Abstract
Plasmodium falciparum enolase (Pfeno) localizes to the cytosol, nucleus, cell membrane and cytoskeletal elements, suggesting multiple non-glycolytic functions for this protein. Our recent observation of association of enolase with the food vacuole (FV) in immuno-gold electron microscopic images of P. falciparum raised the possibility for yet another moonlighting function for this protein. Here we provide additional support for this localization by demonstrating the presence of Pfeno in purified FVs by immunoblotting. To examine the potential functional role of FV-associated Pfeno, we assessed the ability of Pfeno to complement a mutant Saccharomyces cervisiae strain deficient in enolase activity. In this strain (Tetr-Eno2), the enolase 1 gene is deleted and expression of the enolase 2 gene is under the control of a tetracycline repressible promoter. Enolase deficiency in this strain was previously shown to cause growth retardation, vacuolar fragmentation and altered expression of certain vacuolar proteins. Expression of Pfeno in the enolase-deficient yeast strain restored all three phenotypic effects. However, transformation of Tetr-eno2 with an enzymatically active, monomeric mutant form of Pfeno (Δ5Pfeno) fully restored cell growth, but only partially rescued the fragmented vacuolar phenotype, suggesting that the dimeric structure of Pfeno is required for the optimal vacuolar functions. Bioinformatic searches revealed the presence of Plasmodium orthologs of several yeast vacuolar proteins that are predicted to form complexes with Pfeno. Together, these observations raise the possibility that association of Pfeno with food vacuole in Plasmodium may have physiological function(s).
Keywords: Plasmodium, Enolase, Food vacuole, Complementation, Yeast Vps63p
Introduction
Malaria is one of the most severe infectious diseases, resulting in ~250 million cases and ~1 million deaths annually [1]. In the intraerythrocytic stages, which are responsible for morbidity and mortality, the parasite feeds extensively on host cell hemoglobin. The degradation of hemoglobin occurs in the food vacuole (FV) [2, 3], an organelle that is similar to the lysosomal compartment in eukaryotic cells. The amino acids so produced are used for rapid growth and asexual proliferation of the parasite, while heme is converted into a crystalline malarial pigment called hemozoin [4-6]. In addition to hemoglobin degradation and heme polymerization, the parasite FV is also the site for the detoxification of oxygen free radicals, offering a multitude of opportunities for antimalarial drug targeting [7]. Indeed, drugs that interfere with the vacuolar processes are known to cause parasite death[8-10].
The fact that the FV contains proteases that degrade hemoglobin within an acidic environment [9, 11, 12] suggests it to be similar to eukaryotic lysosomes. However, our knowledge about the biogenesis of FV in the parasite is rather poor and there is no clear morphological evidence for the presence of classical eukaryotic endosome-lysosome-like system in Plasmodium [4-6]. Electron microscopic studies on single thin sections of Plasmodium suggest that endocytic vesicles may originate from the parasite surface via a cytoskeletal ring called cytostome [13, 14] and receive degradative enzymes from the parasite’s secretory system to cytostomal vesicles [15]. Thus the substrate hemoglobin and digesting enzymes may be carried together through these cytostomal endocytic vesicles that ultimately fuse with the FV. Evidence for mechanisms involving the transport of hemoglobin to the FV through a vesicle independent pathway have also been provided [16]. In a recent paper, it was proposed that cytosol and hemoglobin filled cytostomal vesicles invaginate, forming acidified peripheral structures where digestion of hemoglobin can occur along with the formation of hemozoin. With the maturation of the parasite, such hemozoin containing vesicles can fuse to form a single acidic FV that can be further fed by the hemoglobin filled vesicles [17]. An alternative mechanism has been proposed in which the FV initially arises as a result of a single event in the ring stage, called the ‘Big gulp’, wherein the parasite folds onto itself to engulf a large volume of hemoglobin. This event is proposed to be the first step in the biogenesis of the parasite’s lysosomal compartment, the FV [18]. Among the multiple pathways that have been proposed for the formation of FV, those that involve generation of small hemoglobin vesicles (SHVs) from the cytostome are more akin to classical endocytosis where vesicles are small and bear the early endosomal regulatory protein Rab5a. Feeding via the cytostome and consequent generation of SHVs is seen in the trophozoite stage [17, 18]. Thus there appear to be multiple pathways for hemoglobin ingestion and transport to the FV, some of which may be unique to the parasite while others resemble vacuolar trafficking in higher eukaryotes, such as Saccharomyces cerevisiae, where the molecular components of the vacuolar machinery are relatively better understood.
One surprising component of the Saccharomyces cerevisiae vacuolar fusion machinery is the glycolytic enzyme enolase [19]. Yeast enolase associates with lysosomes and is required homotypic vacuolar fusion in vitro. In enolase-deficient yeast, lysosomes become fragmented. This moonlighting function of yeast enolase causing vacuolar fusion does not depend on its known enzymatic activity. Intriguingly, P. falciparum enolase (Pfeno) was recently found associated with the FV in immuno-gold electron microscopic images of infected red blood cells (rbcs) [20]. Pfeno was most extensively associated with the FV in the early trophozoite stage, with a gradual decrease in mature trophozoite and schizont stages. Interestingly, feeding of hemoglobin through SHVs also occurs maximally in the trophozoite stage [18]. These observations raised the possibility that Pfeno may be involved in FV development and/or functions. Thus, to explore the functions of Pfeno, we examined whether Pfeno could complement the metabolic and moonlighting functions of yeast enolase as has been observed for PfP0 and PfHSP90 [21, 22]. For these experiments, we took advantage of an existing yeast strain in which the ENO1 gene has been deleted and the ENO2 gene is expressed from a tetracycline repressible promoter (hereafter referred to as Tetr-Eno2) [19]. When grown in the absence of tetrcycline / doxycycline, this strain has ~5% enolase activity compared to the parental strain, contains severely fragmented vacuolar phenotype [19] and has an altered protein expression profile; when Eno2 expression was repressed by adding doxycycline, Tetr-Eno2 failed to grow on medium containing glucose as the energy source [19]. Our results showed that expressing Pfeno in this mutant strain restored all the phenotypic effects induced by enolase deficiency. We further demonstrated that enolase specifically associated with the purified FV preparation from P. yoelii. Bioinformatics searches showed that parasite has orthologs of the molecular components involved in yeast vacuole formation. These results, in conjunction with our immuno-gold electron microscopic imaging [20], suggest that the pathways involved in vacuolar development in yeast and Plasmodium may be similar.
Materials and Methods
Materials
Anti-S. cerivisiae enolase antibody was obtained from Santa Cruz Biotechnology (cat. no. SC-7455), Santa Cruz, CA, USA. FM4-64 and anti-CPY antibody (Monoclonal 10A5) were supplied by Invitrogen Molecular Probes. Anti IgG (mouse/goat/rabbit) labelled with FITC were purchased from Sigma Chemical Co., St. Louis, USA. Anti-Pfeno was raised as described earlier [23]. Polyclonal anti-PfHU (a bacterial histone like protein involved in DNA compaction in apicoplast) antisera was a gift from Dr. S. Habib [24]. Anti-VDAC1 (porin) antibody (cat #ab15895 was supplied by abcam; www.abcam.com). Sequencing grade trypsin was bought from Roche Diagnostics.
Cloning and expression of Pfeno in yeast
The parental yeast strain R11258 (URA::CMV-tTA MATa his3-1 leu2-0 met15-0) and the yeast enolase double mutant strain BDY4 (referred to as Tetr-Eno2 in this manuscript; pENO2::kanR-tet07-TATA URA3::CMV-tTA MATa his3-1 leu2-0 met15-0 pho8::leu eno1::met15) were kindly provided by Dr. W. T. Wickner (Dartmouth Medical School, Hanover, NH, USA). Yeast was maintained according to standard methods [25, 26]. The wild type Pfeno gene and a deletion mutant (Δ5Pfeno) lacking the sequence encoding 104EWGWS108 were PCR-amplified from the plasmids pQE30-Pfeno [23] and pGEX4T1 [27], respectively, and inserted into the yeast expression plasmid p423GPD [28] by homologous recombination in R11258 [29]. Correct insertion was confirmed by PCR and sequencing. R11258 and Tetr-Eno2 yeast were then transformed with the empty parental plasmid p423GPD, p423GPD-Pfeno, and p423GPD-Δ5Pfeno and maintained on synthetic growth medium lacking histidine (SD–His).
SDS PAGE, western blotting and quantification of expressed enolase protein
Protein extracts from the above yeast strains were prepared as described earlier [30] and subjected to electrophoresis on a 10% SDS polyacrylamide gel. Pfeno and ScEno were detected by western blotting with anti-rPfeno antibody (1:1000 dilution) [23] or anti-S.cerevisiae enolase antibody (1:1000 dilution), followed by probing with FITC-conjugated secondary antibodies (1:1000 dilution). The blot was imaged on a Typhoon Trio Scanner (GE Healthcare) and the fluorescence of the protein bands was quantified using Image Quant TL (GE Healthcare). The amounts of ScEno and Pfeno in each strain were estimated using the standard curves generated with known amounts of purified yeast enolase (Sigma) and purified rPfeno, respectively. 2-Dimensional gel electrophoresis was performed as described earlier [31].
In-gel digestion and mass spectrometry
Trypsin digestion and extraction of peptides from the protein bands excised from a gel was performed using standard protocols [32, 33]. Automated MS/MS spectra were acquired on an Agilent 1100 Series LC/MSD Trap XCT Plus System. Spectra were extracted using Spectrum Mill Software (Agilent) and searched against NCBI database.
Measurement of yeast growth
Yeast strains were grown in liquid culture to mid log phase, diluted to an OD600 of 1.0 and subjected to 4-fold serial dilutions in liquid SD–His medium. 5 μL of each dilution was spotted on solid SD–His medium in the presence or absence of doxycyline (10 μg / mL). Similarly, growth of yeast strains in liquid medium in the absence or presence of doxycycline (10 μg / mL) was monitored by measuring the OD600 every hour for 72 hours on an automated Tecan Magellan plate reader maintained at 30°C. The liquid yeast cultures were started with an initial OD600 of 0.1.
Visualization and scoring of vacuoles
Yeast was grown at 30°C in SD–His medium to an OD600 of 1.0 in the absence of doxycycline. Yeast from approximately 1.4 mL of culture were collected by centrifugation, resuspended in 1 ml YPD medium containing 10 μg /ml of doxycycline, and incubated 1 h with shaking. To visualize vacuoles, 2 μL of 8 mM FM4-64, which labels only the vacuolar membrane [34], was added and the culture incubated for 60 minutes in the dark at 30°C. The cells were centrifuged and washed with 1 mL YPD (yeast extract-peptone-dextrose) medium. The cells were resuspended in 1 mL YPD medium and incubated at 30°C for 30 minutes on a shaker. Live cells were imaged on a confocal microscope (Zeiss 710 Meta). Since in steady state, most normal yeast strains contain one to five vacuoles [35], cells with more than 5 vacuolar lumens were considered to possess fragmented vacuoles, while the cells with 1 to 5 vacuolar lumens were taken to be normal. At least 300 cells from each strain were scored for their vacuolar phenotype.
Enolase enzyme assay
Yeast cells from a culture grown overnight to stationary phase were collected by centrifugation and lysed by vortexing with glass beads in 50 mM Tris-HCl, pH 7.4, 0.1% Triton X 100 reduced (Sigma), plus protease inhibitors cocktail (Roche Diagnostics) and 1 mM PMSF (phenylmethanesulfonyl fluoride). Samples were subject to 5-6 cycles of vortexing for 30 sec followed by 30 sec on ice. Cell debris were pelleted by centrifugation at 20,000 x g and the supernatant was assayed for enolase enzyme activity by monitoring the decrease in substrate (Phospho-en-ol-pyruvate) concentration per minute at OD240 [23].
Yeast vacuole isolation
Yeast vacuoles were isolated as described earlier [19, 36]. Briefly, yeast cells were grown in YPD medium and harvested by centrifugation. The cells were washed with Wash Buffer (100 mM Tris-HCl pH 9.4, 10 mM DTT) and then treated with zymolase (450 μL of a 100 mg/mL stock to 800 OD600/mL equivalent cells), followed by dextran sulfate solution (200 μL of a 25 mg/mL stock). The cell-lysate was suspended in a 15% Ficoll-Sorbitol solution, and layered in an Oak-ridge tube. 8%, 4% and 0% Ficoll was subsequently layered carefully over this. These step gradients were then subjected to ultracentrifugation at 80,000 xg for 1.5 hours. Vacuoles were collected from the 0 % - 4 % Ficoll interface. The isolated vacuoles were either frozen at −80°C or denatured with SDS containing sample buffer and run on a 10% polyacrylamide gel for their protein composition.
Growth and isolation of Plasmodium yoelii parasites
Plasmodium yoelii infected mouse rbc stocks (~106 cells) were injected in 8-10 weeks old Swiss mice. The mice were sacrificed when parasitemia reached 30-40% and the blood was collected in acid citrate dextrose (136 mM glucose, 41.6 mM citric acid and 74.8 mM sodium citrate). Rbcs were collected by centrifuging the blood samples at 290 x g in an Eppendorf 5810 R centrifuge, resuspened in 1X PBS (phosphate buffer saline, pH 7.4) containing 1 mM PMSF plus protease inhibitor cocktail and lysed with 0.05% saponin. The released parasites were washed several times with 1X PBS, snap frozen in liquid nitrogen, and stored at −80°C.
Isolation of the parasite FV
FV were purified from P. yoelii essentially as described by Saliba et al [37]. Briefly, 100 mg of P. yoelii cells were suspended in 1 ml of ice cold water (pH 4.5, containing protease inhibitor cocktail). The cell suspension was subjected to trituration for 5 times with a 26.5 gauge needle. The triturate was centrifuged at 18000 x g on an Eppendorf centrifuge 5810 R for 2 minutes. The supernatant was discarded. The pellet was resuspended in 1mL of ‘Uptake Buffer’, pH 7.4 containing 2 mM MgSO4, 100 mM KCl, 10 mM NaCl, 25 mM HEPES, 25 mM NaHCO3, and 5 mM sodium phosphate, to which was added 10 μL of 5 mg / mL of DNAse I [37] and incubated at 37°C for 5 minutes. This was again centrifuged at 18000 xg, and the supernatant was discarded. The pellet was resuspended in 100 μL of ice cold Uptake Buffer and 1.3 mL ice cold 42% Percoll with 0.25 M sucrose and 1.5 mM MgCl2, pH 7.4. This suspension was passed twice through a 26.5 gauge needle and centrifuged at 18000 xg at 4°C for ten minutes. The lowermost dark band on the gradient, which contained the FVs, was isolated, washed with Uptake buffer, and either frozen at −80°C or used immediately for experiments.
Gel filtration chromatography
Gel filtration chromatography on soluble cellular proteins was performed using an AKTA FPLC system (Amersham-Pharmacia Biotech) for the determination of oligomeric state of P. falciparum and yeast enolases as described elsewhere [38]. Typically 1 mL of soluble yeast extract containing 1 mg of protein was loaded on a 120 ml Superdex-200 Column equilibrated in 1X PBS. 2 mL fractions were collected at a flow rate of 1mL/minute and concentrated using Millipore Centricon tubes (5 kDa cut-off) to a volume of ~50 to 100 μL. 10 μL from μL of 5X SDS sample buffer and used for SDS-PAGE followed by western blotting.
Results
Expression of Pfeno in an enolase-deficient yeast strain
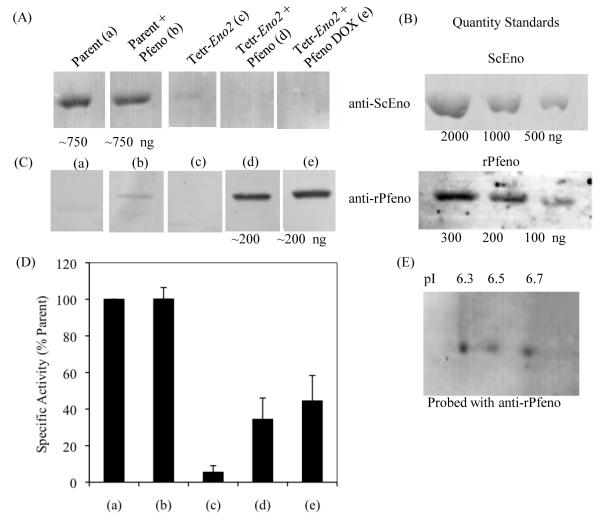
The previously observed colocalization of enolase with the FV in immuno gold electron microscopic images [20] is reminiscent of the association of enolase with yeast vacuoles [19]. Yeast express two enolase proteins, Eno1 and Eno2, both of which co-purify with vacuoles [19]. In S. cerevisiae mutants with reduced expression of enolase, the vacuoles are fragmented and lack several key proteins. Thus in yeast, enolase is required for an as yet unidentified step that affects vacuolar fusion and protein trafficking to the vacuole. To begin to investigate whether the observed association of Pfeno with the FV was functionally relevant, we used Pfeno to genetically complement an S. cerevisiae strain deficient in enolase. In this strain (Tetr-Eno2), ENO1 was deleted and ENO2 was expressed from a tetracycline repressible promoter; ENO2 is essential for yeast viability and cannot be knocked out. Tetr-Eno2 and the parental yeast strain R11258 were transformed with a Pfeno expression plasmid and empty vector. P. falciparum and yeast enolase protein levels in these strains in the presence or absence of 10 μg / mL doxycycline were assessed by western blotting (Fig. 1). Using an antibody that recognized both yeast enolase proteins (ScEno) but not Pfeno, strong expression of yeast enolase was observed in the parental cell line transformed with the empty vector or the Pfeno expression plasmid. In contrast, ScEno was barely detectable in the Tetr-Eno2 strain transformed with empty vector and grown in the absence of doxycycline, indicating that the tetracycline repressible promoter is much weaker than the enolase promoter (Fig. 1A (c)). The addition of 10 μg / mL doxycline to the culture media reduced ScEno levels below the limit of detection (Fig. 1A (e)). Surprisingly, ScEno was also undetectable in Tetr-Eno2 transformed with the Pfeno expression plasmid (Fig. 1A (d)). Pfeno was detected in the parental yeast strain transformed with the Pfeno expression plasmid, but much higher levels were observed in Tetr-Eno2 grown in the absence or presence of 10 μg / mL doxycycline (Fig. 1C (b,d and e)). As expected, no bands were detected with the Pfeno antisera in lysates from yeast transformed with the empty vector, confirming the specificity of the anti-rPfeno antibodies for Pfeno (Fig. 1C (a and c)). To estimate the amounts of endogenous yeast Eno2p and P. falciparum enolase (Pfeno) produced in the various yeast strains, we performed western blots with increasing amounts of purified ScEno and rPfeno (Fig. 1B). From this we estimated that samples from parental yeast strains contained approximately 750 ng of enolase, whereas the Tetr-Eno2 sample contained about 200 ng of Pfeno. Since equivalent amounts of proteins were loaded in each lane, this suggests that the relative amount of Pfeno expressed in Tetr-Eno2 yeast is about 25% of yeast enolase levels in parental yeast strains. We independently corroborated this value by measuring enolase enzymatic activity in soluble extracts from the various yeast strains (Fig. 1D). Relative to the parent strain, Tetr-Eno2 grown in the absence of doxycycline had ~5% activity while the Tetr-Eno2 expressing Pfeno had ~30-40% activity.
Figure 1.
Quantification of enolase in (a) Parent strain, (b) Parent strain transformed with P. falciparum enolase (Parent + Pfeno), (c) Tetr-Eno2 strain, (d) Tetr-Eno2 strain transformed with Pfeno and grown in the absence and (e) in the presence of 10 μg/mL doxycycline (DOX). Western blot analysis was performed for the quantification of the expressed enolase (ScEno or rPfeno) in cell extracts of different strains of yeast. (A) Western blot of yeast enolase; (B) Western blot of pure ScEno or rPfeno used as standards; (C) western blot of Pfeno. Equivalent amounts of protein were loaded in each lane. (D) Specific activity of enolase in various cell extracts plotted as the percent of the parent strain and (E) Western blot of two dimensional gel electrophoresis showing the variant profile of Pfeno in Tetr-Eno2 + Pfeno (DOX) yeast cells. Anti-rPfeno antibody was used to probe the membrane.
As Pfeno is known to undergo several post-translational modifications in Plasmodium [31], we subjected extracts from Tetr-Eno2 expressing Pfeno (grown in the presence of 10 μg / mL doxycycline) to two-dimensional gel electrophoresis and immunoblot analysis with anti-rPfeno antibodies. Three variants of Pfeno were present in the transformed yeast (Fig. 1E), indicating that Pfeno expressed in yeast is also modified. However, this profile lacked two of the acidic forms of Pfeno (pI=5.9 and 6.1) that may correspond to multi-site phosphorylations [31].
Pfeno restored growth of enolase-deficient yeast
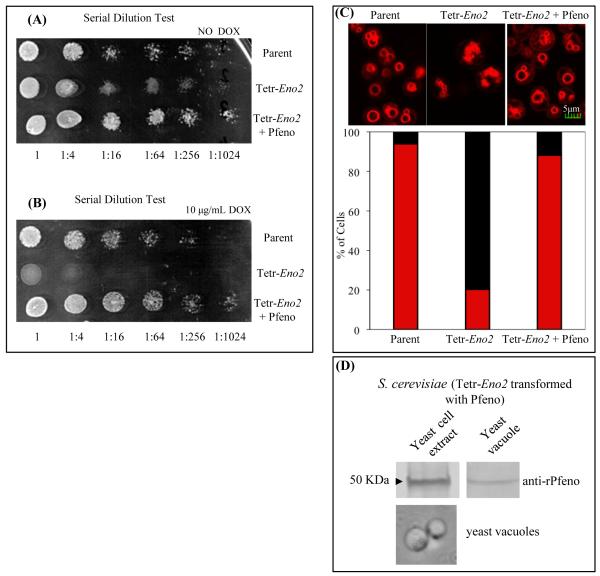
Enolase expression is required for optimal growth of yeast on medium containing glucose as the energy source. The effect of Pfeno expression on the growth of parental and enolase-deficient yeast was monitored in liquid medium (Supple. Fig. 1) and on solid agar (Fig. 2). The Tetr-Eno2 strain, which has ~5% of the enolase activity of the parent strain, exhibited only a slight growth retardation (Supple. Fig. 1A). However, when Eno2 expression was completely inhibited by adding doxycycline to the culture medium, no growth was evident in liquid or solid medium (Supple. Fig. 1B; Fig. 2B). Expression of Pfeno in Tetr-Eno2, restored the growth to wild type levels in the absence or presence of doxycycline, demonstrating the ability of Pfeno to fully complement the growth phenotype in yeast.
Figure 2.
Transformation of enolase-deficient yeast mutant with Pfeno restores growth. Serial dilution growth assay for Parent, Tetr-Eno2 and Tetr-Eno2 + Pfeno strains in (A) absence and (B) presence of doxycycline. (C) Fragmented vacuolar phenotype in enolase-deficient yeast cells was also restored on transformation with Pfeno. Confocal microscopic images of FM4-64 labeled Parent, Tetr-Eno2 and Tetr-Eno2 + Pfeno yeast strains. Cultures were grown at 30°C. Sections shown are of 0.5 μm optical thickness. Lower panel presents percent distribution of cells having fragmented (black bar) and normal (red bar ) vacuoles. At least 300 cells from each strain were scored of vacuolar morphology. (D) Western blot analysis for the expression of Pfeno in the Tetr-Eno2 + Pfeno yeast strain. Bright field image of purified yeast vacuoles is shown in the lower panel.
Pfeno rescued the vacuolar fragmentation phenotype in enolase-deficient yeast
As discussed above, yeast enolase is associated with vacuoles and has a nonenzymatic role in homotypic vacuolar fusion. Yeast strains with reduced expression of enolase have defects in vacuolar fusion and exhibit fragmented vacuoles [19]. To determine if Pfeno could rescue the vacuolar defects in enolase-deficient yeast strains, we stained yeast vacuoles with the fluorescent dye FM4-64 and counted the number of vacuoles per cell. As previously reported, the Tetr-Eno2 strain displayed fragmented vacuoles, with ~80% of cells having more than 5 vacuoles per cell (Fig. 2C). In contrast, most parental cells displayed a single large vacuole and less than 10% contained 5 or more smaller vacuoles. Expressing Pfeno in the Tetr-Eno2 strain increased the number of cells with large, well formed vacuoles and reduced the number of fragmented vacuoles to nearly the same level as observed in the parental strain, indicating that Pfeno restored vacuolar fusion. Consistent with this observation, Pfeno was detected in preparations of yeast vacuoles by immunoblotting (Fig. 2D), as has been observed for yeast enolase [19].
Pfeno restored protein targetting to vacuoles in enolase-deficient yeast
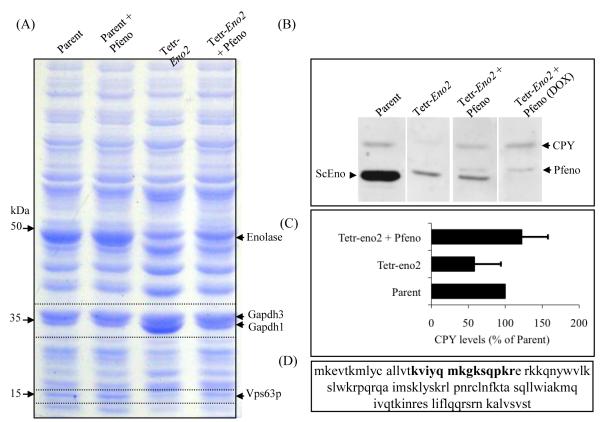
In yeast strains with reduced enolase expression, protein targetting to the vacuole is altered and several proteins found in vacuoles from parental strains are not detected [19]. In order to examine the effect of enolase deficiency on the expressed protein profile of yeast, soluble extracts from parental and Tetr-Eno2 strains were analysed on SDS-PAGE. Visual inspection of the coommassie stained gel revealed protein bands with altered intensity in Tetr-Eno2 as compared to the parent (Fig. 3A). To test whether the parasite enzyme can complement these altered levels of protein in the Tetr-Eno2 yeast, extract from Pfeno-expressing Tetr-Eno2 yeast was also analysed. A band at 50 kDa (the expected size for yeast and Plasmodium enolase) showed a decreased intensity in the Tetr-Eno2 strain that was only partially restored by Pfeno expression. This is consistent with the observation that Pfeno levels on western blots and in enzymatic assays were only 30–40 % of the levels in the parental strain. A band at ~36 kDa increased in intensity in the Tetr-Eno2 strain, but was reduced to parental levels when Pfeno was expressed. Mass spectrometric identification of proteins in this band showed it was most likely glyceraldehyde-3-phosphate dehydrogenase 1 (Gapdh1) based on sequencing of nine distinct peptides that had ~25% of sequence coverage for Gapdh1. Gapdh3 was identified as the protein band just above Gapdh1 (25 peptides sequenced with 54% coverage) and showed no significant variation in parental and Tetr-Eno2 cell extracts (Fig. 3A). A third protein at ~15 kDa showed a marked decrease in its levels in the Tetr-Eno2 strain and returned to normal in yeast expressing Pfeno. Mass spectrometric analysis of tryptic digests identified a peptide ‘(k)viyqmkgksqpkr’ which covers 11% of Vps63 (YLR261C, Fig. 3D) [39]. VPS63 is a hypothetical ORF that is currently annotated as “dubious” because it overlaps extensively with YPT6 and is translated in a different reading frame (Supplementary Figure 2). Though this represents low coverage of Vps63, the size of the band is also consistent with the predicted molecular weight of Vps63 (12.9 kDa). To our knowledge, this represents the first experimental evidence that Vps63 is produced in yeast. Taken together, these data support the view that transformation of enolase-deficient Tetr-Eno2 strain with Pfeno is able to restore normal protein expression profile in yeast. Mutations in vacuolar protein sorting (VPS) genes disrupt the trafficking of proteins such as carboxypeptidase-Y (CPY) to the vacuole [39-41]. In these strains, CPY is secreted rather than transported to the lumen of the lysosome. To determine if intracellular levels of CPY are affected in enolase-deficient and Pfeno-expressing yeast strains, we performed immunoblotting with anti-CPY antibodies on cellular extracts from parental, Tetr-Eno2 and Tetr-Eno2 expressing Pfeno (Fig. 3B). Although we observed considerable variation in multiple measurements, intracellular CPY levels were reduced in Tetr-Eno2 compared to the parental strain (p<0.05, Fig. 3B and C). However, in each case expression of Pfeno in Tetr-Eno2 restored the CPY level to that of the parental strain. Based on similar effects caused by VPS mutants, the reduction in CPY levels in enolase-deficient yeast is likely due to the improper sorting and secretion of CPY out of the cell. Regardless of the molecular mechanism underlying this observation, expression of Pfeno was able to completely reverse this defect.
Figure 3.
Altered levels of Gapdh1, Vps63 and CPY in enolase-deficient yeast are restored upon transformation with Pfeno. (A) SDS-PAGE analysis of soluble proteins from cell extracts of various yeast strains. Equivalent amounts of protein were loaded in each lane. Proteins that showed altered coommassie intensity (indicated by arrows), were identified by mass spectrometry. (B) Western blot analysis for the presence of CPY in the soluble extracts of various yeast strains and (C) quantification of CPY levels in Tetr-Eno2 and Tetr-Eno2+ Pfeno strains as percent of the Parent strain (n=5). (D) Vps63 sequence. The peptide sequenced from the tryptic digest of ~15kDa band from the Parent strain that led to identification of this band as Vps63 is marked in bold.
Dimeric Pfeno is required to fully complement the vacuolar defect in Tetr-Eno2
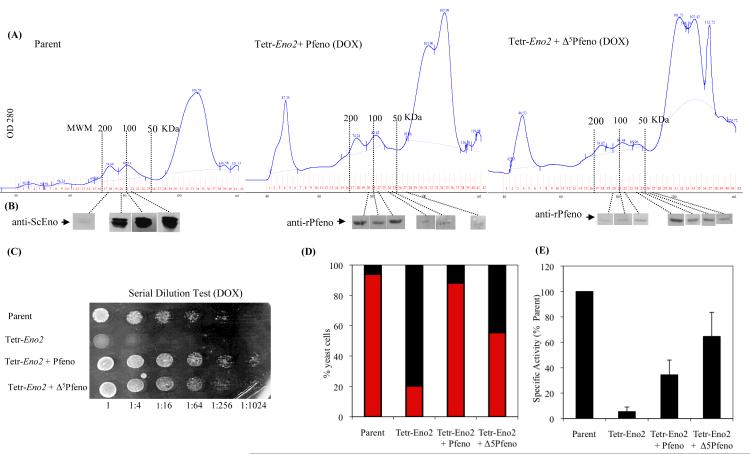
To test whether monomeric Pfeno is as effective as the dimeric form in complementing the phenotypes observed in enolase-deficient yeast, we expressed a mutant version of Pfeno (Δ5Pfeno) in Tetr-Eno2. The plant specific pentapeptide insert (104EWGWS108) is deleted in Δ5Pfeno, resulting in a protein that is largely monomeric and has much less activity than Pfeno [27]. To determine the oligomeric state of ScEno, Pfeno and Δ5Pfeno when expressed in yeast, we prepared soluble cellular extracts from the parental yeast strain and Tetr-Eno2 expressing either Pfeno or Δ5Pfeno, and subjected the samples to gel filtration chromatography. Each chromatographic fraction was then analyzed by SDS-PAGE and western blotting for the presence of enolase (Fig. 4A). Yeast enolase and Pfeno expressed in Tetr-Eno2 were largely dimeric with a molecular mass of ~100 kDa, whereas Δ5Pfeno was primarily found in the monomeric form. Δ5Pfeno restored growth of Tetr-Eno2 yeast to nearly the same level as Pfeno on solid agar (Fig. 4C) and in liquid medium (data not shown). Enolase enzymatic activity was much greater in Tetr-Eno2 yeast expressing Δ5Pfeno compared to those expressing Pfeno (Fig. 4E). Since Δ5Pfeno has significantly lower specific activity, the higher levels of activity observed in cellular extracts suggest that greater amounts of Δ5Pfeno protein were being produced. However, despite potentially being expressed at higher levels, Δ5Pfeno only partially rescued the fragmented vacuolar phenotype in Tetr-Eno2 cells (~60% of cells showed < 5 vacuoles in Tetr-Eno2 expressing Δ5Pfeno as compared to ~90% of the cells expressing Pfeno) (Fig. 4D). Thus, although the monomeric Δ5Pfeno could fully restore cellular growth in the Tetr-Eno2 strain, it rescued the vacuolar fragmentation phenotype with lower efficiency indicating that the dimeric structure of Pfeno is functionally more efficient for vacuolar fusion processes.
Figure 4.
(A) Superdex-200 gel filtration profiles of ScEno, Pfeno and Δ5Pfeno expressed in yeast. (B) Western blots for the presence of enolase in each fraction from gel filtration chromatography. Comparison of (C) growth in serial dilution test; (D) vacuolar phenotype (fragmented (black bar) and normal (red bar)) and (E) Specific activity of enolase in Parent, Tetr-Eno2, Tetr-Eno2+ Pfeno and Tetr-Eno2+ Δ5Pfeno strains of yeast.
Enolase is present in purified preparations of Plasmodium FV
In an earlier study on sub-cellular distribution of Pfeno in Plasmodium infected rbcs, close association of enolase with vacuolar membrane and hemozoin crystals as well as vacuolar lumen was observed in early and mid stage trophozoites [20]. In the mature trophozoites and schizonts, the amount of enolase associated with the FV seemed to decrease, suggesting a role for Pfeno in early stages of vacuolar development or hemozoin formation. Pfeno was also reported to interact directly with ferriprotoporphyrin IX, a precursor of hemozoin [42].
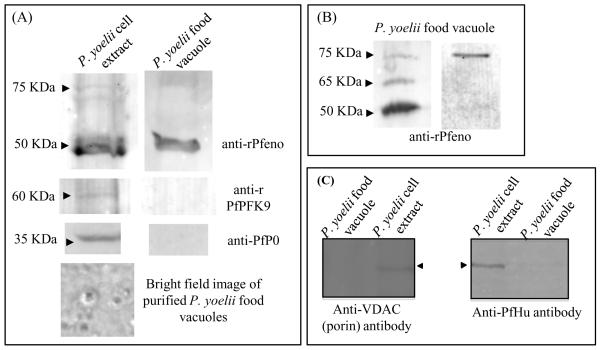
In order to obtain further evidence for the vacuolar association of enolase as observed in the in situ immuno-gold electron microscopic (IEM) images, we analyzed purified preparations of parasite FV by western blotting for the presence of enolase. FVs were isolated from P. yoelii by centrifugation on a percoll gradient. Bright field images of the purified FVs revealed vacuoles with a distinct dark structure inside corresponding to hemozoin (Fig. 5A). No intact parasite cells were observed in these preparations. Western analysis of whole cell extracts and purified FVs showed the presence of the 50 kDa enolase band in both preparations (Fig. 5A). To rule out contamination from cytosol, the vacuolar preparation was tested for the presence of two other cytosolic proteins, PfPFK9 (phosphofructokinase) and PfP0 (ribosomal phosphoprotein P0). Both PfPFK9 and PfP0 were detected in the whole cell extract with anti-PfPFK9 and anti-PfP0 antibodies, respectively, but not in the vacuolar preparation (Fig. 5A), indicating that the FV preparation was free from any significant cytosolic contaminations. The possibility of contamination of the vacuolar preparation with apicoplast and mitochondria was ruled out by similar western analyses on parasite whole cell extract and purified FV preparation using anti-VDAC (porin) (mitochondrial marker) and anti-PfHu (apicoplast marker) antibodies (Fig. 5C). Thus, we conclude that Pfeno is stably and specifically associated with the FV.
Figure 5.
Enolase is associated with purified food vacuoles (FV) from Plasmodium yoelii. (A) Western blot of P. yoelii whole cell extract and of purified FVs using anti-rPfeno antibody. Similar blots were probed with anti-PfPFK9 and anti-PfP0 antibodies that detects PFK9 and P0 in soluble whole cell extract but not in FV preparation (middle panel). Bright field image of purified FV (lower panel). (B) Some preparations of the purified FVs from P. yoelii showed higher molecular mass protein bands (at ~65 and 75 KDa) along with expected 50 KDa band in western blots, reacting with anti-rPfeno antibodies. (C) Western blot of P. yoelii cell extracts and purified FVs using anti-VDAC1 (anti-porin antibody, a mitochondrial marker) and anti-PfHu (an apicoplast marker). Typically ~10 μg of whole cell extract proteins and ~2 μg of FV proteins were used for each lane.
Although the 50 kDa band corresponding to monomeric Pfeno was the predominant form detected with anti-rPfeno antibodies in the FV preparations, two higher molecular weight species of ~65 and ~75 kDa were also occasionally observed (Fig. 5B). These variants apear to be unique to FV and have not been observed with other parasite cell fractions [31]. The cause of this reduced mobility is not yet known. However, the ~12-15 kDa size shift is consistent with the possibility of conjugation of Pfeno with ubiquitin. In the FV proteome of the P. falciparum, a ubiquitin ribosomal fusion protein uba52 homolog (PF13_0346) with MW~14.6 kDa has been identified [43]. Further analysis of high molecular bands (observed in the Western blots) is needed to determine whether some fraction of vacuolar enolase gets ubiquitinated.
Discussion
Yeast expresses two enolase isozymes, of which Eno2 is essential for survival while Eno1 is dispensable [44, 45]. Apart from catalyzing inter-conversion between 2-phosphoglycerate and phopshoenolpyruvate for glycolysis and gluconeogenesis, functional involvement of enolases in mitochondria [46] and vacuole [19] has been reported in yeast. Here we provide the first evidence that Pfeno can function in vacuolar fusion. Since yeast and P. falciparum enolases exhibit ~60% identity, we performed genetic complementation experiments in an enolase-deficient yeast mutant ( Tetr-Eno2). Tetr-Eno2 yeast grow slowly compared to the parental strain due to the lower level of ENO2 expression from the Tetr promoter. As expected, this strain failed to grow in the presence of 10 μg/mL doxycycline. The cells also have fragmented vacuoles and an altered protein expression profile ([19] and Fig. 1-3). Transformation of the Tetr-Eno2 yeast strain with a Pfeno expression plasmid resulted in full complementation of all the observed phenotypes, despite the fact that Pfeno expression levels were ~30-40% of ScEno in the parent strain, both in terms of activity as well as the amount of protein. Rescue of yeast growth did not depend on the oligomeric state of Pfeno, as the dimerization-defective Δ5Pfeno mutant, which lacks the five amino-acid insert 104EWGWS108, stimulated Tetr-Eno2 growth to the same degree as Pfeno. However, dimerization was required to rescue the vacuolar defects in Tetr-Eno2. The molecular basis of this effect is not understood. However, as the role of enolase in vacuolar fusion does not depend on the known enzymatic activity of enolase, it is possible that such variation between monomeric and dimeric forms of Pfeno in the ability to induce vesicular fusion may arise due to their differences in binding to other vacuolar proteins.
Based on this report and our previous study [20], we now have three lines of evidence that suggest a possible involvement of Pfeno in FV function: (i) Pfeno was associated with the Plasmodium FV in IEM images [20]; (ii) western blot analysis demonstrated that enolase co-purified with FV preparations (Fig. 5); and (iii) Pfeno rescued the vacuolar fragmentation defect in enolase-deficient yeast. The validity of the first two lines of evidence critically depends on the quality of the anti-Pfeno antibodies used in these experiments. The specificity of the anti-Pfeno antibodies for Pfeno has been extensively demonstrated by our earlier studies [47] and by the work of Foth et al, who used the anti-Pfeno antibodies to probe western blots of two-dimensional gels and validated the reacting proteins using mass spectrometry [48]. In addition, the specificity of the association of Pfeno with purified FVs was further supported by western blots with antibodies against cytosolic, mitochondrial and apicoplast proteins that indicated the FV preparation was not contaminated with proteins from these compartments. However, although the localization data suggests a role in FV function and the yeast complementation assay suggests involvement in vesicle fusion, definitive evidence supporting such a function for Pfeno in the parasite remains to be ascertained.
A model for S. cerevisiae and P. falciparum enolase function in vacuolar fusion
Of the multiple mechanisms for targeting proteins to FV that have been described, the endocytic-like pathway would seem to be the most likley one to involve Pfeno. This pathway utilizes small Rab5a-marked vesicles that emerge from the cytostome and fuse with the FV in a manner that resembles the endocytic pathway in higher eukaryotes [18]. Alternatively, Pfeno may traffic to the FV in a manner simlar to MSP1-19. As the parasite feeds on the hemoglobin, transport vesicles coated with MSP1-19 reach the FV and persist till the end of schizont stage [49]. Similar to MSP1-19, enolase is also present on the surface of the merozoite and is also observable in transport vesicles to the FV [20]. Other pathways, such as the “Big Gulp” and the ingestion of parasite proteins such as heme detoxification protein (HDP) after being secreted into the erythrocyte cytosol [50] can be ruled out by the absence of Pfeno in the rbc cytosol [20].
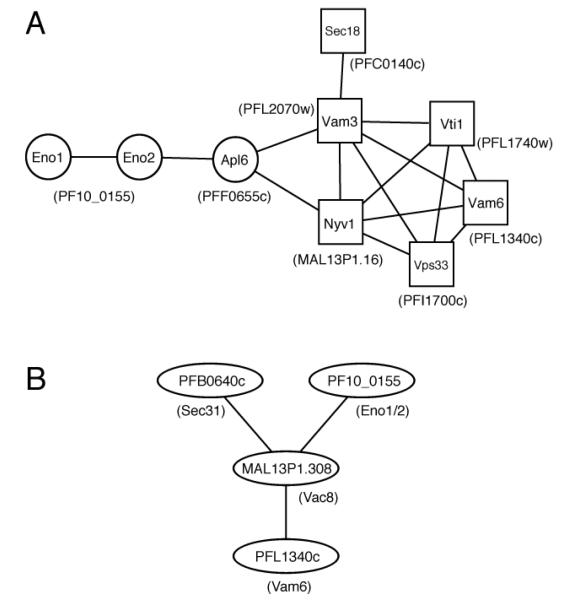
The ability of Pfeno to complement the vacuolar functions of yeast enolase suggests that the underlying molecular interactions may be conserved. However, the precise role of ScEno in vacuolar fusion is not understood. Adding purified enolase to yeast vacuoles from enolase-expressing yeast stimulated fusion, but purified enolase did not promote fusion of vacuoles from enolase-deficient yeast, indicating that the lack of enolase caused other defects in the vacuoles [19]. Consistent with this hypothesis, several vacuolar fusion proteins fail to localize to the yeast vacuole when the cell is deficient in enolase, suggesting that enolase is also important for proper trafficking or retention of a subset of vacuolar proteins. A search of ScEno physical interactions in the Biogrid database identified APL6 as the only potential interacting protein with known functions in the vacuole [51, 52]. Apl6 is a beta3-like subunit of the yeast AP-3 complex that is involved in transporting proteins to the vacuole via an alternate pathway that bypasses multi-vesicular bodies [53-55]. Interestingly, APL6 interacts directly or indirectly with the six yeast proteins that are missing from vacuoles in enolase-deficient yeast, at least one of which, Vam3, is known to be transported to the vacuolar membrane by this alternate pathway [56]. Thus, ScEno may be mediating its vacuolar effects through APL6. These interactions are shown in Fig 6A.
Figure 6.
Protein interactions of enolase and vacuolar proteins. (A) A protein interaction map of Eno1, Eno2, Apl6, and the six proteins missing from vacuoles in enolase-deficient yeast. All interactions involving these protein were downloaded from the BioGrid database and visualized in Cytoscape [58, 59]. This diagram shows only the interactions between these proteins. Additional interactions with other yeast proteins can be found in Supplementary Figure 3. Proteins missing from vacuoles in enolase-deficient yeast are indicated by squares. P. falciparum orthologs are shown in parentheses. (B) Protein interaction map of putative vacuolar proteins linked to Pfeno. Interactions from [57] were downloaded from PlasmoDB and visualized in Cytoscape. Yeast orthologs are shown in the parentheses.
To determine if a similar pathway may be functioning in P. falciparum, we searched PlasmoDB for potential orthologs of the yeast proteins in Fig. 6A. Putative orthologs for each protein were identified and are indicated in parentheses next to the homologous yeast protein. To determine if there was any evidence for these interactions among P. falciparum proteins, we searched the large-scale yeast two-hybrid interactions from an earlier report [57]. Although these interactions were not identified, an interaction between Pfeno and Mal13P1.308 was found (Fig. 6B). MAL13P1.308 is an armadillo domain-containing protein with low homology to S. cerevisiae Vac8, a vacuolar protein that makes extensive interactions with the proteins in Fig. 6A (Supplementary Fig. 3). MAL13P1.308 in turn interacted with two other putative vacuolar proteins: PFB0640c, an ortholog of Sec31; and PFL1340c, an ortholog of Vam6. Future studies are needed to validate these putative interactions and to determine if the orthologous interactions predicted from yeast also occur in Plasmodium. If such interactions are detected, it will raise another possibility for the involvement of enolase in the fusion of vesicles generated from endoplasmic reticulum-Golgi pathway.
Supplementary Material
Supplementary Figure 1: Growth curves for various yeast strains (Parent, Parent + Pfeno, Tetr-Eno2 and Tetr-Eno2 + Pfeno) in liquid medium. (A) in absence and (B) presence of 10 μg/mL doxycycline.
Supplementary Figure 2: Overlapping sequences of the “dubious” VPS63 ORF and the YPT6 ORF. (A) Genetic features around YPT6 in Chromosome XII show overlap of YPT6 and VPS63 gene sequences. This figure has been taken from the Saccharomyces Genome Database (www.yeastgenome.org). (B) mRNA sequence for VPS63 (red) has been shown as a part of the mRNA sequence for YPT6 (blue). The sequence that is common to both has been marked in purple. VPS63 is a frame shifted ORF that overlaps up to 90% with YPT6. Since a peptide sequence corresponding to VPS63 protein is identified by mass spectrometry, this protein is indeed expressed in S. cerevisiae.
Supplementary Figure 3. Extended protein interaction map of Eno1, Eno2, Apl6, and the six proteins missing from vacuoles in enolase-deficient yeast. Interactions with Eno1, Eno2, Apl6, Sec18, Vti1, Nyv1, Vam3, Vps33 Sec18, and Vam6 were downloaded from the BioGrid database and visualized in Cytoscape. Proteins that have at least two interactions with these nine proteins are shown. Interactions between proteins other than Eno1, Eno2, Apl6, Sec18, Vti1, Nyv1, Vam3, Vps33 Sec18, and Vam6 in this network were not included. The six proteins missing from vacuoles in enolase-deficient yeast are indicated by squares. Vac8, whose P. falciparum ortholog MAL13P1.308 interaction with Pfeno, is indicated by a diamond.
Acknowledgements
We thank Prof. S. Sharma of TIFR, Mumbai for providing anti-PfPFK9, anti-PfP0 antibodies and Dr. S. Habib, CDRI, Lucknow, India for a gift of anti-PfHU antibody. The enolase mutant strain (Δeno1 eno2tetR ) was a kind gift from Dr. W. Wickner. We also thank Mr. Rubul Mout for help with the gel filtration experiments. D.J.L. acknowledges support from the National Institute of General Medical Sciences (GM092829).
Abbreviations
- anti-rPfeno
polyclonal antibody raised against recombinant Pfeno
- anti-ScEno
anti-S. cerevisiae enolase antibody
- CPY
carboxypeptidase-Y (EC: 3.4.16.1)
- ENO2p
yeast enolase 2 protein
- FV
food vacuole
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase (EC 1.2.1.12)
- Pfeno
Plasmodium falciparum enolase (EC 4.2.1.11)
- rbc
red blood cell
- SHV
small hemoglobin vesicle
- ScEno
S. cerevisiae enolase (EC 4.2.1.11)
- Tetr-Eno2, S. cerevisiae strain in which ENO1 was deleted and ENO2 was expressed from a tetracycline repressible promoter, transformed with the empty vector p423GPD
Tetr-Eno2 + Pfeno, Tetr-Eno2 strain transformed with p423GPD-Pfeno
- Vps63p
vacuolar protein sorting 63 protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].World Health Organization World malaria report. 2009 < http://www.who.int/malaria/publications/atoz/9789241563901/en/index.html>.
- [2].Ginsburg H. Some reflections concerning host erythrocyte-malarial parasite interrelationships. Blood Cells. 1990;16(2-3):225–35. [PubMed] [Google Scholar]
- [3].Goldberg DE, Slater AF, Cerami A, Henderson GB. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc Natl Acad Sci U S A. 1990;87(8):2931–5. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hempelmann E, Egan TJ. Pigment biocrystallization in Plasmodium falciparum. Trends Parasitol. 2002;18(1):11. doi: 10.1016/s1471-4922(01)02146-8. [DOI] [PubMed] [Google Scholar]
- [5].Scholl PF, Tripathi AK, Sullivan DJ. Bioavailable iron and heme metabolism in Plasmodium falciparum. Curr Top Microbiol Immunol. 2005;295:293–324. doi: 10.1007/3-540-29088-5_12. [DOI] [PubMed] [Google Scholar]
- [6].Slater AF, Cerami A. Inhibition by chloroquine of a novel haem polymerase enzyme activity in malaria trophozoites. Nature. 1992;355(6356):167–9. doi: 10.1038/355167a0. [DOI] [PubMed] [Google Scholar]
- [7].Olliaro PL, Goldberg DE. The Plasmodium digestive vacuole: metabolic headquarters and choice drug target. Parasitol Today. 1995;11(8):294–7. doi: 10.1016/0169-4758(95)80042-5. [DOI] [PubMed] [Google Scholar]
- [8].Banerjee R, Goldberg DE. In: The Plasmodium food vacuole: In Antimalarial Chemotherapy: Mechanism of Action, Resistance, and New Directions in Drug Discovery. Rosenthal PJ, editor. Humana Press; Totawa, NJ: 2001. pp. 43–63. [Google Scholar]
- [9].Francis SE, Sullivan DJ, Jr., Goldberg DE. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu Rev Microbiol. 1997;51:97–123. doi: 10.1146/annurev.micro.51.1.97. [DOI] [PubMed] [Google Scholar]
- [10].Banerjee RaG DE. In: The Plasmodium food vacuole: In Antimalarial Chemotherapy: Mechanism of Action, Resistance, and New Directions in Drug Discovery. Rosenthal PJ, editor. Humana Press; Totawa, NJ: 2001. pp. 43–63. [Google Scholar]
- [11].Rosenthal PJ. Cysteine proteases of malaria parasites. Int J Parasitol. 2004;34(13-14):1489–99. doi: 10.1016/j.ijpara.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [12].Saliba KJ, Allen RJ, Zissis S, Bray PG, Ward SA, Kirk K. Acidification of the malaria parasite’s digestive vacuole by a H+-ATPase and a H+-pyrophosphatase. J Biol Chem. 2003;278(8):5605–12. doi: 10.1074/jbc.M208648200. [DOI] [PubMed] [Google Scholar]
- [13].Aikawa M, Huff CG, Spinz H. Comparative feeding mechanisms of avian and primate malarial parasites. Mil Med. 1966;131(9)(Suppl):969–83. [PubMed] [Google Scholar]
- [14].Langreth SG, Jensen JB, Reese RT, Trager W. Fine structure of human malaria in vitro. J Protozool. 1978;25(4):443–52. doi: 10.1111/j.1550-7408.1978.tb04167.x. [DOI] [PubMed] [Google Scholar]
- [15].Klemba M, Beatty W, Gluzman I, Goldberg DE. Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J Cell Biol. 2004;164(1):47–56. doi: 10.1083/jcb200307147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lazarus MD, Schneider TG, Taraschi TF. A new model for hemoglobin ingestion and transport by the human malaria parasite Plasmodium falciparum. J Cell Sci. 2008;121(Pt 11):1937–49. doi: 10.1242/jcs.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bakar NA, Klonis N, Hanssen E, Chan C, Leann T. Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of Plasmodium falciparum. J Cell Sci. 2010;123:441–450. doi: 10.1242/jcs.061499. [DOI] [PubMed] [Google Scholar]
- [18].Elliott DA, McIntosh MT, Hosgood HD, 3rd, Chen S, Zhang G, Baevova P, et al. Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A. 2008;105(7):2463–8. doi: 10.1073/pnas.0711067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Decker BL, Wickner WT. Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J Biol Chem. 2006;281(20):14523–8. doi: 10.1074/jbc.M600911200. [DOI] [PubMed] [Google Scholar]
- [20].Bhowmick IP, Kumar N, Sharma S, Coppens I, Jarori GK. Plasmodium falciparum enolase: stage-specific expression and sub-cellular localization. Malar J. 2009;8:179. doi: 10.1186/1475-2875-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Aruna K, Chakraborty T, Rao PN, Santos C, Ballesta JP, Sharma S. Functional complementation of yeast ribosomal P0 protein with Plasmodium falciparum P0. Gene. 2005;357(1):9–17. doi: 10.1016/j.gene.2005.04.007. [DOI] [PubMed] [Google Scholar]
- [22].Wider D, Peli-Gulli MP, Briand PA, Tatu U, Picard D. The complementation of yeast with human or Plasmodium falciparum Hsp90 confers differential inhibitor sensitivities. Mol Biochem Parasitol. 2009;164(2):147–52. doi: 10.1016/j.molbiopara.2008.12.011. [DOI] [PubMed] [Google Scholar]
- [23].Pal-Bhowmick I, Sadagopan K, Vora HK, Sehgal A, Sharma S, Jarori GK. Cloning, over-expression, purification and characterization of Plasmodium falciparum enolase. Eur J Biochem. 2004;271(23-24):4845–54. doi: 10.1111/j.1432-1033.2004.04450.x. [DOI] [PubMed] [Google Scholar]
- [24].Ram EV, Naik R, Ganguli M, Habib S. DNA organization by the apicoplast-targeted bacterial histone-like protein of Plasmodium falciparum. Nucleic Acids Res. 2008;36(15):5061–73. doi: 10.1093/nar/gkn483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guthrie C, Fink GR. Guide to Yeast genetics and molecular biology. Academic Press; San Diego, CA: 1981. [Google Scholar]
- [26].Colby D, Leboy PS, Guthrie C. Yeast tRNA precursor mutated at a splice junction is correctly processed in vivo. Proc Natl Acad Sci U S A. 1981;78(1):415–9. doi: 10.1073/pnas.78.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vora HK, Shaik FR, Pal-Bhowmick I, Mout R, Jarori GK. Effect of deletion of a plant like pentapeptide insert on kinetic, structural and immunological properties of enolase from Plasmodium falciparum. Arch Biochem Biophys. 2009;485(2):128–38. doi: 10.1016/j.abb.2009.02.012. [DOI] [PubMed] [Google Scholar]
- [28].Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156(1):119–22. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- [29].Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–16. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- [30].Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–60. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [31].Pal-Bhowmick I, Vora HK, Jarori GK. Sub-cellular localization and post-translational modifications of the Plasmodium yoelii enolase suggest moonlighting functions. Malar J. 2007;6:45. doi: 10.1186/1475-2875-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chait BT, Wang R, Beavis RC, Kent SB. Protein ladder sequencing. Science. 1993;262(5130):89–92. doi: 10.1126/science.8211132. [DOI] [PubMed] [Google Scholar]
- [33].Ha GH, Lee SU, Kang DG, Ha NY, Kim SH, Kim J, et al. Proteome analysis of human stomach tissue: separation of soluble proteins by two-dimensional polyacrylamide gel electrophoresis and identification by mass spectrometry. Electrophoresis. 2002;23(15):2513–24. doi: 10.1002/1522-2683(200208)23:15<2513::AID-ELPS2513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [34].Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128(5):779–92. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wickner W. Yeast vacuoles and membrane fusion pathways. Embo J. 2002;21(6):1241–7. doi: 10.1093/emboj/21.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Haas A, Conradt B, Wickner W. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J Cell Biol. 1994;126(1):87–97. doi: 10.1083/jcb.126.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Saliba KJ, Folb PI, Smith PJ. Role for the Plasmodium falciparum digestive vacuole in chloroquine resistance. Biochem Pharmacol. 1998;56(3):313–20. doi: 10.1016/s0006-2952(98)00140-3. [DOI] [PubMed] [Google Scholar]
- [38].Pal-Bhowmick I, Krishnan S, Jarori GK. Differential susceptibility of Plasmodium falciparum versus yeast and mammalian enolases to dissociation into active monomers. FEBS J. 2007;274(8):1932–45. doi: 10.1111/j.1742-4658.2007.05738.x. [DOI] [PubMed] [Google Scholar]
- [39].Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13(7):2486–501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Burd CG, Babst M, Emr SD. Novel pathways, membrane coats and PI kinase regulation in yeast lysosomal trafficking. Semin Cell Dev Biol. 1998;9(5):527–33. doi: 10.1006/scdb.1998.0255. [DOI] [PubMed] [Google Scholar]
- [41].Mullins C, Bonifacino JS. The molecular machinery for lysosome biogenesis. Bioessays. 2001;23(4):333–43. doi: 10.1002/bies.1048. [DOI] [PubMed] [Google Scholar]
- [42].Famin O, Ginsburg H. The treatment of Plasmodium falciparum-infected erythrocytes with chloroquine leads to accumulation of ferriprotoporphyrin IX bound to particular parasite proteins and to the inhibition of the parasite’s 6-phosphogluconate dehydrogenase. Parasite. 2003;10(1):39–50. doi: 10.1051/parasite/2003101p39. [DOI] [PubMed] [Google Scholar]
- [43].Lamarque M, Tastet C, Poncet J, Demettre E, Jouin P, Vial H, Dubremetz J-F. Food vacuole proteome of the malaria paraiste Plasmodium falciparum. Proteomics Clin. Appl. 2008;2:1361–1374. doi: 10.1002/prca.200700112. [DOI] [PubMed] [Google Scholar]
- [44].McAlister L, Holland MJ. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982;257(12):7181–8. [PubMed] [Google Scholar]
- [45].Holland MJ, Holland JP, Thill GP, Jackson KA. The primary structures of two yeast enolase genes. Homology between the 5′ noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981;256(3):1385–95. [PubMed] [Google Scholar]
- [46].Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20(12):1609–20. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pal-Bhowmick I, Mehta M, Coppens I, Sharma S, Jarori GK. Protective properties and surface localization of Plasmodium falciparum enolase. Infect Immun. 2007;75(11):5500–8. doi: 10.1128/IAI.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Foth BJ, Zhang N, Mok S, Preiser PR, Bozdech Z. Quantitative protein expression profiling reveals extensive post-transcriptional regulation and post-translational modifications in schizont-stage malaria parasites. Genome Biol. 2008;9(12):R177. doi: 10.1186/gb-2008-9-12-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dluzewski AR, Ling IT, Hopkins JM, Grainger M, Margos G, Mitchell GH, et al. Formation of the food vacuole in Plasmodium falciparum: a potential role for the 19 kDa fragment of merozoite surface protein 1 (MSP1(19)) PLoS One. 2008;3(8):e3085. doi: 10.1371/journal.pone.0003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jani D, Nagarkatti R, Beatty W, Angel R, Slebodnick C, Andersen J, et al. HDP-a novel heme detoxification protein from the malaria parasite. PLoS Pathog. 2008;4(4):e1000053. doi: 10.1371/journal.ppat.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D637–40. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- [54].Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- [55].Pokrzywa W, Guerriat B, Dodzian J, Morsomme P. Dual sorting of the Saccharomyces cerevisiae vacuolar protein Sna4p. Eukaryot Cell. 2009;8(3):278–86. doi: 10.1128/EC.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kim J, Scott SV, Klionsky DJ. Alternative protein sorting pathways. Int Rev Cytol. 2000;198:153–201. doi: 10.1016/s0074-7696(00)98005-7. [DOI] [PubMed] [Google Scholar]
- [57].LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438(7064):103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- [58].Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Growth curves for various yeast strains (Parent, Parent + Pfeno, Tetr-Eno2 and Tetr-Eno2 + Pfeno) in liquid medium. (A) in absence and (B) presence of 10 μg/mL doxycycline.
Supplementary Figure 2: Overlapping sequences of the “dubious” VPS63 ORF and the YPT6 ORF. (A) Genetic features around YPT6 in Chromosome XII show overlap of YPT6 and VPS63 gene sequences. This figure has been taken from the Saccharomyces Genome Database (www.yeastgenome.org). (B) mRNA sequence for VPS63 (red) has been shown as a part of the mRNA sequence for YPT6 (blue). The sequence that is common to both has been marked in purple. VPS63 is a frame shifted ORF that overlaps up to 90% with YPT6. Since a peptide sequence corresponding to VPS63 protein is identified by mass spectrometry, this protein is indeed expressed in S. cerevisiae.
Supplementary Figure 3. Extended protein interaction map of Eno1, Eno2, Apl6, and the six proteins missing from vacuoles in enolase-deficient yeast. Interactions with Eno1, Eno2, Apl6, Sec18, Vti1, Nyv1, Vam3, Vps33 Sec18, and Vam6 were downloaded from the BioGrid database and visualized in Cytoscape. Proteins that have at least two interactions with these nine proteins are shown. Interactions between proteins other than Eno1, Eno2, Apl6, Sec18, Vti1, Nyv1, Vam3, Vps33 Sec18, and Vam6 in this network were not included. The six proteins missing from vacuoles in enolase-deficient yeast are indicated by squares. Vac8, whose P. falciparum ortholog MAL13P1.308 interaction with Pfeno, is indicated by a diamond.