvRGL365 were located in the P. vulgaris physical map FPC_Contig137. Surprisingly, PvRGL083 and PvRGL236 have a match to different regions of the same BAC-end sequence PV_GBa0056B11.r (EI454956). Another seven PvRGLs matched to a Genome Survey Sequence (GSS) with E value less than 1e-50 from other common bean genotypes in GenBank.
Mapping PvRGLs to G. max pseudomolecules and P. vulgaris genetic map
In order to take advantage of the synteny blocks between common bean and soybean as per McClean et al. [48], we used the BLASTn algorithm to search against soybean genome with the same criteria as McClean et al. [48]. Given that nearly every common bean locus can match to two soybean loci, the best two soybean hits were selected. Out of 365 PvRGLs, 360 (98.63%) PvRGLs could be mapped to the G. max genome pseudomolecules. Among the 360 PvRGLs, 343 (95.28%) have soybean hits with E value less than 1e-50. The 360 PvRGLs are broadly distributed among the 20 G. max pseudomolecules. Among the 713 soybean loci matched to 360 PvRGLs (7 PvRGLs with only one hit from soybean), 689 (96.63%) have transcripts. The annotation of these transcripts indicates most of them show high similarity to sequences containing NBS, LRR, PK, TM and toxin reductase domains (See Additional file 4: Mapping PvRGLs to G. max pseudomolecules and P. vulgaris genetic map). Based on the comparison of PvRGLs to the soybean genome sequences and the conserved blocks between common bean and soybean [48], a total of 237 (64.93%) PvRGLs can be located in the P. vulgaris genetic map. The PvRGLs are broadly distributed among the 11 P. vulgaris linkage groups, but the distribution was not even. P. vulgaris chromosome 8 (Pv8) contained the largest number of PvRGLs, 41; while Pv4 contained the least number of PvRGLs, 9 (See Additional file 4: Mapping PvRGLs to G. max pseudomolecules and P. vulgaris genetic map).
RGL-tagged molecular markers
Ninety primer pairs were designed based on the 105 PvRGLs matching to BAC-end sequences. Eighty two of ninety primer pairs (91.11%) have specific products among the six genotypes. All of the PCR products were sequenced three times from both the 5' and 3' ends to ensure polymorphisms were not due to sequencing errors. The average fragment size was about 590 base pairs, while the range was from 309 to 1,549 base pairs. Approximately 291 kb of sequence data was collected from six genotypes for sequence analysis. Sequence alignments indicated single nucleotide polymorphisms (SNPs) and small insertions/deletions (InDels) occurred frequently. Twenty three SNPs were related to restriction enzyme sites, and 11 small InDels were identified.
Based on small InDels, 11 RGL-tagged STS markers gave rise to distinct and polymorphic PCR bands among six genotypes (Table 2). Likewise, 19 RGL-tagged CAPS markers have been obtained from those SNPs related to restriction sites (Table 3), and each CAPS marker showed unambiguous polymorphic bands following digestion with its corresponding restriction endonuclease.
Table 2.
Primer sequences and sizes of InDels for the RGL-tagged STS markers
| PvRGL | Forward primer | Reverse primer | InDel Sizes (bp)* |
|---|---|---|---|
| PvRGL011 | GAGGTCATTCACATTATCGGTTT | TTCAAGTCACCACTGGCAAAG | 14/G |
| PvRGL016 | TGCACCATGACTGTTCTCCACC | AATACCACCTCCAGCCCAAGA | 6/G |
| PvRGL041 | GCCTCCGTTTGAAAGTTGCTC | CACGAAATATCTTACGTTTGTTTGC | 7/G |
| PvRGL049 | CACATTAGTAGCCTTTATGTCCC | CTTCGGTCCTGTCTACAAGGT | 10/S, O, B, P, A |
| PvRGL084 | TGTAATCATTGACAAGCGTGGAG | TTGCTTGAGCATTGATGTATTG | 6/G |
| PvRGL153 | AGTGATTTCCATTTATACGAGG | AAAAGCCAAGTAACATTCTTCT | 8/S, O, B, P, A |
| PvRGL155 | GAACCAGCAATTTGTTACCGAG | TAGTTGTTGAGCAACCTTTAGC | 6/S, O, G, B, P |
| PvRGL189 | AAGGGCAGATAACACCTCATTC | GATAGTGGTTAAATTTCACCAAGG | 13/S, G, B, P, A |
| PvRGL193 | CAGCTATAATAACTTTGGTGGGAG | CATTTAACAATGATGCAGGGAC | 4/S, O, G, P |
| PvRGL272 | CATCAGTCACGTTGAAGGAAGG | TATGTCCCAACAATTCTTCCCG | 3/G |
| PvRGL297 | TTGGAGCAATGAGAAAATAGGGG | TCCAGCAGAAGCCCTGTGATG | 3/P |
*The genotypes shown in this column have insertions with the corresponding InDel size
Table 3.
Primer sequences and restriction enzymes for the RGL-tagged CAPS markers
| PvRGL | Forward primer | Reverse primer | Restriction enzymes* |
|---|---|---|---|
| PvRGL016 | TGCACCATGACTGTTCTCCACC | AATACCACCTCCAGCCCAAGA | HindIII/G |
| PvRGL039 | CTACTCCCTCAAAGGAATCACC | TGAGAAAGTTTTTGCTCAATGG | DraI/B, A |
| PvRGL040 | TCTTAAAGGACGCACCTACACG | ATTGGCCTTCAAGACGGTTAT | SnaBI/G |
| PvRGL041 | GCCTCCGTTTGAAAGTTGCTC | CACGAAATATCTTACGTTTGTTTGC | AgeI/G |
| PvRGL049 | GAACAATGTTAGTAGTACTTCGCC | GGCTTCCTCTTGAGAGTACTTTG | PflMI/S, O, G, B, P |
| PvRGL050 | CACATTAGTAGCCTTTATGTCCC | CTTCGGTCCTGTCTACAAGGT | HindIII/G |
| PvRGL065 | CGCTTAGGAGTTGGTAGAGTAACAC | TCAAGGAAACACCGACAGAATG | NsiI/S, O, B, P, A |
| PvRGL073 | CCAAAATCAGAAACTTTGGCATG | AGAGCCCTGGGCAGGAAAGAC | AflII/G |
| PvRGL093 | TGGGTTACTGTTGTGACGATGT | TATGTGCCCATGATCCTGGTAG | DraI/O, P |
| PvRGL099 | CTTCGCCATCTCCTTGTCTATC | GCCCTATGCTCAGCCTTTCTAG | BglII/B, G |
| PvRGL102 | TGAAGCCCTTATGCACAGGTCC | TCATCATCGCCAAACCCCAAC | XmnI/S, O, P |
| PvRGL153 | AGTGATTTCCATTTATACGAGG | AAAAGCCAAGTAACATTCTTCT | SphI/O |
| PvRGL155 | GAACCAGCAATTTGTTACCGAG | TAGTTGTTGAGCAACCTTTAGC | NdeI/O, B, P |
| PvRGL173 | GGATCATTGCCTGTATTCGAGG | TGGCACTGTCCCTGATAAACTG | EcoRI/G, B, A |
| PvRGL246 | CAGCCACCTTGGGTTGGTAATC | GACGCAGTTGCTAAATGGACACC | HindIII/S, O, P |
| PvRGL264 | GTAACTCGTGTCCTCCTCATCT | GATACTTGAGAAGGTAAAGGCTG | SpeI/B |
| PvRGL297 | TTGGAGCAATGAGAAAATAGGGG | TCCAGCAGAAGCCCTGTGATG | SpeI/G |
| PvRGL302 | CTCTTAAAGGACGCACCTACAC | TATTGGCCTTCAAGACGGTTAT | SnaBI/G |
| PvRGL308 | TAAGATGTGTCAATAAATATGCTG | CAGGAAGTTGCTGTAAAGAAGT | BspMI/G |
*The genotypes shown in this column can be digested by the corresponding restriction enzyme
Discussion
As a component of the gene-for-gene resistance mechanism in plants, resistance genes play an important role in recognizing products encoded by specific avirulence genes of a pathogen [49]. In this study, 365 tentative RGLs from common bean were successfully identified by data-mining based on the availability of 454-derived sequences in our lab and common bean ESTs in GenBank. About 60% (218) of identified PvRGLs were from 454-derived sequences; moreover, 166 (76.15%) of the 218 PvRGLs were new transcripts.
ESTs are highly valuable for genome annotation and gene structure prediction [50]. The 454 sequencing is a faster and more cost-effective method of producing sequence data than the Sanger method and is capable of producing a 400 to 600 million base pairs per run with 400 to 500 base pair read length [51]. It has been successfully used to maximize gene discovery, improve gene predictions, and detect SNPs and mutations [52,53]. In the past few years, data-mining approaches have been successfully used to isolate RGLs or RGAs from sugarcane [36], wheat [37] and maize [25]. As reported previously, Dilbirligi and Gill [37] adopted four different data-mining methods including domain search, individual and multiple motif searches, consensus sequence search, and individual full-length search to mine R-gene-like wheat sequences, and showed that the individual full-length search was the most successful method. There were 243 NBS-LRRs in addition to 101 other types of expressed R-gene candidates which were then isolated via an individual full-length search using a low E value of e-1 [37]. Xiao et al. [25] used three methods including modified AFLP, RACE and data-mining to isolate RGAs and R-gene-like ESTs from maize and found that the data-mining method is the most efficient way. A total of 186 expressed RGAs were recovered from 550,000 maize ESTs using a moderate E value of e-10 or better [25]. Rossi et al. revealed 88 RGLs from sugarcane ESTs by using a very stringent E value of e-50 or better and represented three major classes of R genes, namely NBS-LRR, LRR-TM and PK [36]. The above three research reports showed that different E values have a great effect on the number of resulting RGA or RGLs. In the present study, the moderate E value of e-10, similar to that in maize, was used to mine common bean RGLs and a total of 365 tentative PvRGLs were identified. Of the 365 tentative PvRGLs, 29 belonged to NBS-LRR type, 96 belonged to LRR, LRR-TM, and LRR-PK type, 229 belonged to PK type, six and five contained sequences with similar to putative TM domains and Toxin reductase domains, respectively. The number of RGLs identified in the present study was about two times more than those in maize due to three reasons. Firstly, 1.77 million 454-derived sequences and common bean ESTs were screened to identify RGLs in common bean, about three times more EST sequences than those in maize (550,000 ESTs). Secondly, 454 sequencing can generate more sequence data than the Sanger sequencing. Therefore, transcripts at extremely low levels can be detected [51]. Finally, some identified PvRGLs match the same R genes or RGLs. For example, PvRGL083 and PvRGL236 match different regions of the same BAC-end sequence.
RT-PCR was used to examine the expression of the PvRGLs in this study. Results indicated that all of the selected PvRGLs were actually expressed in the leaves of genotype Sierra (Figure 1). In contrast, many RGLs or RGAs amplified from genomic DNA using degenerate primers or mined from whole genome are not expressed [33]. Previously, eight classes of disease-resistance related sequences were amplified from common bean DNA using degenerate primers based on the conserved NBS domain [33]. Expression analysis indicated that three RGAs (SB1, SB3 and SB8) were not expressed [33]. In Lotus, 62 NBS-encoding sequences were considered as pseudogenes due to encoding of incomplete protein sequences [41]. In Arabidopsis, at least 12 NBS-LRR-encoding genes were predicted to be pseudogenes due to frame shift and nonsense mutations [10].
The PvRGLs discovered in this study correspond to most of the 25 previous common bean R genes or RGAs in the PRGdb database [2]. PvRGL266 and PvRGL275 have strong hits to the P. vulgaris TL5601 disease resistance protein gene with amino acid identity of 90% and 85%, respectively. TL5601, located in the I locus of common bean, controls resistance to Bean Common Mosaic Virus [54]. PvRGL262 matched to coiled-coil NBS-LRR (CNL)-B11 with amino acid identity of 94%. CNL-B11 was mapped in the B4 R gene cluster which contained at least three R genes (Co-9, Co-y, and Co-z) and QTL effective against anthracnose, and Bean golden yellow mosaic virus [55,56]. PvRGL309 was a part of polygalacturonase-inhibiting protein (PGIP) gene. PGIP can inhibit fungal endopolygalacturonases and is considered to be an important factor for plant resistance to phytopathogenic fungi [57]. PvRGL294 was the same as, but much longer in the 5' end than the previous RGL SB3 and OB9 [33].
So far, most of the known R genes have been cloned by map-based cloning and transposon tagging approaches [58]. Therefore, mapping RGLs and RGAs to genomes and/or genetic maps is very important and will facilitate R gene cloning. In this study, 105 PvRGLs could be integrated into the common bean FPC physical map by comparison of PvRGLs to P. vulgaris BAC-end sequences. Additionally, we were able to anchor 237 PvRGLs to the common bean genetic map by using conserved syntenic blocks between common bean and soybean. The PvRGLs are broadly, but unevenly distributed among the 11 linkage groups of common bean with a strong tendency of clustering. For example, 41 PvRGLs, the largest number, were mapped to Pv8, while 17 of them clustered at the bottom of Pv8. David et al. (2008) also found that many specific R genes against various pathogens cluster together: for example, in the B4 resistance gene cluster, 73 BAC clones (FI159954 - FI160067) were identified by using a NBS probe PRLJ1 to screen a common bean BAC library [56]. In the present study, PvRGL173 was anchored at the top of Pv4 by in silico mapping. Meanwhile, PvRGL173 shows high similarity (92.56% and 96.79%) with BAC-end sequence FI160023 (5e-153) and FI159996 (1e-101), respectively. PvRGL173 should be located in the B4 resistance gene cluster. The B4 resistance gene cluster contains various R genes resistant to different pathogens, such as Colletotrichum lindemuthianum (anthracnose), Uromyces appendiculatus (rust), and Pseudomonas syringae pv. phaseolicola (halo blight), in addition to QTLs for resistance to BGYMV and anthracnose; therefore further work will be needed to determine the role of PvRGL173. In Medicago, two superclusters of disease resistance genes were identified. One is located at the top of Mt3 containing 73 CNL and 9 TNL encoding sequences. The other is located at the bottom of Mt6 containing 57 TNL encoding sequences [38]. Clustering of R genes facilitates the genetic variation of R genes and benefits in the evolution of new R genes. Bertioli et al. found that retrotransposons were associated with the evolution of some resistance gene clusters via analysis of the synteny among Arachis, Lotus, and Medicago [59]. Several other hypotheses such as duplication, gene conversion, and unequal crossing-over have been proposed to elucidate R gene cluster and evolution [1,38]. Therefore, further studies will be needed to better understand common bean R gene evolution.
Conclusions
In this study, 365 PvRGLs were identified from 454-derived sequences and common bean ESTs in GenBank using data-mining. As a result, 105 and 237 PvRGLs were mapped to the P. vulgaris FPC physical map and genetic map, respectively. RGL-tagged markers were developed for 25 unique PvRGLs, including 11 STS and 19 CAPS markers. These methods and results will help develop more RGL-tagged molecular markers that can be used further for genetic mapping and the isolation of resistance genes from common bean in the future.
Methods
Plant material
Plant genotypes used in this study included P. vulgaris cv. Sierra (S), Olathe (O), G19833 (G), Bat93 (B), Pinto114 (P), and Aurora (A). All of the genotypes were planted in a greenhouse. Leaves of each genotype were harvested from two-week old seedlings for DNA extraction. Additionally, two-week old leaves from Sierra were collected for RNA extraction.
Data mining of common bean RGLs
The amino-acid sequences of 50 known R genes (Table 1), covering the five major R gene classes, were used to search for (tBLASTn) homologues in 454-derived sequences and common bean ESTs in GenBank http://www.ncbi.nlm.nih.gov. Those sequences with scores more than or equal to 100 and E values less than or equal to 1e-10 were clustered to develop unigenes, and all of the unigenes were considered as putative PvRGLs [25]. The resulting unigenes were in turn used to search the GenBank databases by BLASTX to confirm their putative R gene-like functions. In addition, all tentative PvRGLs were used to identify possible genomic sequences in the Phaseolus genome database http://phaseolus.genomics.purdue.edu/.
Mapping of PvRGLs to G. max pseudomolecules and P. vulgaris linkage groups
The 20 pseudomolecules of the soybean genome were downloaded from ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v7.0/Gmax/[60]. BLASTn algorithm was used to search soybean genome as per McClean et al. [48]. Firstly, we selected the hits with a cutoff E value less than 1e-10 and overlaps of at least 150 bp. Secondly, the best two soybean hits for PvRGLs were retrieved for further analysis (See Additional file 4 for all of PvRGLs that matched to soybean with these criteria). The PvRGLs were mapped to common bean linkage groups based on the syntenic blocks between common bean and soybean according to McClean et al. [48].
RNA extraction and expression analysis
Total RNA was isolated from two-week old leaves with Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Total RNA was treated with DNaseΙ two times to remove all DNA contamination. cDNA synthesis was carried out from 1 μg of RNA using the MMLV reverse transcriptase (New England Biolabs, Ipswich, MA). The RT-PCR primers were designed and synthesized at Integrated DNA Technologies (IDT, Coralville, IA). The RT-PCR was performed with the BIO-RAD PCR system (Hercules, CA) with the following cycling parameters: 94°C for 3 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 60 s, and a final elongation at 72°C for 5 min.
PCR amplification and sequence analysis
DNA was isolated from the above six genotypes according to the methods described in Doyle and Doyle [61]. PCR amplification was carried out by designing forward and reverse primers from PvRGLs having a hit to BAC-end sequences. The PCR products were purified using QIAquick® Gel Extraction kit (Germantown, MD) and sequenced at the Macrogen USA Corp (Rockville, MD). The software DNAMAN version 4.0 was used for sequence assembly and multiple sequence alignment to confirm either identities or similarities.
Development of RGL-tagged molecular markers
PvRGL sequences amplified from the above six genotypes were carefully analyzed by the DNAMAN program to identify InDels and SNPs, and those with InDels and SNPs were taken as candidate RGL markers. For STS markers, two 200-bp sequences flanking the InDel site were extracted and used to design PCR primers. Polymorphic PCR bands from the six genotypes were observed on 6% polyacrylamide gels. For CAPS markers, polymorphic PCR bands were observed among the six genotypes following digestion with a particular restriction endonuclease.
Authors' contributions
ZL participated in conceiving the research, sequence analysis, developing molecular markers, PvRGL mapping, and writing the manuscript. MC participated in material preparation, developing molecular markers, and contributed to writing of the manuscript. AT participated in PvRGL sequencing and contributed to the writing of the manuscript. VK participated in conceiving the research, providing the 454 sequences, and contributed to writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sequences of 365 tentative PvRGLs.
Characterization of 365 tentative PvRGLs.
105 PvRGLs having a hit with a BAC-end sequence.
Mapping PvRGLs to G. max pseudomolecules and P. vulgaris genetic map.
Contributor Information
Zhanji Liu, Email: zliu@uga.edu.
Mollee Crampton, Email: mcrampto@udel.edu.
Antonette Todd, Email: atodd@desu.edu.
Venu Kalavacharla, Email: vkalavacharla@desu.edu.
Acknowledgements
VK acknowledges funding by NSF grant EPS-0814251. ZL was supported by USDA grant 2008-02675 to VK. MC and AT were supported by NSF grant EPS-0814251 to VK.
References
- Ellis J, Dodds P, Pryor T. The generation of plant disease resistance gene specificities. Trends Plant Sci. 2000;5:373–379. doi: 10.1016/S1360-1385(00)01694-0. [DOI] [PubMed] [Google Scholar]
- Sanseverino W, Roma G, Simone MD, Faino L, Melito S, Stupka E, Frusciante L, Ercolano MR. PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2009;38:D815. doi: 10.1093/nar/gkp978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal GS, Briggs SP. Reductase activity encoded by the Hm1 disease resistance gene in maize. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- Whitham S, McCormick S, Baker B. The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc Natl Acad Sci USA. 1996;93:8776–8781. doi: 10.1073/pnas.93.16.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG, Lane C. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat protein. Cell. 1996;84:451–459. doi: 10.1016/S0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- Bent AF. Plant disease resistance genes: function meets structure. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313X.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JDG. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansavini S. Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol Plant Microbe Interact. 2001;14:508–515. doi: 10.1094/MPMI.2001.14.4.508. [DOI] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, DieRGLarde P, Groenendijk J, Topsch S, Vos P, Salamini F, Schulze-Lefert P. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/S0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- Botella MA, Coleman MJ, Hughes DE, Nishimura MT, Jones JDG, Somerville SC. Map positions of 47 Arabidopsis sequences with sequence similarity to disease resistance genes. Plant J. 1997;12:1197–1211. doi: 10.1046/j.1365-313X.1997.12051197.x. [DOI] [PubMed] [Google Scholar]
- Aarts MGM, Hekkert BL, Holub EB, Beynon JL, Stiekema WJ, Pereira A. Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact. 1998;11:251–258. doi: 10.1094/MPMI.1998.11.4.251. [DOI] [PubMed] [Google Scholar]
- Kanazin V, Mareck L, Shoemaker P. Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA. 1996;93:11746–11750. doi: 10.1073/pnas.93.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MA, Marek LF, Lohnes D, Cregan P, Shoemaker RC. Expression and genome oRGLnization of resistance gene analogs in soybean. Genome. 2000;43:86–93. doi: 10.1139/g99-107. [DOI] [PubMed] [Google Scholar]
- Yu YG, Buss GR, Maroof MA. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA. 1996;93:11751–11756. doi: 10.1073/pnas.93.21.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mago R, Nair S, Mohan M. Resistance gene analogues from rice: cloning, sequencing and mapping. Theor Appl Genet. 1999;99:50–57. doi: 10.1007/s001220051207. [DOI] [Google Scholar]
- Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A. The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact. 1998;11:968–978. doi: 10.1094/MPMI.1998.11.10.968. [DOI] [PubMed] [Google Scholar]
- Xiao WK, Xu ML, Zhao JR, Wang FG, Li JS, Dai JR. Genome-wide isolation of resistance gene analogs in maize (Zea mays L.) Theor Appl Genet. 2006;113:63–72. doi: 10.1007/s00122-006-0272-8. [DOI] [PubMed] [Google Scholar]
- Seah S, Sivasithamparam K, Karakousis A, Lagudah ES. Cloning and characterization of a family of disease resistance gene analogs from wheat and barley. Theor Appl Genet. 1998;97:937–945. doi: 10.1007/s001220050974. [DOI] [Google Scholar]
- Wan HJ, Zhao ZG, Malik A, Qian CT, Chen JF. Identification and characterization of potential NBS-encoding resistance genes and induction kinetics of a putative candidate gene associated with downy mildew resistance in Cucumis. BMC plant Biol. 2010;10:186. doi: 10.1186/1471-2229-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YL, Xu ZL, Jiao FC, Yu HQ, Xiao BG, Li YP, Lu XP. Cloning, structural features, and expression analysis of resistance gene analogs in tobacco. Mol Biol Rep. 2010;37:345–354. doi: 10.1007/s11033-009-9749-2. [DOI] [PubMed] [Google Scholar]
- Huettel B, Santra D, Muehlbauer J, Kahl G. Resistance gene analogues of chickpea (Cicer arietinum L.): isolation, genetic mapping and association with a Fusarium resistance gene cluster. Theor Appl Genet. 2002;105:479–490. doi: 10.1007/s00122-002-0933-1. [DOI] [PubMed] [Google Scholar]
- Nair RA, Thomas G. Isolation, characterization and expression studies of resistance gene candidates (RGCs) from Zingiber spp. Theor Appl Genet. 2007;116:123–134. doi: 10.1007/s00122-007-0652-8. [DOI] [PubMed] [Google Scholar]
- Bertioli DJ, Leal-Bertioli SC, Lion MB, Santos VL, Pappas G, Cannon SB, Guimaraes PM. A large scale analysis of resistance gene homologues in Arachis. Mol Gen Genomics. 2003;270:35–45. doi: 10.1007/s00438-003-0893-4. [DOI] [PubMed] [Google Scholar]
- Leal-Bertioli SC, Jose AC, Alves-Freitas DM, Moretzsohn MC, Guimaraes PM, Nielen S, Vidigal BS, Pereira RW, Pike J, Favero AP, Parniske M, Varshney RK, Bertioli DJ. Identification of candidate genome regions controlling disease resistance in Arachis. BMC Plant Biol. 2009;22:112. doi: 10.1186/1471-2229-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin MI, Vallejos CE, McClean PE. Disease-resistance related sequences in common bean. Genome. 1999;42:41–47. doi: 10.1139/g98-097. [DOI] [PubMed] [Google Scholar]
- Lopez CE, Acosta IF, Jara C, Pedraza F, Gaitan-Solis E, Gallego G, Beebe S, Tohme J. Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathol. 2003;93:88–95. doi: 10.1094/PHYTO.2003.93.1.88. [DOI] [PubMed] [Google Scholar]
- Kalavacharla V, Stavely JR, Myers JR, McClean PE. Crg, a gene required for Ur-3-mediated rust resistance in common bean, maps to a resistance gene analog cluster. Mol Plant Microbe Interact. 2000;13:1237–1242. doi: 10.1094/MPMI.2000.13.11.1237. [DOI] [PubMed] [Google Scholar]
- Rossi M, Auaujo PG, Paulet F, Garsmeur O, Dias VM, Chen H, Van Sluys MA. D' Hont A: Genomic distribution and characterization of EST-derived resistance gene analogs (RGAs) in sugarcane. Mol Gen Genomics. 2003;269:406–419. doi: 10.1007/s00438-003-0849-8. [DOI] [PubMed] [Google Scholar]
- Dilbirligi M, Gill KS. Identification and analysis of expressed resistance gene sequences in wheat. Plant Mol Biol. 2003;53:771–787. doi: 10.1023/B:PLAN.0000023663.55701.5f. [DOI] [PubMed] [Google Scholar]
- Zhu H, Cannon SB, Young ND, Cook DR. Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact. 2002;15:529–539. doi: 10.1094/MPMI.2002.15.6.529. [DOI] [PubMed] [Google Scholar]
- Monsi B, Wisser RJ, Pennill L, Hulbert SH. Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet. 2004;109:1434–1447. doi: 10.1007/s00122-004-1758-x. [DOI] [PubMed] [Google Scholar]
- Ameline-Torregrosa C, Wang BB, O'Bleness MS, Deshpande S, Zhu HY, Roe B, Young ND, Cannon SB. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008;146:5–21. doi: 10.1104/pp.107.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Cheng Y, Ma W, Zhao Y, Jiang HY, Zhang M. Identification and characterization of NBS-encoding disease resistance genes in Lotus japonicus. Plant Syst Evol. 2010;289:101–110. doi: 10.1007/s00606-010-0331-0. [DOI] [Google Scholar]
- Shendure J, Ji HL. Next-generation DNA sequencing. Nature Biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Broughton W, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus ssp.) model food legumes. Plant and Soil. 2003;252:55–128. [Google Scholar]
- Shi C, Chaudhary S, Yu KF, Park SJ, Navabi A, McClean PE. Identification of candidate genes associated with CBB resistance in common bean HR45 (Phaseolus vulgaris L.) using cDNA-AFLP. Mol Biol Rep. 2011;38:75–81. doi: 10.1007/s11033-010-0079-1. [DOI] [PubMed] [Google Scholar]
- Thibivilliers S, Joshi T, Campbell KB, Scheffler B, Xu D, Coopers B, Nguyen HT, Stacey G. Generation of Phaseolus vulgaris ESTs and investigation of their regulation upon Uromyces appendiculatus infection. BMC Plant Biol. 2009;9:46. doi: 10.1186/1471-2229-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalavacharla V, Liu Z, Meyers BC, Thimmapuram J, Melmaiee K. Identification and analysis of common bean (Phaseolus vulgaris L.) transcriptomes by massively parallel pyrosequencing. BMC Plant Biol. 2011;11:135. doi: 10.1186/1471-2229-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Goicoechea JL, Collura K, Gill N, Lin JY, Yu Y, Kudrna D, Zuccolo A, Vallejos CE, Munoz-Torres M, Blair MW, Tohme J, Tomkins J, McClean P, Wing RA, Jackson SA. BAC-end sequence analysis and a draft physical map of the common bean (Phaseolus vulgaris L.) genome. Tropical Plant Biol. 2008;1:40–48. doi: 10.1007/s12042-007-9003-9. [DOI] [Google Scholar]
- McClean PE, Mamidi S, McConnell M, Chikara S, Lee R. Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics. 2010;11:184. doi: 10.1186/1471-2164-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. The complementary genic systems in flax and flax rust. Adv Genet. 1956;8:29–54. [Google Scholar]
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Song QJ, Fickus EW, Quigley CV, Lim JS, Choi IY, Hwang EY, Pastor-Corrales M, Cregan PB. High-throughput SNP discovery and assay development in common bean. BMC Genomics. 2010;11:475. doi: 10.1186/1471-2164-11-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos CE, Astua-Monge G, Jones V, Plyler TR, Sakiyama NS, Mackenzie SA. Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics. 2006;172:1229–1242. doi: 10.1534/genetics.105.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffroy V, Macadre C, David P, Pedrosa-Harand A, Sevignac M, Dauga C, Langin T. Molecular analysis of a large subtelomeric nucleotide-binding-site-leucine-rich-repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics. 2009;181:405–419. doi: 10.1534/genetics.108.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P, Sevignac M, Thareau V, Catillon Y, Kami J, Gepts P, Langin T, Geffroy V. BAC end sequences corresponding to the B4 resistance gene cluster in common bean: a resource for markers and synteny analyses. Mol Genet Genomics. 2008;280:521–533. doi: 10.1007/s00438-008-0384-8. [DOI] [PubMed] [Google Scholar]
- Toubart P, Desiderio A, Salvi G, Cervone F, Daroda L, Lorenzo GD, Bergmann C, Darvill AG, Albersheim P. Cloning and characterization of the gene encoding the endopolygalacturonase-inhibiting protein (PGIP) of Phaseolus vulgaris L. Plant J. 1992;2:367–373. doi: 10.1046/j.1365-313x.1992.t01-35-00999.x. [DOI] [PubMed] [Google Scholar]
- Xiao WK, Zhao J, Fan SC, Li L, Dai JR, Xu ML. Mapping of genome-wide resistance gene analogs (RGAs) in maize (Zea mays L.) Theor Appl Genet. 2007;115:501–508. doi: 10.1007/s00122-007-0583-4. [DOI] [PubMed] [Google Scholar]
- Bertioli DJ, Moretzsohn MC, Madsen LH, Sandal N, Leal-Bertioli SC, Guimaraes PM, Hougaard BK, Fredslund J, Schauser L, Nielsen AM, Sato S, Tabata S, Cannon SB, Stougaard J. An analyses of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;10:45. doi: 10.1186/1471-2164-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of 365 tentative PvRGLs.
Characterization of 365 tentative PvRGLs.
105 PvRGLs having a hit with a BAC-end sequence.
Mapping PvRGLs to G. max pseudomolecules and P. vulgaris genetic map.