Abstract
We tested the fitness consequences of introgression of fast-growing domesticated fish into a wild population. Fry from wild and domesticated rainbow trout (Oncorhynchus mykiss) crosses, F1 hybrids, and first- and second-generation backcrosses were released into two natural lakes. Parentage analysis using microsatellite loci facilitated the identification of survivors, so fitness was estimated in nature from the first-feeding stage. Results indicated that under certain conditions, domesticated fish survived at least as well as wild fish within the same environment. Relative growth and survival of the crosses, however, were highly dependent on environment. During the first summer, fastest-growing crosses had the highest survival, but this trend was reversed after one winter and another summer. Although the F1 hybrids showed evidence of outbreeding depression because of the disruption of local adaptation, there was little evidence of outbreeding depression in the backcrosses, and the second-generation backcrosses exhibited a wild-type phenotype. This information is relevant for assessing the multigenerational risk of escaped or released domesticated fish should they successfully interbreed with wild populations and provides information on how to minimize detrimental impacts of a conservation breeding and/or management programme. These data also further understanding of the selection pressures in nature that maintain submaximal rates of growth.
Keywords: aquaculture, fisheries management, hybridization, phenotypic plasticity, predator prey interactions
Introduction
Global rates of extinction are at an all time high and increasing because of degradation of habitat, climate change, interactions with invasive species and other human-induced impacts (Woodruff 2001). There are many examples of threatened populations that require management intervention for preservation, either through conservation of the genetic integrity and history of the population or through supplementation to regenerate a self-sustaining population protected from inbreeding depression or a genetic bottleneck. There is extensive debate on the efficacy of management interventions in preserving the genetic integrity of wild populations (Hindar et al. 1991; for example). It is still not clear whether the potential benefits of incorporating increased levels of genetic diversity in a threatened population would overcome potential negative impacts on fitness through outbreeding depression, a reduction in fitness that may arise through either loss of local adaptation or through disruption of locally adapted gene complexes. In fact, collection of empirical data on the occurrence of outbreeding depression and evaluation of predictors of risk of fitness impacts has been presented as the number one top priority area in conservation genetics where additional data are required (Frankham 2010).
For outbreeding depression to occur, there must be genetically based differences among the two populations or crosses that are interbred. During management intervention, crosses will often be reared in captivity to conserve the genetic lineage of threatened populations or provide a source of supplementation. Genetic adaptation to captivity, or domestication, has been documented in a large number of plants, animals and bacteria, and although these phenotypic traits optimize fitness within a culture environment (Price 1997), these adaptations are usually assumed to be disadvantageous in the natural environment (Frankham 2008). This assumption assumes that if hybridization between domesticated and wild populations occurs, alleles associated with domestication should decrease in frequency over time as selection acts on the population in nature. Empirical data for this assumption is minimal; however, recent studies with sunflowers have found that traits associated with domestication can actually help to speed the rate of introgression of new alleles into the wild population (Mercer et al. 2007).
Salmonids represent an important model for tests of outbreeding depression, not only because there are clear and measurable genetically based differences between domesticated and wild strains, but also because many wild salmonid populations are threatened owing to risk from fishing and associated population supplementation activities, as well as from unintentional interactions with highly domesticated farm strains. As supplementation or captive breeding is increasingly being applied to conserve native fish populations, and with a rapidly growing aquaculture industry, there are concerns about the impact of genetic interactions between native populations and strains of fish reared in culture (Araki et al. 2007; Utter and Epifanio 2002; Utter et al. 1993). Domestication in salmon through adaptation to a culture environment has been shown to be associated with large changes in gene expression (Roberge et al. 2008; Devlin et al. 2009; Tymchuk et al. 2009). Phenotypically, domestication alters behavioural responses to risk of predation with domesticated fish showing a reduced response to predators relative to wild fish (Johnsson and Abrahams 1991; Fleming and Einum 1997; Johnsson et al. 2001; Tymchuk et al. 2006; Houde et al. 2010). In general, prey in the wild will respond behaviourally to risk of predation by, for example, increasing their cryptic behaviour or reducing foraging effort (Sih 1987; Lima and Dill 1990). Because of a decreased response to risk of predation, the domestic individuals that survive would likely maintain high foraging activity and faster growth. Indeed, many studies have found that domesticated salmonids, as well as parallel transformations generated by growth hormone transgenesis (Devlin et al. 2004), are faster growing and tend to have a lower response to risk of predation than wild fish (Johnsson and Björnsson 1994; Johnsson et al. 1996; Fleming and Einum 1997; Abrahams and Sutterlin 1999; Sundström et al. 2004; Biro et al. 2004a, 2006; Tymchuk et al. 2006, 2007) although this is not always the case (Houde et al. 2010) and may vary according to species, population and life history.
According to optimal foraging theory (Emlen 1966; MacArthur and Pianka 1966), the ability to obtain food resources from the environment is an important component of fitness upon which selection will act. Often, however, an individual cannot maximize extraction of food resources from the environment without facing a concurrent increase in risk of predation (Werner et al. 1983; Werner and Gilliam 1984; Lima and Dill 1990; Werner and Anholt 1993). Indeed, trade-offs between foraging and predation can shape optimal life-history strategies of a species. The relative selective pressure of predation and competition, however, may vary depending on environmental conditions. For example, there may be a fitness advantage to growing fast and potentially out-competing conspecifics when predation threat is low and/or if future resources will be limited, even though there may be an initial cost of increased mortality because of predation (Biro et al. 2004a; Tymchuk et al. 2007). Alternatively, under high risk of predation and/or very low food availability, slow growth may provide increased fitness by reducing costs associated with foraging, such as energy expenditure and predation risk. For many animals, and fish in particular, it appears that species have evolved a suite of linked life-history, metabolic and behavioural traits integrated to optimize fitness within their native environment (Biro and Stamps 2008, 2010; Careau et al. 2010).
In the present study, we test the fitness consequences of altered rates of growth and behaviour under risk of predation within a natural environment. Specifically, we used crosses of rainbow trout (Oncorhynchus mykiss) known to express different rates of growth and behavioural phenotypes within both culture and semi-natural environments (Tymchuk and Devlin 2005). This experimental model makes use of pure wild (slow-growing) and domesticated (fast-growing) crosses, first generation domesticated by wild hybrids (F1), the first backcross generation (BC1) produced by crossing F1 hybrids with the wild parental line, and a second backcross generation (BC2) produced by crossing BC1 families again to the wild parental line. Impacts of fitness attributable to outbreeding depression may arise from loss of local adaptation in the first generation of hybridization between the wild and non-native populations (the F1 cross), whereas outbreeding depression attributable to disruption of locally adapted gene complexes cannot occur until the backcross generations BC1 and BC2 after chromosomal recombination had occurred in F1 and BC1 parents, respectively (Edmands 2007). Therefore, this study provides an important contribution to our understanding of outbreeding depression because we were able to measure fitness over three generations of interaction following introgression of domesticated alleles into the wild strain. The offspring from these crosses were released into two natural experimental research lakes. Previous work using these lakes found that pure crosses of fast-growing domesticated fish suffer increased mortality relative to wild fish when risk of predation is high, but not when risk of predation is low (Biro et al. 2004a, 2006). This research extends previous work by testing for fitness costs, in nature, of introgression of domesticated alleles into a wild genomic background over multiple generations. This information is of relevance for understanding the impacts of domestication on fitness of wild populations and assessing the multigenerational risk of escaped or released domesticated fish should they successfully interbreed with wild populations.
Materials and methods
Fish
We used crosses of rainbow trout known to express different rates of growth found to be largely controlled by additive genetic variance (Tymchuk and Devlin 2005; Tymchuk et al. 2007). The fast-growing domesticated fish (D) originated from the Spring Valley Trout Farm (Langley, British Columbia, Canada) and the wild trout (W) were collected from nature from Pennask Lake (British Columbia) for all generations (Fig. 1). The parental series was generated over several years (Fig. 1) to allow all cross types to mature and be crossed in the same year to generate the full series of crosses representing an introgression event. On 7 June 2005, five single-pair, pure domesticated (D) crosses were fertilized, using five females and five males. On 9 June 2005, sperm was collected from domesticated (D), wild (W), domesticated x wild hybrid (F1), and F1 × wild backcross (BC1) males (six different males for each genotype) reared at Fisheries and Oceans Canada Laboratory (UBC/DFO Centre for Aquaculture and Environmental Research) in West Vancouver, BC (Tymchuk and Devlin 2005). Eggs were collected from 240 wild-caught Pennask Lake females and fertilized in single-pair crosses, with each female crossed with one of the four possible male cross types (D, W, F1, BC1). In total, the following crosses were produced (Fig. 1): pure domesticated (D, n = 5), pure wild (W, n = 49), domesticated x wild hybrids (F1, n = 68), F1 × wild backcrosses (BC1, n = 67) and BC1 × wild backcrosses (BC2, n = 56). Fertilized eggs were incubated at the laboratory in Heath trays with flow-through well water at 10°C until they had absorbed their yolk. Upon reaching the first-feeding stage, the fry were measured and stocked into the lakes the following day (28 July 2005).
Figure 1.
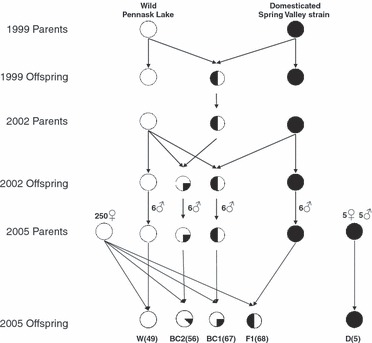
A flow diagram of the cross design. Families are pure wild (W; 0% domesticated alleles), second-generation backcross (BC2; 12.5% domesticated alleles), first-generation backcross (BC1, 25% domesticated alleles), F1 hybrid (F1; 50% domesticated alleles) and pure domesticated (D; 100% domesticated alleles). Numbers to the left of sex symbols in the 2005 parents indicate the number of parents that were used overall, and numbers in brackets in the 2005 Offspring indicate the number of females fertilized as single-pair crosses (each female was fertilized with one male).
Growth and survival in natural lakes
We used two small experimental lakes (PPH and CPH) with similar characteristics (3.3 and 4.1 ha respectively, maximum depth for both lakes is 18 m) located adjacent to each other in south-central BC, Canada (49°50′–49°56′N, 120°33′–120°34′W; Fig. 2). Both lakes were assessed on 11 June 2005 to verify that they contained adult rainbow trout, which are by far the most significant predator on young trout in these lakes (Beckmann et al. 2006). This assessment revealed the presence of many yearling and older rainbow trout. Trout cannot naturally spawn within these lakes, and therefore any trout within the lake were from previous releases. A more comprehensive estimate of predator density and size was completed during the fall sampling, and these data are included in the results section. Each genotype was stocked 28 July 2005 at a density of 1000 fry/ha except for the domesticated fry that were stocked at 200 fry/ha (because of low availability of eggs when crosses were performed). Fry were left undisturbed within these lakes to experience natural selective and competitive forces until 8 October 2005 when we began lethal gillnet sampling for five consecutive nights, using standardized gangs of graded mesh gillnets and constant netting effort to estimate the population size and growth of the fry that survived over the summer (details in Post et al. 1999; Biro et al. 2003). As vulnerability to gillnetting increases with body size, the total catch was adjusted for size dependence in recapture probability by an established size-specific vulnerability function determined from previous mark–recapture experiments in the lakes (Biro et al. 2003, 2004a,b). The adipose fin was clipped from each fish and stored in ethanol. Following the same netting protocol, the lakes were sampled again 1 year later (23–26 October 2006) to compare relative growth and survival as yearlings, after one winter and another summer of growth. Survival estimates for the yearlings represent the proportion of fish surviving from the previous sampling.
Figure 2.
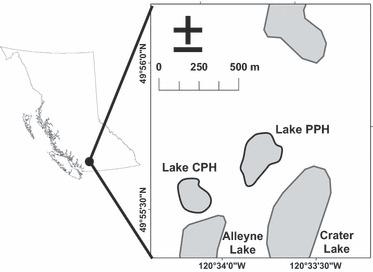
Map showing the location of the two research lakes, Crater Pothole (CPH) and Pete's Pothole (PPH), near Merritt, British Columbia.
Parentage assignments
DNA was extracted from fins using DNeasy 96 Tissue kit (QIAGEN, Valencia, CA, USA) as described by the manufacturer. Genotypes of the survivors were determined by identification of parents based on 12 polymorphic microsatellite DNA loci: OMM1234, OMM1231 and OMM1270 (Rexroad and Palti 2003); Ssa407, Ssa408, Ssa410 and Ssa417 (Cairney et al. 2000); omy77 (Morris et al. 1996); ots3 (Banks et al. 1999); OMM1128 (Rexroad et al. 2001); and ots104 and ots107 (Nelson and Beacham 1999). These loci were selected based on nonlinkage with fitness-related loci, good amplification and high diversity within our samples. For OMM1234, OMM1270, Ssa407, Ssa408, Ssa 410 and Ssa417, microsatellite amplifications via polymerase chain reactions (PCRs) were performed in 10-μL reactions containing 1× reaction buffer [20 mm Tris-HCl (pH 8.4), 50 mm KCl], 1.5 mm MgCl2, 0.2 mm each dNTP, 0.55 μm each primer, 0.25 units of Taq polymerase (Invitrogen) and ∼10 ng of template DNA. Samples were amplified using an Applied Biosystems Inc. (ABI; Foster City, CA, USA) GeneAmp® PCR System 2700 thermocycler using touchdown PCR: one cycle of 95°C for 3 min; 10 cycles of 95°C for 30 s, 63°C for 1 min (−0.5°C per cycle), and 72°C for 1 min; 20 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s; one cycle of 72°C for 20 min. The remaining loci were amplified in two multiplex PCRs using the QIAGEN Multiplex PCR Kit (QIAGEN). The first multiplex consisted of the loci Ots104 and Ots107 and the second multiplex consisted of OMM1128, OMM1231, Ots3 and Omy77. Each multiplex PCR was performed in a 20 uL volume containing 100–150 ng of genomic DNA, 10 uL of QIAGEN Multiplex PCR Master Mix (consisting of 1 U HotStar Taq DNA polymerase, 10× QIAGEN Multiplex PCR buffer with a final concentration of 3 mm MgCl2 and 0.2 mm of each dNTPs) and 0.2 um of fluorescent-labelled forward primer (fluorescent dyes were FAM, VIC, Ned and PET) and nonlabelled reverse primer. PCR protocols were one cycle of 95°C for 10 min; 38 cycles of 94°C for 30 s, 50°C (Ots104, Ots107) or 54°C (OMM1128, OMM1231, Ots3 and Omy77) for 30 s, 72°C for 30 s; and a final; extension of 72°C for 10 min. PCR products were run on an ABI 3730 genetic analyzer (Applied Biosystems), and genotypes were visualized with the genotyping software, GeneMapper® 3.7 (Applied Biosystems).
A simulated set of 400 offspring was randomly generated based on the genotypes of the parents used for the crosses. Using CERVUS 3.0, a maximum-likelihood programme that calculates logarithm-of-odds scores for candidate parents from simulations based on given allele frequencies (Kalinowski et al. 2007), parentage was assigned to these simulated offspring to estimate a correctness rate, or the proportion of offspring correctly assigned. For this data set, simulations were run within CERVUS based on the assumptions that 100% of the sires and dams were captured and with a 1% genotyping error rate (as suggested by the user manual). Based on parentage assignment of a simulated population of offspring, the correctness of CERVUS was estimated to be 89.3% when family was assigned (by identifying both sire and dam), whereas the correctness increased to 95.5% when assigned to cross type (based only on the genotype of the sire). Therefore, for this study, paternity was assigned to offspring to identify their cross type (W, BC2, BC1, F1, or D), using the same settings within CERVUS for the allele frequency simulations (100% of the sires/dams captured and 1% error rate).
Line cross analysis
Joint-scaling regression technique was used to test for additive and additive-dominance (AD) models of gene action and their contribution to the phenotypic divergence between the wild and domesticated trout, following the procedure outlined in Lynch and Walsh (1998) and Tymchuk et al. (2006, 2007). The joint-scaling test uses least-squares regression to estimate the model parameters and then compares the observed line means with the predicted means (Cavalli 1952; Hayman 1960). This analysis was carried out on lake means for mass, specific growth rate in mass (SGRmass) and survival.
The following model was fit to the observed line means,
where zi is the ith line mean, μ0 is the mean of all line means, αi is the additive genetic effect, δi is the dominance effect and εi is the residual error. The error term incorporates deviation of the observed and predicted line means in addition to any epistatic effects that could not be estimated directly because of a limited number of degrees of freedom. With M as the matrix of coefficients, the linear model becomes
where 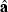 is a vector containing estimates of the model parameters, mean (μ), additive effects (α) and dominance effects (δ). The weighted least-squares solution is
is a vector containing estimates of the model parameters, mean (μ), additive effects (α) and dominance effects (δ). The weighted least-squares solution is
with V as a diagonal matrix of squared standard errors of the means, and 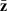 a vector of line means. The sampling variances and covariances of the parameter estimates are given by the covariance matrix
a vector of line means. The sampling variances and covariances of the parameter estimates are given by the covariance matrix
The additive and AD genetic models were tested by sequential model fitting, beginning with the simpler additive model. The additive model was tested with a goodness-of-fit test statistic
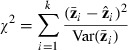 |
with degrees of freedom equal to the number of lines (four) minus the number of estimated parameters. For the additive model, df = 2. The AD model was then tested, with df = 1. The likelihood-ratio test was used to see whether the AD model was a significant improvement over the additive model (A)
with df = 1.
Statistical analyses
One-way anova with cross as a fixed factor was used to test for differences among the line crosses in size over the first summer and after one winter of growth. Tukey's honest significance test was used for unplanned pairwise comparisons between crosses. Relative survival of crosses within each lake was compared with a G-test using the pure wild crosses as a reference with the null hypothesis that there is no difference in survival among crosses. Bonferroni adjustment was applied to the P-values to account for multiple test comparisons; differences were significant at P < 0.05. Mortality of corresponding groups raised under culture conditions was minimal, so no corrections were made to survival measured in the wild. Multiple linear regression in SigmaStat (Systat Software Inc., San Jose, CA, USA) was used to test for a relationship between size and survival in each of the two lakes and for the two growth periods (summer and winter).
Results
After release into the natural lakes in July 2005, a total of 76 (0.4% of total released) and 234 (1.5% of total) fry were captured in October 2005 from CPH and PPH, respectively. For CPH, 75 individuals (99%) were assigned to cross type at 80% confidence level, and 234 individuals from PPH (100%) were likewise assigned. The following year, 90 additional fish were collected from PPH and 86 (96%) were assigned to cross type. No fish were captured from CPH in 2006. Any unassigned fish or fish clipped for mark–recapture estimates were excluded from estimates of survival. Although multiple individuals were often captured from the same family within a cross, it is unlikely that the results reflect only family variation. With the exception of BC2 in CPH, all crosses were represented by multiple families from at least three different males. The majority of recaptured families were represented by <20 offspring, and only two families from PPH had a greater number of offspring (a BC2 family with 25 offspring and a BC2 family with 78).
Both summer and winter survival of the fry in the experimental lakes varied according to genotype (Fig. 3). The estimated number of predators within CPH (n = 555, mean length = 26.3 ± 8.1 cm) was approximately three times higher than the number in PPH (n = 190, mean length = 27.4 ± 8.1 cm) as determined by gillnetting during the fall sampling period, and there was an associated lower survival rate in CPH (0.4%) relative to PPH (1.5%). Within CPH, the rank order of survival over summer was D (13) > F1 (25) > BC1 (12) > BC2 (3) = W (3). The numbers in brackets indicate the number of recaptured fish assigned to the cross. The rank order of survival in PPH over the summer followed a similar pattern, with the exception of the F1 hybrids: D (29) = BC1 (116) > BC2 (32) > W (17) = F1 (21). Relative over-winter survival (Fig. 3B) was the highest for the W and BC2 crosses, the lowest for the BC1 cross and the D and F1 strains had similar and intermediate survival rates. Over-summer survival of the crosses in CPH and PPH, and survival over the subsequent year in PPH, did not fit an additive nor dominance model of gene action, indicating epistasis (all χ2 > 17.129 and all P < 0.0002). Over-summer survival of all hybrid crosses (F1, BC1 and BC2) was lower than would be expected by an additive model. Within PPH, however, the over-summer survival of the BC1 and BC2 crosses was higher than would be expected by an additive model. The F1 cross had lower survival than expected, as observed in CPH. After another season within PPH, all the hybrid crosses had lower survival than would be expected based on an additive model of gene action.
Figure 3.
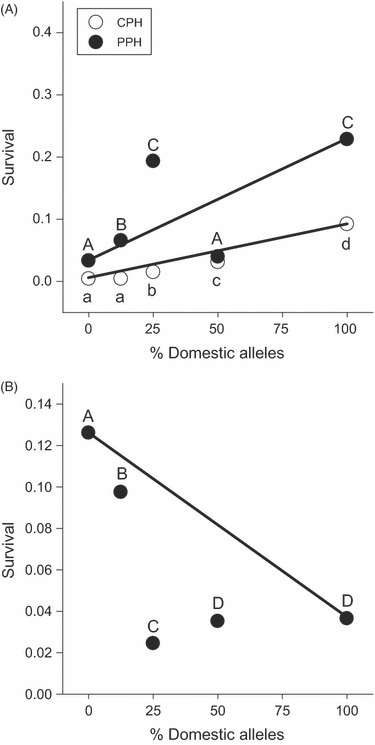
Estimated summer (A) and winter (B) survival of the fry released into the two natural lakes, CPH and PPH. Points represent the proportion of fish surviving from those released 20 July 2005. Letters indicate significant differences in survival among the genotypes (G-test, all P < 0.05). Strains are pure wild (W; 0% domesticated alleles), second-generation backcross (BC2; 12.5% domesticated alleles), first-generation backcross (BC1, 25% domesticated alleles), F1 hybrid (F1; 50% domesticated alleles) and pure domesticated (D; 100% domesticated alleles). The line connecting the D and W means indicates the a priori expectation of additive genetic effects; character means for all hybrids should fall on this line if divergence was because of genes with only additive effects.
A random sample of fry was collected from the pooled crosses and measured prior to release into the lakes at 50 days postfertilization (dpf); the D fry were smaller than the other crosses (F4 = 11.826, P < 0.001). After growing throughout the summer (121–126 dpf), there was a significant difference in size among the crosses from PPH (F4,210 = 9.850, P < 0.001) but not from CPH (F4,51 = 0.278, P = 0.891). In PPH, the W and BC2 crosses were the smallest, and the D and BC1 crosses were the largest (Fig. 4A). The F1 hybrids were intermediate to and not significantly different from any of the cross types. The size differences among the strains from PPH were no longer present at the end of their second growing season (F4,85 = 1.327, P = 0.267, Fig. 4B). Additive genetic effects adequately explained the mean size of the crosses after one summer in CPH (χ2 = 2.342, P = 0.505) but size of the crosses in PPH at this time did not fit either an additive or dominance model of gene action. In particular, the BC2 cross was smaller than would be expected by an additive model, whereas the BC1 cross was larger than expected. After another season of growth in PPH, however, the mean mass of the crosses fit an additive model (χ2 = 2.393, P = 0.495).
Figure 4.
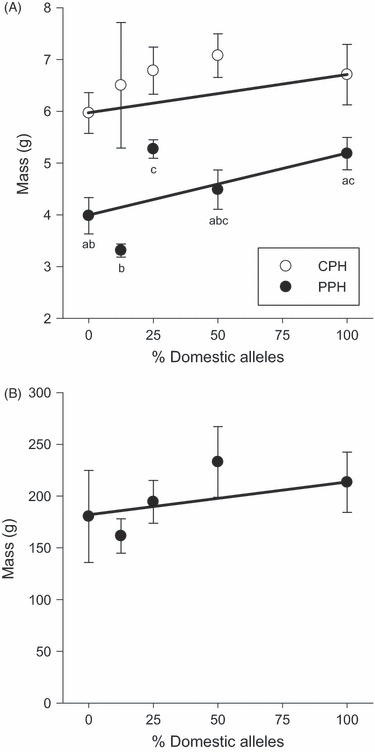
Mean mass (g) of the crosses reared over summer (A) and over winter (B) in two natural lakes, CPH and PPH. Points represent the cross means and associated standard error, and letters indicate significant differences among cross types. Only the fish sampled from PPH at the end of summer showed significant differences in size. Crosses are pure wild (W; 0% domesticated alleles), second-generation backcross (BC2; 12.5% domesticated alleles), first-generation backcross (BC1; 25% domesticated alleles), F1 hybrid (F1; 50% domesticated alleles) and pure domesticated (D; 100% domesticated alleles). The lines connecting the D and W means indicate a priori expectation of additive genetic effects.
There was a strong relationship between size and survival for both lakes, and at both sampling times, but the shape and direction of the relationship differed between sampling periods. At the end of the first summer, the relationship between size and survival was best explained by a second-order polynomial, in both PPH (R2 = 0.950, P = 0.049) and CPH (R2 = 0.963, P = 0.036, Fig. 5A). The fastest-growing fish had the highest survival within each lake, whereas the lowest survival was not seen in the slowest growing fish, but rather in those growing slightly faster. This relationship was reversed in PPH after growth over winter and a second summer (R2 = 0.935, P = 0.007, Fig. 5B), with the larger fish having lower survival.
Figure 5.
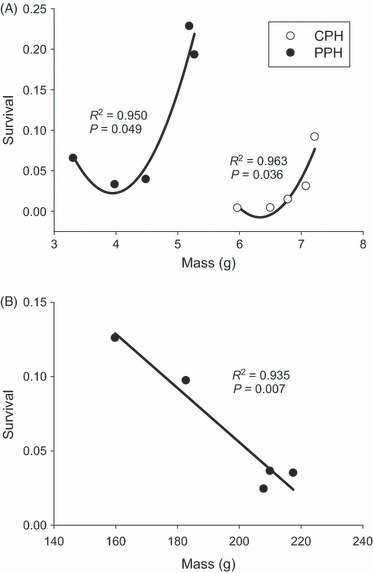
Relationship between mean size (mass) and survival for each cross type after a summer in PPH and CPH (A) and an additional year of growth in PPH (B).
Discussion
Results of this experiment indicated that natural, complex environments alter the relative growth and survival of wild, domesticated and hybrid crosses compared to results obtained under culture conditions (Tymchuk and Devlin 2005) and provided further evidence that the ability to detect heterosis and/or inbreeding depression will depend on the environment in which the hybrids are raised (Tymchuk et al. 2007). It is also evident that not only geographically different natural environments (i.e. different lakes), but even different time periods within the same lake, can cause further variation in relative estimated fitness. In species with a short lifespan, such as Drosophila, it has long been known that rapid natural selection can occur in response to seasonal fluctuations in environmental conditions (Dobzhansky 1943) with changes in the relative frequencies of gene arrangements depending on season. As fish have a longer life cycle that typically encompasses multiple seasons and environments, it is logical to assume that the wild-type phenotype would have been selected for the highest overall fitness across the entire range of environments and the full life history of the animal, and our results suggest the temporal stability of this fitness is variable. In fact, under the conditions tested in this experiment, the wild-type phenotype did not always incur the highest fitness relative to the domesticated cross and overall survival (average of first season multiplied by subsequent season survival) was lower than the overall estimated survival for the domesticated cross.
This study also highlighted an important observation relevant to the debate on the ecological importance of altered rates of growth within populations. Clearly, the ideal rate of growth is highly dependent on the environment, supporting the view that disruption of locally adapted gene complexes could have a significant impact on species such as salmonids that show local adaptation on small geographic scales (Templeton 1986). During the summer, when conditions were conducive to growth (higher levels of food, higher temperatures), there was a positive correlation between growth and survival. However, after a second year of growth that included one winter and another summer, there was a decrease in survival with faster rates of growth. The crosses that had the highest survival over summer suffered the highest mortality over the second season, indicating that the optimum growth is a trade-off between these two different selection regimes. The mortality of the different crosses within the culture environment was negligible from first-feeding onward, so there were no intrinsic differences in viability at this stage that may have caused the trend in mortality observed in the field. However, it is important to note that survival consequences to pure strains of fish selected in nature for different growth rates may differ from those observed in the present experiments where genotypes and consequent growth rates have been generated by repeated introgression.
After the first summer of growth, there were some differences in the rank survival of the strains from PPH and CPH, with the exception of the F1 hybrids. Although overall survival was higher in PPH relative to CPH, the rank orders of survival, from the highest to the lowest, in both lakes were pure domesticated, first-generation backcross, second-generation backcross and pure wild. This suggests that the crosses exhibiting a more wild-type phenotype had lower survival; this trend fits with observations on the same crosses within a semi-natural environment designed for competitive interactions among strains (Tymchuk et al. 2007). Outbreeding depression is evident when fitness of offspring from crosses between two divergent lines is lower than the fitness of offspring from either parental line. Based on this criterion, these data therefore provide little evidence for outbreeding depression in either of the two backcrosses, both of which should have disruption of parental gene complexes (Edmands 2007) that may be coadapted, with the exception of survival of the BC1 cross in PPH after a second year in the lake. Fraser et al. (2008, 2010) also found limited evidence of outbreeding depression after two generations of introgression between locally adapted and nonresident or farmed Atlantic salmon (Salmo salar). The authors present some interesting possible reasons for the lack of detectable outbreeding depression, such as the effect of some residual duplicated gene loci on the length of time required to detect outbreeding depression. Further studies designed to examine the mechanisms regulating hybrid fitness will be important to further our understanding and ability to predict the impact of interaction between native and domesticated cross types.
The domesticated × wild F1 hybrids were also an exception to the observed trend of a linear relationship between the level of introgression and survival. Within PPH, the F1 hybrids had a survival rate close to that of the pure wild fish, whereas in CPH this cross had a survival rate intermediate to the wild and domesticated crosses, as would be expected based on additive gene action. The underlying reasons for the variation in F1 hybrid survival between lakes are not clear, but they may reflect the impact of different environmental conditions on the expression of outbreeding depression versus heterosis; this suggests that disruption of adaptation to distinct natural conditions may be attributable to decreased fitness observed in this study (Edmands 2007). CPH had approximately three times the predator load of PPH, and therefore the lower survival in this lake was expected because larger trout are the main predator on these small fish (Beckmann et al. 2006). With the smaller number of fry in the lake, or because of inherent characteristics of the lake, however, there may have been more size-suitable food available for this cohort, thereby reducing competition for limited resources. In PPH, survival was much higher and therefore food resources may have been more limited relative to CPH. The size of the surviving fish supports this speculation; all fish grew larger in CPH relative to PPH, and in fact there were no significant differences in size among the strains within CPH. The size differences between strains emerged only in the more competitive PPH environment.
After an additional year of growth, there was a general reversal in the relative survival of the crosses in PPH. The pure wild crosses and second-generation backcrosses had the highest survival, and the first-generation backcross had the lowest survival. The pure domesticated and F1 hybrid crosses had similar survival rates that were closer to the lower BC1 survival as opposed to the higher W and BC2 survival. As the source of mortality cannot be known for certain in the wild, these fish may not necessarily have been consumed by a predator, but may have also died due to other causes, such as starvation or pathogens, for example. Although the domesticated fish were significantly larger that the wild fish at the first sampling, there were no longer any differences in size after growth over winter, indicating that food resources were likely severely limited during this time, or that faster-growing fish suffered greater mortality. Previous studies have indicated that over-winter mortality of trout in these lakes tends to be high and size-dependent, indicating this time period acts as a strong selection force on these populations (Biro et al. 2004b). Biro et al. (2004b), however, found that size was positively associated with survival with larger fish having a slower decline in lipid levels, and therefore able to survive longer periods of starvation. A positive correlation between size and over-winter survival may be specific to within-strain comparisons, but other studies have also found that larger fish could have an over-winter survival disadvantage (Carlson et al. 2008). Perhaps faster-growing domesticated strains have obligate increased metabolism that depletes lipid reserves at an even faster rate than the slower-growing wild fish that may have evolved metabolic rates to withstand food shortages. More rapid use of lipid resources in fast-growing transgenic strains compared to wild type supports this idea (Raven et al. 2006; Leggatt et al. 2009).
A previous study comparing growth and mortality between domesticated and wild rainbow trout fingerlings in these same lakes found that the domesticated fish showed a high survival advantage under no risk of predation, a small survival advantage under an intermediate risk of predation and lower survival when predation was high (Biro et al. 2004a). A more recent study indicated that domesticated fry had higher survival than wild fry in lakes without predators, but lower survival than wild fry in lakes with predators (Biro et al. 2006). In the present study, only the F1 hybrids varied in relative survival between lakes experiencing different levels of predation, and the domestic fish had higher survival over the first summer in both lakes. We offer four reasons for the observed differences between these two studies. First, the fry used in this study were released at first-feeding stage, whereas Biro et al. (2004a) reared the fish in culture conditions until they were much larger (approximately 15 cm in length) before release, and thus differences in developmental stage, maternal influences and rearing experience exist between the fish used in that and the present studies. A critical window affecting lifetime survival is during the early fry stage, which was not examined in Biro et al. (2004a). Biro et al. (2006), however, released fry at a stage similar to the present study, although they did experience some feeding within the culture environment before release. Second, the strains of wild and domesticated fish used in these three studies were from different strains and may therefore express different relative phenotypes affecting fitness that are unrelated to their common phenotypes (such as rapid growth or enhanced feeding motivation, for example). Third, the difference in results between these two studies may have arisen from different levels of overall mortality. In this study, the overall survival of the fry was only 1.6% for the high predator lake and 8.9% for the low predator lake, whereas in the study by Biro et al. (2004a), the overall survival under risk of predation was 38%. In similar studies with Atlantic salmon, lower survival of hybrid and backcross offspring between farm (domesticated) and wild fish relative to wild fish has been observed (McGinnity et al. 2003), although in these studies the pure farm parental cross also had reduced survival relative to the wild fish (differing from the present results). Another study found no difference in fitness between farm and wild crosses in the natural environment (Fleming et al. 2000), suggesting that cross type differences and environmental fluctuations are likely playing important roles in influencing the fitness outcomes of introgression between wild and domesticated crosses of salmonids. In fact, the magnitude of difference between parental populations will likely affect the fitness of hybrids in nature (i.e. Fraser et al. 2010; Araki et al. 2007). Finally, while the lakes used in the present study were the same as Biro et al. (2004a) mentioned earlier, the environmental conditions within each lake are not known to be constant and thus may have differed among the studies and affected a range of fitness-related traits we are unaware of.
Evidence of the ability of fish to express faster growth during compensatory growth responses, domestication selection or GH treatment provides further support that growth rates in nature are at levels below the physiological maximum (Donaldson et al. 1979; Ali et al. 2003; Devlin et al. 2004; Tymchuk and Devlin 2005). It has been suggested that if faster rates of growth were advantageous in terms of overall fitness, this characteristic would have evolved within the natural environment and that this has not occurred is because of trade-offs with other fitness costs (Arendt 1997). This argument assumes, however, that any increase in magnitude of growth rate would provide an equivalent magnitude of advantage or disadvantage to an individual. There may in fact be other non-wild-type optimal growth rates that would incur similar levels of lifetime fitness, but perhaps the intermediate steps arriving at that rate actually cause lower fitness. Depending on the shape of the trade-off curve, it may be possible to have more than one optimal solution to the trade-off between fitness advantage and disadvantage (Partridge et al. 1991; Mangel and Stamps 2001) and perhaps a minimum quantum growth shift that must occur before this trade-off is balanced. It has been well established for the fish used in this experiment that there is a linear relationship between the proportion of domesticated alleles within the genotype and growth (Tymchuk and Devlin 2005) under conditions without competition or predation. As the hybrid cross tends to have growth rates intermediate to the parental strains, perhaps these are no longer able to balance growth advantages with associated fitness costs and therefore show reduced survival relative to the parental crosses. Within the conditions tested in this study, the F1 hybrids had the lowest overall survival.
An important piece of information that remains unknown to assess introgression effects as described here is the relative reproductive fitness of both sexes of the different crosses at maturity, within a natural environment, and the role of maternal effects on relative survival. If there is unequal reproductive fitness among the crosses, any genetic threat to wild populations may be altered significantly, particularly if the domesticated fish cannot successfully interbreed with the wild population in the first place (because of sterilization measures, maturation timing among strains, or body size effects) or if the hybrids have such a great reduction in fitness that they are unable to survive to maturity. Regarding maternal effects, all genotypes were produced by crossing wild or hybrid males to wild females from nature; however, the pure domesticated genotype was generated from females reared under culture conditions. It is not known to what extent this may affect egg quality and subsequent survival, and rearing domestic strain fish in nature would be ideal in future studies.
In conclusion, this research shows the relative fitness consequences of introgression beyond first-generation hybrids of domesticated alleles into a wild population of rainbow trout. These experiments allowed estimation of survival fitness in nature from the first-feeding (fry) stage as individual fish were assigned to family based on microsatellite analysis, rather than having to rear fish in culture environment to a size that would allow marking by tagging or fin-clipping. As selection attributed to mortality at this early life-history stage can be quite strong (Elliott 1990), it is important to remove the influence of the culture environment that is known to have strong influences after the first-feeding stage, which this study has done. The present data suggest that first-generation hybridization between domesticated and wild individuals could affect the survival of wild populations if large numbers or sustained releases or escapes of fish occurred and interbred with wild fish. These effects could be further augmented by initial lower viability of F1 eggs, as observed in this study, and because domesticated fish tend to have higher fecundity than wild fish. The extent of the impact on the wild population will depend on the size, frequency, and timing of the release or escape of domesticated fish. In addition, we recognize that many effects of domestication are likely cross, environment and species specific and that additional studies will be required to draw general conclusions. The present data also suggest that the introgression of small numbers of domesticated genotypes into wild populations may be absorbed such that the second-generation backcrosses would be largely indistinguishable from wild-type phenotype. It is likely, however, that some alleles associated with the domesticated strain would still remain within the population, and it is not known whether these may impact the ability of the wild population to genetically adapt to changing environmental conditions.
Applications to conservation and management of threatened populations
Frankham et al. (2011) have proposed that management decisions on the level of gene flow allowed between two populations require a cost-benefit approach whereby overall fitness is the currency, inbreeding depression and outbreeding depression are costs, and genetic rescue or avoidance of outbreeding depression are the benefits. Inbreeding depression seems to be widespread and relatively easy to predict and quantify (Frankham et al. 2011), suggesting that hybridization in general could provide positive impacts on population fitness through heterosis (McClelland and Naish 2007). Recently, however, studies on interactions between divergent populations suggest that outbreeding depression can be severe if it occurs and may cause larger negative impacts on reproductive fitness than inbreeding depression (Araki et al. 2009, Theriault et al. 2011). Results presented here provide useful knowledge for conducting an appropriate cost-benefit analysis on population supplementation for conservation. First, the effect of environment on hybrid fitness is significant and can alter the relative fitness of the parental lines and their hybrid offspring. This supports earlier work with plants (Johnston et al. 1991; Mercer et al. 2006) and sticklebacks (Hatfield and Schluter 1999) and underscores the need to include ecological characteristics as factors within the cost-benefit assessment. Second, the relative fitness of parental lines and their hybrid offspring may change depending on life-history stage. In the present study, very different predictions would arise if data were extrapolated only from fitness effects observed from the first sampling following summer growth, compared to the second sampling that included both summer and overwintering effects. Tests for fitness costs associated with outbreeding depression should ideally be conducted using data from throughout the entire life history of the organism. Third, even though the pure domesticated crosses had the highest overall survival (averaged across the two lakes and multiplied between timepoints), the F1 hybrids had the lowest overall survival, suggesting that outbreeding depression because of loss of adaptation to natural conditions may be an important risk to the conservation of native populations exposed to intentional or unintentional interbreeding with non-native populations. If populations must be supplemented with cultured crosses for conservation purposes, attempts should be made to minimize domestication of the cultured population.
Acknowledgments
We thank M. Lõhmus, F. Sundström, N. Hofs, C. Biagi, J. L. Smith, M. Williams, D. Sakhrani and G. Rigter for their assistance with fertilization of the crosses and fieldwork. We also wish to express our gratitude to the staff at the Fraser Valley Trout Hatchery and the Freshwater Fisheries Society, in particular A. Clarke and T. Godin. The authors appreciate the constructive input of four anonymous reviewers that helped to improve the manuscript. Funding for these experiments was provided by the Canadian Regulatory System for Biotechnology to RHD.
Data archiving statement
Data for this study are available at Dryad (DOI: 10.5061/dryad.np208d8b).
Literature cited
- Abrahams MV, Sutterlin A. The foraging and antipredator behaviour of growth-enhanced transgenic Atlantic salmon. Animal Behaviour. 1999;58:933–942. doi: 10.1006/anbe.1999.1229. [DOI] [PubMed] [Google Scholar]
- Ali M, Nicieza AG, Wooton RJ. Compensatory growth in fishes: a response to growth depression. Fish and Fisheries. 2003;4:147–190. [Google Scholar]
- Araki H, Cooper B, Blouin MS. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 2007;318:100–103. doi: 10.1126/science.1145621. [DOI] [PubMed] [Google Scholar]
- Araki H, Cooper B, Blouin MS. Carry-over effect of captive breeding reduces reproductive fitness of wild-born descendants in the wild. Biology Letters. 2009;5:621–624. doi: 10.1098/rsbl.2009.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt JD. Adaptive intrinsic growth rates: an integration across taxa. Quarterly Reviews of Biology. 1997;72:149–177. [Google Scholar]
- Banks MA, Blouin MS, Baldwin BA, Rashbrook VK, Fitzgerald HA, Blankenship SM, Hedgecock D. Isolation and inheritance of novel microsatellites in Chinook salmon (Oncorhynchus tschawytscha. Journal of Heredity. 1999;90:281–288. [Google Scholar]
- Beckmann C, Biro PA, Post JR. Asymmetric effect of bird predation on fish population size-structure in whole-lake experiments. Canadian Journal of Zoology. 2006;84:1584–1593. [Google Scholar]
- Biro PA, Stamps JA. Are animal personality traits linked to life-history productivity? Trends in Ecology & Evolution. 2008;23:361–368. doi: 10.1016/j.tree.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Biro PA, Stamps JA. Do consistent individual differences in metabolic rate promote consistent individual differences in behaviour? Trends in Ecology & Evolution. 2010;25:653–659. doi: 10.1016/j.tree.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Biro PA, Post JR, Parkinson EA. From individuals to populations: prey fish risk-taking mediates mortality in whole-system experiments. Ecology. 2003;84:2419–2431. [Google Scholar]
- Biro PA, Abrahams MV, Post JR, Parkinson EA. Predators select against high growth rates and risk-taking behaviour in domestic trout populations. Proceedings of the Royal Society of London Series B. 2004a;271:2233–2237. doi: 10.1098/rspb.2004.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biro PA, Morton AE, Post JR, Parkinson EA. Over-winter lipid depletion and mortality of age-0 rainbow trout (Oncorhynchus mykiss. Canadian Journal of Fisheries and Aquatic Sciences. 2004b;61:1513–1519. [Google Scholar]
- Biro PA, Abrahams MV, Post JR, Parkinson EA. Behavioural trade-offs between growth and mortality explain the evolution of submaximal growth rates. Journal of Animal Ecology. 2006;75:1165–1171. doi: 10.1111/j.1365-2656.2006.01137.x. [DOI] [PubMed] [Google Scholar]
- Cairney M, Taggart JB, Høyheim B. Characterization of microsatellite and minisatellite loci in Atlantic salmon (Salmo salar L.) and cross-species amplification in other salmonids. Molecular Ecology. 2000;9:2155–2234. [PubMed] [Google Scholar]
- Careau V, Réale D, Humphries MM, Thomas DW. The pace of life under artificial selection: personality, energy expenditure, and longevity are correlated in domestic dogs. American Naturalist. 2010;175:753–758. doi: 10.1086/652435. [DOI] [PubMed] [Google Scholar]
- Carlson SM, Olsen EM, Vøllstad LA. Seasonal mortality and the effect of body size: a review and an empirical test using individual data on brown trout. Functional Ecology. 2008;22:663–673. [Google Scholar]
- Cavalli LL. An analysis of linkage in quantitative inheritance. In: Reeve ECR, Waddington CH, editors. Quantitative Inheritance. London: Her Majesty's Stationery Office; 1952. pp. 135–144. [Google Scholar]
- Devlin RH, D'Andrade M, Uh M, Biagi CA. Population effects of GH transgenic salmon are dependent upon food availability and genotype by environmental interactions. Proceedings of the National Academy of Science. 2004;101:9303–9308. doi: 10.1073/pnas.0400023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Sakhrani D, Tymchuk W, Goh B. Domestication and growth hormone transgenesis cause similar changes in gene expression profiles in coho salmon. Proceedings of the National Academy of Science. 2009;106:3047–3052. doi: 10.1073/pnas.0809798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky TG. Genetics of natural populations IX: temporal changes in the composition of populations of Drosophila pseudoobscura. Genetics. 1943;28:162–186. doi: 10.1093/genetics/28.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson EM, Fagerlund UH, Higgs DA, McBride JR. Hormonal enhancement of growth in fish. In: Hoar WS, Randall JD, Brett JR, editors. Fish Physiology Vol VIII Bioenergetics and Growth. New York: Academic Press; 1979. pp. 455–597. [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Molecular Ecology. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- Elliott JM. Mechanisms responsible for population regulation in young migratory trout, Salmo trutta III. The role of territorial behaviour. Journal of Animal Ecology. 1990;59:803–818. [Google Scholar]
- Emlen JM. The role of time and energy in food preference. American Naturalist. 1966;100:611–617. [Google Scholar]
- Fleming IA, Einum S. Experimental tests of genetic divergence of farmed from wild Atlantic salmon due to domestication. ICES Journal of Marine Science. 1997;54:1051–1063. [Google Scholar]
- Fleming IA, Hindar K, Mjoelneroed IB, Jonsson B, Balstad T, Lamberg A. Lifetime success and interactions of farm salmon invading a native population. Proceedings of the Royal Society of London Series B. 2000;267:1517–1523. doi: 10.1098/rspb.2000.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Genetic adaptation to captivity in species conservation programs. Molecular Ecology. 2008;17:325–333. doi: 10.1111/j.1365-294X.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- Frankham R. Where are we in conservation genetics and where do we need to go? Conservation Genetics. 2010;11:661–663. [Google Scholar]
- Frankham R, Ballou JD, Eldridge MDB, Lacy RC, Ralls K, Dudash MR, Fenster CB. Predicting the probability of outbreeding depression. Conservation Biology. 2011;25:465–475. doi: 10.1111/j.1523-1739.2011.01662.x. [DOI] [PubMed] [Google Scholar]
- Fraser DJ, Cook AM, Eddington JD, Bentzen P, Hutchings JA. Mixed evidence for reduced local adaptation in wild salmon resulting from interbreeding with escaped farmed salmon: complexities in hybrid fitness. Evolutionary Applications. 2008;1:502–512. doi: 10.1111/j.1752-4571.2008.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DJ, Houde ALS, Debes PV, O'Reilly P, Eddington JD, Hutchings JA. Consequences of farmed-wild hybridization across divergent wild populations and multiple traits in salmon. Ecological Applications. 2010;20:935–953. doi: 10.1890/09-0694.1. [DOI] [PubMed] [Google Scholar]
- Hatfield T, Schluter D. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution. 1999;53:866–873. doi: 10.1111/j.1558-5646.1999.tb05380.x. [DOI] [PubMed] [Google Scholar]
- Hayman BI. The separation of epistatic from additive and dominance variation in generation means. Genetica. 1960;31:133–146. doi: 10.1007/BF01984430. [DOI] [PubMed] [Google Scholar]
- Hindar K, Ryman N, Utter F. Genetic effects of cultured fish on natural fish populations. Canadian Journal of Fisheries and Aquatic Sciences. 1991;48:945–957. [Google Scholar]
- Houde ALS, Fraser DJ, Hutchings JA. Reduced anti-predator responses in multi-generational hybrids of farmed and wild Atlantic salmon (Salmo salar L.) Conservation Genetics. 2010;11:785–794. [Google Scholar]
- Johnsson JI, Abrahams MV. Domestication increases foraging under threat of predation in juvenile steelhead trout (Oncorhynchus mykiss): an experimental study. Canadian Journal of Fisheries and Aquatic Sciences. 1991;48:243–247. [Google Scholar]
- Johnsson JI, Björnsson BT. Growth hormone increases growth rate, appetite and dominance in juvenile rainbow trout, Oncorhynchus mykiss. Animal Behavior. 1994;48:177–186. [Google Scholar]
- Johnsson JI, Petersson E, Jönsson E, Björnsson BTh, Järvi T. Domestication and growth hormone alter antipredator behaviour and growth patterns in juvenile brown trout, Salmo trutta. Canadian Journal of Fisheries and Aquatic Sciences. 1996;53:1546–1554. [Google Scholar]
- Johnsson JI, Hojesjo J, Fleming IA. Behavioural and heart rate responses to predation risk in wild and domestic Atlantic salmon. Canadian Journal of Fisheries and Aquatic Sciences. 2001;58:788–794. [Google Scholar]
- Johnston JA, Grise DJ, Donovan LA, Arnold ML. Environment-dependent performance and fitness of Iris brevicaulis, I. fulva (Iridaceae), and hybrids. American Journal of Botany. 1991;88:933–938. [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Leggatt RA, Raven PA, Mommsen TP, Sakhrani D, Higgs D, Devlin RH. Growth hormone transgenesis influences carbohydrate, lipid and protein metabolism capacity for energy production in coho salmon (Oncorhynchus kisutch. Comparative Biochemistry and Physiology. Part B, Biochemistry and Molecular Biology. 2009;154:121–133. doi: 10.1016/j.cbpb.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Lima SL, Dill LM. Behavioral decisions made under the risk of predation: a review and prospectus. Canadian Journal of Zoology. 1990;68:619–640. [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates, Inc; 1998. [Google Scholar]
- MacArthur RH, Pianka ER. On the optimal use of patchy environment. American Naturalist. 1966;100:603–609. [Google Scholar]
- Mangel M, Stamps J. Trade-offs between growth and mortality and the maintenance of individual variation in growth. Evolutionary Ecology Research. 2001;3:583–593. [Google Scholar]
- McClelland EK, Naish KA. What is the fitness outcome of crossing unrelated fish populations? A meta-analysis and an evaluation of future research directions. Conservation Genetics. 2007;8:397–416. [Google Scholar]
- McGinnity P, Prodöhl P, Ferguson A, Hynes R, ó Maoiléidigh N, Baker N, Cotter D, et al. Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proceedings of the Royal Society of London Series B. 2003;270:2443–2450. doi: 10.1098/rspb.2003.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KL, Wyse DL, Shaw RG. Effects of competition on the fitness of wild and crop-wild hybrid sunflower a diversity of wild populations and crop lines. Evolution. 2006;60:2044–2055. [PubMed] [Google Scholar]
- Mercer KL, Andow DA, Wyse DL, Shaw RG. Stress and domestication traits increase the relative fitness of crop-wild hybrids in sunflower. Ecology Letters. 2007;10:383–393. doi: 10.1111/j.1461-0248.2007.01029.x. [DOI] [PubMed] [Google Scholar]
- Morris DB, Richard KR, Wright JM. Microsatellites from rainbow trout (Oncorhynchus mykiss) and their use for genetic study of salmonids. Canadian Journal of Fisheries and Aquatic Sciences. 1996;53:120–126. [Google Scholar]
- Nelson RJ, Beacham TD. Isolation and cross species amplification of microsatellite loci useful for study of Pacific salmon. Animal Genetics. 1999;30:225–244. doi: 10.1046/j.1365-2052.1999.00404-4.x. [DOI] [PubMed] [Google Scholar]
- Partridge L, Sibly R, Beverton RJH, Hill WG. Constraints in the evolution of life histories. Philosophical Transactions of the Royal Society of London Series B. 1991;332:3–13. [Google Scholar]
- Post JR, Parkinson EA, Johnston NT. Density-dependent processes in structured fish populations: assessment of interaction strengths in whole-lake experiments. Ecological Monographs. 1999;69:155–175. [Google Scholar]
- Price EO. Behavioral genetics and the process of animal domestication. In: Grandin T, editor. Genetics and the Behaviour of Domestic Animals. San Diego: Academic Press; 1997. pp. 31–65. [Google Scholar]
- Raven PA, Devlin RH, Higgs DA. Influence of dietary digestible energy content on growth, protein and energy utilization and body composition of growth hormone transgenic and non-transgenic coho salmon (Oncorhynchus kisutch. Aquaculture. 2006;254:730–747. [Google Scholar]
- Rexroad CE, III, Palti Y. Development of ninety-seven polymorphic microsatellite markers for rainbow trout. Transactions of the American Fisheries Society. 2003;132:1214–1221. [Google Scholar]
- Rexroad CE, Coleman RL, Martin AM, Hershberger WK, Killefer J. Thirty-five polymorphic microsatellite markers for rainbow trout (Oncorhynchus mykiss. Animal Genetics. 2001;32:317–319. doi: 10.1046/j.1365-2052.2001.0730b.x. [DOI] [PubMed] [Google Scholar]
- Roberge C, Normandeau E, Einum S, Guderley H, Bernatchez L. Genetic consequences of interbreeding between farmed and wild Atlantic salmon: insights from the transcriptome. Molecular Ecology. 2008;17:314–324. doi: 10.1111/j.1365-294X.2007.03438.x. [DOI] [PubMed] [Google Scholar]
- Sih A. Predators and prey lifestyles: an evolutionary and ecological overview. In: Kerfoot WC, Sih A, editors. Predation: Direct and Indirect Impacts on Aquatic Communities. Hanover, New Hampshire: University of New England Press; 1987. pp. 203–224. [Google Scholar]
- Sundström LF, Lôhmus M, Johnsson JI, Devlin RH. Growth hormone transgenic salmon pay for growth potential with increased predation mortality. Proceedings of the Royal Society of London Series B. 2004;271(Suppl. 5):S350–S352. doi: 10.1098/rsbl.2004.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Coadaptation and outbreeding depression. In: Soulé ME, editor. Conservation Biology: The Science of Scarcity and Diversity. Sunderland, MA: Sinauer; 1986. pp. 105–116. [Google Scholar]
- Theriault V, Moyer GR, Jackson LS, Blouin MS, Banks MA. Reduced reproductive success of hatchery coho salmon in the wild: insights into most likely mechanisms. Molecular Ecology. 2011;20:1860–1869. doi: 10.1111/j.1365-294X.2011.05058.x. [DOI] [PubMed] [Google Scholar]
- Tymchuk WE, Devlin RH. Growth differences among first and second generation hybrids of domestic and wild rainbow trout (Oncorhynchus mykiss. Aquaculture. 2005;245:295–300. [Google Scholar]
- Tymchuk WE, Biagi CA, Withler RE, Devlin RH. Growth and behavioural consequences of introgression of a domesticated aquaculture genotype into a native strain of coho salmon (Oncorhynchus kisutch. Transactions of the American Fisheries Society. 2006;135:442–445. [Google Scholar]
- Tymchuk WE, Sundström LF, Devlin RH. Growth and survival trade-offs and outbreeding depression in rainbow trout (Oncorhynchus mykiss. Evolution. 2007;61:1225–1237. doi: 10.1111/j.1558-5646.2007.00102.x. [DOI] [PubMed] [Google Scholar]
- Tymchuk WE, Sakhrani D, Devlin RH. Domestication causes large-scale effects on gene expression in rainbow trout: analysis of muscle, liver and brain transcriptomes. General and Comparative Endocrinology. 2009;164:175–183. doi: 10.1016/j.ygcen.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Utter F, Epifanio J. Marine aquaculture: genetic potentialities and pitfalls. Reviews in Fish Biology and Fisheries. 2002;12:59–77. [Google Scholar]
- Utter F, Hindar K, Ryman N. Genetic effects of aquaculture and natural salmonid populations. In: Heen K, Monahan RL, Utter F, editors. Salmon Aquaculture. Oxford, UK: Fishing News Books; 1993. pp. 144–165. [Google Scholar]
- Werner EE, Anholt BR. Ecological consequences of the trade-off between growth and mortality rates mediated by foraging activity. American Naturalist. 1993;142:242–272. doi: 10.1086/285537. [DOI] [PubMed] [Google Scholar]
- Werner EE, Gilliam JF. The ontogenetic niche and species interactions in size-structured populations. Annual Reviews of Ecology, Evolution and Systematics. 1984;15:393–425. [Google Scholar]
- Werner EE, Gilliam JF, Hall DJ, Mittelbach GG. An experimental test of the effects of predation risk on habitat use in fish. Ecology. 1983;64:1540–1548. [Google Scholar]
- Woodruff DS. Declines of biomes and biotas and the future of evolution. Proceedings of the National Academy of Sciences. 2001;98:5471–5476. doi: 10.1073/pnas.101093798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data for this study are available at Dryad (DOI: 10.5061/dryad.np208d8b).


