Abstract
Biodiversity is increasingly subjected to human-induced changes of the environment. To persist, populations continually have to adapt to these often stressful changes including pollution and climate change. Genetic erosion in small populations, owing to fragmentation of natural habitats, is expected to obstruct such adaptive responses: (i) genetic drift will cause a decrease in the level of adaptive genetic variation, thereby limiting evolutionary responses; (ii) inbreeding and the concomitant inbreeding depression will reduce individual fitness and, consequently, the tolerance of populations to environmental stress. Importantly, inbreeding generally increases the sensitivity of a population to stress, thereby increasing the amount of inbreeding depression. As adaptation to stress is most often accompanied by increased mortality (cost of selection), the increase in the ‘cost of inbreeding’ under stress is expected to severely hamper evolutionary adaptive processes. Inbreeding thus plays a pivotal role in this process and is expected to limit the probability of genetically eroded populations to successfully adapt to stressful environmental conditions. Consequently, the dynamics of small fragmented populations may differ considerably from large nonfragmented populations. The resilience of fragmented populations to changing and deteriorating environments is expected to be greatly decreased. Alleviating inbreeding depression, therefore, is crucial to ensure population persistence.
Keywords: anthropogenic stress, changing environments, cost of inbreeding, genetic drift, genetic variation, habitat fragmentation, inbreeding depression, population persistence
Background
Anthropogenic stress and stress responses
Biodiversity has become increasingly exposed to human alterations of natural habitats, and abiotic and biotic environments are both changing rapidly, often unpredictably, and species and populations are progressively more subjected to stressful environmental conditions. Industrial pollution and the use of pesticides have shown to affect biodiversity dramatically (Carson 1962; MacNair 1997; Rattner 2009). The emission of greenhouse gases is thought to be responsible for a gradual increase in ambient temperatures worldwide, while locally more extreme and variable temperatures are expected. Consequently, many populations will increasingly experience temperatures that are near to their physiological limits (Chown et al. 2010), leading often to changes in the distributional range of species (Thomas et al. 2004; Parmesan 2006). Such range shifts will result in changes in the complex interactions between species, thereby potentially causing biotic stress on the resident community. Clearly, all these anthropogenic changes of the natural environment will rapidly change selection pressures (Wilkinson 2001; Sgrò et al. 2011) and endanger the persistence of populations.
When faced with new stressful conditions and increased selection pressures, organisms can respond in several ways. If they are not able to adapt, they will either go extinct or they have to avoid the stressful conditions: through changes in local behavior, as has been observed in response to DDT treatment (Roberts and Andre 1994) and temperature stress (Dahlgaard et al. 2001), or by migration to areas that are less stressful. In response to climate change, shifts in the distribution of many species have been documented (Parmesan and Yohe 2003; Thomas et al. 2004; Hitch and Leberg 2006; Parmesan 2006).
Organisms can also adjust to the new and changing conditions, either through phenotypic plasticity or through changes in genetic composition or both. Phenotypic plasticity is the ability of an organism to adjust its phenotype in response to the altered environmental conditions, thereby improving its tolerance to these changes (Schlichting 1986; Pigliucci 2005; but see also Huey et al. 1999), even though it has to be realized that plastic responses to environmental change are not necessarily adaptive (Grether 2005; Ghalambor et al. 2007). Plastic responses can be variable and include behavioral, morphological, physiological, demographic, and life history changes. They are observed regularly and can be costly (Nussey et al. 2007; Leimu et al. 2010). Moreover, plastic responses are often either limited through architectural constraints or restricted in terms of resource allocation (Auld et al. 2010; Chevin et al. 2010; Leimu et al. 2010). Therefore, plastic responses might often provide a more short-term and partly ‘emergency’ solution to cope with the stress, while a longer-term response might require evolutionary adaptation.
Owing to natural selection, allele frequency changes can occur that increase the number of more tolerant individuals in the population, enabling the population to track environmental changes genetically. In the past, pesticide resistance and heavy metal tolerance have been shown to develop rapidly (Bishop and Cook 1981; MacNair 1997). However, not all species or populations do show rapid adaptive genetic responses, most probably because they do not necessarily possess the mutations that underlie resistance (MacNair 1997). The development of resistance is in most cases based on the presence of specific alleles that are already present in a population in low frequency prior to the occurrence of the stress (MacNair 1997; McKenzie and Batterham 1998). More recently, also rapid genetic changes have been reported resulting from climate change (Bradshaw and Holzapfel 2006; Franks et al. 2007; Reusch and Wood 2007; but see Gienapp et al. 2008). Also with respect to adaptation to climate change, evidence exists that evolutionary responses do not always occur because the necessary genetic variation is not present in natural populations (Bradshaw and McNeilly 1991; Kellerman et al. 2006). Realizing that the onset of adaptation relies mostly on the presence of beneficial variants already present in the stressed population and not on the production of new variants by mutation (Orr and Unckless 2008; Teotónio et al. 2009) implies that the evolutionary stress response is positively related to the amount of standing genetic variation (Lynch and Lande 1993; Blows and Hoffmann 2005). Thus, the ability to cope with changing and stressful environmental conditions depends on both how well individuals can phenotypically adjust to the altered conditions and the genetic variation present in the population for evolutionary adaptation.
Habitat fragmentation and genetic erosion
Apart from the mentioned anthropogenic stresses, human interference with nature has other major implications. Large-scale destruction of natural habitats has caused large populations of many species to become fragmented, resulting in small ‘remnant’ populations that become increasingly isolated. Subdivision of large populations in combination with limited gene flow between the fragments has significant ecological and genetic consequences. Ecologically, habitat fragmentation will have demographic effects as small populations are progressively more affected by demographic and environmental stochasticity greatly increasing their extinction probability (Lande 1993; Chevin et al. 2010; Leimu et al. 2010).
From a population genetics perspective, small relatively isolated populations become increasingly subject to genetic drift and inbreeding, resulting in loss of genetic variation and a decrease in fitness, a process here referred to as genetic erosion.
Genetic drift will cause allele frequencies to fluctuate, which over time leads to random loss and fixation of alleles and an increase in homozygosity. When selection coefficients are smaller than 1/2Ne, genetic drift becomes stronger than natural selection, and the variation is driven by the same dynamics as neutral genetic variation independent of whether the alleles have deleterious or beneficial effects on fitness (Kimura 1983:45). On the other hand, deleterious alleles with large fitness effect, such as recessive lethals and detrimentals, will be effectively selected against and removed from the population when becoming homozygous (purging) (Hedrick 1994). The probability of an allele to become fixed through genetic drift equals its initial frequency (Kimura 1983:45). This means that rare alleles have the lowest probability to get fixed and thus the highest probability to get lost. As most stress resistance alleles have generally low frequencies in populations under benign conditions (MacNair 1997), these would be easily lost from small populations, making them less able to adapt genetically when subjected to stresses. Even though low-frequency deleterious alleles also would have a high probability to get lost by chance, still a significant proportion of these will get fixed as many loci carry mildly deleterious alleles: estimates for Drosophila are on the order of 5000 loci (Lande 1995). Because the force of genetic drift increases with decreasing population size, the potential to respond to natural selection will, in general, decrease with decreasing population size, even though this relation in practice will be confounded by selection and dispersal. (Willi et al. 2006).
At the same time, in small isolated populations the inbreeding coefficient, f, increases over time as most parents will share ancestors (biparental inbreeding). The detrimental effects of inbreeding, particularly in normally outbreeding species, are well documented and do increase the extinction probability of populations (Bijlsma et al. 2000; Hedrick and Kalinowski 2000; Frankham et al. 2002; Reed 2005). Inbreeding depression has not only been observed in captive, laboratory and domestic species (Ralls et al. 1988; Frankham et al. 2002; Kristensen and Sørensen 2005), but also evidence for the occurrence of inbreeding depression in wild populations is accumulating (Crnokrak and Roff 1999; Hedrick and Kalinowski 2000; Keller and Waller 2002). Moreover, inbreeding depression has been shown to be often more severe in the wild compared to benign captive conditions (Jiménez et al. 1994; Keller 1998; Crnokrak and Roff 1999; Kristensen et al. 2008).
Although the genetic basis of inbreeding depression is still under discussion, it is currently accepted to be mainly due to increased homozygosity for (partly) recessive, mildly deleterious alleles (Charlesworth and Charlesworth 1987; Charlesworth and Willis 2009). This would also explain why inbreeding depression is significantly greater for traits directly related to fitness (life history traits) than for morphological traits, as the former exhibit more directional dominance (a prerequisite for the occurrence of inbreeding depression) while the latter show mostly additive gene action (DeRose and Roff 1999; Wright et al. 2008).
In short, whereas sufficient tolerance and levels of genetic variation are required for populations to cope with the ongoing deterioration of natural environments, fragmentation of habitats and the concomitant genetic erosion are expected to significantly impede adaptive responses. In the following, we focus on the consequences of genetic drift, inbreeding, and inbreeding depression for adaptive responses and the persistence of biodiversity under stressful conditions.
Stress tolerance and plastic responses
Inbreeding and stress perception
Inbreeding affects most fitness-related traits negatively. However, the magnitude of inbreeding depression generally is found to vary considerable according to species, population, trait, and environmental and ecological conditions (Keller and Waller 2002; Armbuster and Reed 2005; Cheptou and Donohue 2011; Kristensen et al. 2011). Given the rapid anthropogenic changes of natural environments, the environmental dependency of the magnitude of inbreeding depression is of crucial importance. The magnitude of inbreeding depression generally increases under adverse environmental conditions. For instance, for Drosophila, an increase in inbreeding depression was observed for both cold and heat stress under both laboratory and natural conditions (Kristensen et al. 2008; Joubert and Bijlsma 2010). For example, Fig. 1 shows that the viability of inbred lines decreases relatively more at extreme temperatures than that of noninbred populations of Drosophila melanogaster. Such interactions between environment and the magnitude of inbreeding depression have been observed for various taxa, for example insects (Bijlsma et al. 1999, 2000; Dahlgaard and Hoffmann 2000), crustaceans (Haag et al. 2002), plants (Koelewijn 1998; Cheptou et al. 2000), birds (Keller et al. 2002), and mammals (Ross-Gillespie et al. 2007), although there are exceptions (Waller et al. 2008). The meta-analysis by Fox and Reed (2011) shows clearly that the magnitude of inbreeding depression significantly and positively correlates with the stressfulness of the environment. Several mechanisms have been proposed to explain this apparent interaction between inbreeding and environment. Many of these involve specific genotype-by-environment (G×E) interactions, for example increased expression of deleterious mutations or the expression of additional deleterious loci (Bijlsma et al. 1999; Keller and Waller 2002; Cheptou and Donohue 2011; Fox and Reed 2011).
Figure 1.
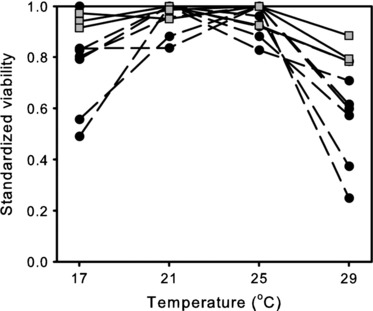
Viability of inbred (black circles, broken lines) and noninbred (gray squares, solid lines) populations of Drosophila melanogaster at four different temperatures. For each population, the viability is scaled for each temperature relative to the highest viability observed for that population. The highest viability was set at 1 for each population (from Joubert and Bijlsma 2010).
We here discuss two possible mechanisms to explain the apparent G×E interactions. Figure 2 shows the mortality in the pupal stage for nine inbred lines (F = 0.6) of D. melanogaster when exposed to 29°C during their whole preadult development. Whereas at 25°C the mortality in the pupal stage for these lines is <10% (data not shown), several inbred lines show increased mortality at 29°C. Two lines, however, show a striking high mortality of 90–100% when exposed to 29°C. As the inbreeding was performed at 25°C where the highly detrimental effect is not expressed, the deleterious allele could easily become fixed under inbreeding at a permissive temperature and will cause immediate extinction when temperatures rise above the threshold (Bakker et al. 2010; Bijlsma et al. 2010). It is only when environmental conditions suddenly change that deleterious effects will become expressed. It is important to realize that if the inbreeding would have been performed under the high-temperature stress conditions, these nearly lethal alleles would have been purged from the populations (and led to increased local adaptation). Such conditional, highly deleterious alleles have been regularly observed in many species. In Drosophila, for example, they have been observed for different life history traits, for example viability (Dobzhansky et al. 1955; Bijlsma et al. 1999), lifespan (Vermeulen and Bijlsma 2004a,b) and male fertility (Pedersen et al. 2011). In fact, pesticide resistance and disease resistance loci carry conditional expressed alleles, and in these cases, the normal nonresistant allele becomes highly deleterious under pesticide or disease stress.
Figure 2.
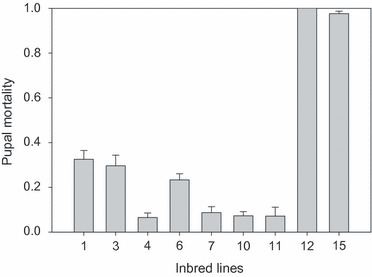
Mortality during the pupal stage (fraction noneclosed pupae) at 29°C for nine independent inbred lines of Drosophila melanogaster. For each inbred line, mean pupal survival (±SE) is based on five replicates started with 100 eggs each (R. Bijlsma, unpublished data).
However, the magnitude of inbreeding depression can also increase with increasing stress levels without assuming G×E interactions, as outlined in Fig. 3 (top). We assume that mean fitness for a given trait is lower for inbred than for outbred individuals and that also the variance among individuals is greater for inbred than for outbred ones. As there is little information about the real fitness distribution, we have assumed normal distributions; however, other fitness distributions would not change the reasoning. We further assume hard selection, that is, individuals need a vigor above a certain threshold in order to survive. If the intensity of selection increases from benign to high stress, the minimum vigor needed to survive increases. Consequently, the fraction of individuals that does not survive increases, particularly so for the inbreds. Under this scenario, it is clear that (i) inbreds show a much lower tolerance to increased stress levels and (ii) the level of inbreeding depression (bottom figure) increases when stress levels increase. This model would also explain the observation that inbreeding depression increases under environmental stress.
Figure 3.
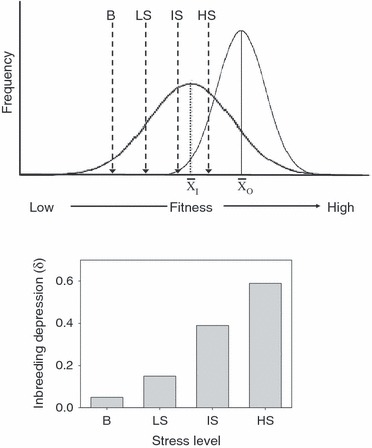
Top: Schematic diagram depicting fitness distributions for inbred individuals (left curve with mean xi) and outbred individuals (right curve with mean xo). The vertical lines represent the threshold values for the hard selection below which individuals do not survive for four different stress levels: benign (B), low stress (LS), intermediate stress (IS) and high stress (HS). Bottom: Amount of inbreeding depression (δ) expected at the four stress levels, B, LS, IS and HS. From the top figure survival rates were estimated to be 0.95, 0.85, 0.60 and 0.35 for the inbred individuals and 1.00, 1.00, 0.99 and 0.85 for the outbred individuals for the four respective stress levels, and these rates were used to calculate the expected level of inbreeding depression as: δ = (survival outbreds − survival inbreds)/(survival outbreds).
Whatever underlying mechanism explains the observations best, the finding that inbreeding increases the sensitivity to stress for many fitness traits seems to be quite general (Armbuster and Reed 2005; Fox and Reed 2011; Cheptou and Donohue 2011; but see Waller et al. 2008). This has important consequences for the persistence of populations. Bijlsma et al. (2000) showed that the extinction probability of small populations significantly increased because of inbreeding and that the probability also greatly increased under stress conditions. More importantly, they also observed that the extinction probability under stress increased with increasing inbreeding coefficient. Figure 4 shows that the same level of stress is experienced differently: the higher the inbreeding level, the greater the stress impact (for details, see Bijlsma et al. 2000). Moreover, the relation was found to be different between ethanol stress (near linear) and high-temperature stress (more exponential), indicating that different genes and different G×E interactions underlie the response to both stresses.
Figure 4.
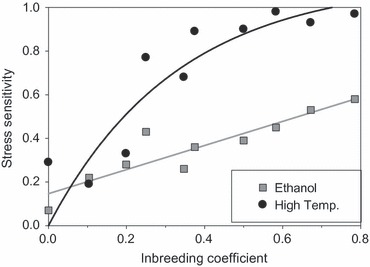
Stress sensitivity of Drosophila melanogaster populations in relation to their inbreeding coefficient for high-temperature stress and ethanol stress. Stress sensitivity is expressed as the decrease in survival probability due to the stress factor corrected for the survival probability observed under benign conditions for the same populations: stress sensitivity = (survival benign − survival stress)/(survival benign) (redrawn after Bijlsma et al. 2000).
All in all, genetic erosion caused by fragmentation decreases individual and population fitness and at the same time increases the sensitivity to stress conditions. The environmental dependency of inbreeding depression emphasizes that human-induced environmental changes, such as climate change, will impact strongly and negatively on fitness. Consequently, species that in recent time have suffered from habitat fragmentation and did become inbred could be much more vulnerable to human-induced environmental changes than species that still exist in large populations.
Inbreeding and plasticity
Generally, genetically eroded populations will have decreased levels of genetic variability and lower evolutionary potential (see next section). Consequently, their persistence might to a larger extent be dependent on the capability of the organism to respond to environmental challenges by phenotypic plasticity that can augment the evolutionary potential of a population (Bradshaw 1965; Pigliucci 2005). Thus, the presence of plastic responses may significantly affect the persistence of populations in a changing world (Pertoldi et al. 2007; Auld and Relyea 2010; Chevin et al. 2010; Beldade et al. 2011).
As phenotypic plasticity has a genetic basis and genetic variation for plasticity is generally observed (Pigliucci 2005), genetic erosion might also hamper plastic responses. Moreover, plastic responses can be costly (Auld et al. 2010; Leimu et al. 2010). Inbreeding has been observed to increase the amount of energy needed for maintenance significantly, leaving less energy to be available for allocation to other processes, such as plasticity (Ketola and Kotiaho 2009). This possibly could explain the results of Auld and Relyea (2010), as the inducible response to predators, increased shell thickness, in the freshwater snail Physa acuta might be costly. Lower energy levels because of inbreeding, therefore, might hamper this plastic response. Several studies on plant species have indicated that individuals from small populations showed lower plastic responses than individuals from large populations (Fischer et al. 2000; Paschke et al. 2003; Pluess and Stöcklin 2004). This was, in general, correlated with decreased genetic variability in the small populations, suggesting that genetic erosion does disrupt plastic responses. On the other hand, studies that examined directly the effect of inbreeding on plasticity showed mixed results: whereas Maynard Smith et al. (1955), Schiegg et al. (2002) and Auld and Relyea (2010) found a significant decrease in adaptive plasticity upon inbreeding, other studies found little effect (Schlichting and Levin 1986; Kristensen et al. 2011; Luquet et al. 2011). Given the importance of phenotypic plasticity for many small populations of conservation concern (Pertoldi et al. 2007), further research on these issues is clearly needed. If phenotypic plasticity is hampered by inbreeding, this would render genetically eroded populations even more at risk in changing environments.
Genetic responses
Population size and levels of genetic variation
Genetic diversity is a prerequisite for adaptive evolution. Only when the rate of evolution at least matches the rate of continuous environmental change, populations may be able to persist (Lynch and Lande 1993; Bürger and Lynch 1995). For abrupt environmental change, the situation might be more complex as, in addition to the evolutionary processes, demographic processes increase in importance (Gomulkiewicz and Holt 1995; Bell and Gonzalez 2009, 2011). Genetic drift is expected to decrease genetic diversity in small populations at a rate proportional to the population size (Wright 1931). This is well supported by the rate of loss observed for neutral variation both in experimental and natural populations (Frankham et al. 2002; Johnson et al. 2004; DiBattista 2008; Hoeck et al. 2010). Fragmentation causes the variability present in a once-undivided large population to become redistributed from within populations to among (sub)populations (Wright 1931, 1951). This is also true for quantitative genetic variation for which the additive genetic variation (Va) also decreases with decreasing population size. When strictly additive, the variance decreases at the same rate as neutral variation with a factor 1 − f per generation, where f is the inbreeding coefficient (see Willi et al. 2006). Several studies indicated that bottlenecks can inflate Va in the short term (Bryant et al. 1986; Van Buskirk and Willi 2006; and references therein). However, the importance of the phenomenon of increased Va after a bottleneck is thought to be questionable for several reasons (Barton and Turelli 2004; Van Buskirk and Willi 2006).
If genetic variation is compromised by the genetic erosion process, there are two ways this can be counteracted. First, the variation can be replenished by gene flow between population fragments. If gene flow is sufficient, it can restore the within-population variation to normal levels. To achieve this, the number of migrants per generation (Nm) should be substantial (Keller and Waller 2002). In addition, gene flow might also promote the spread of new beneficial mutations (Bell and Gonzalez 2011). However, high levels of gene flow might disrupt patterns of local adaptation, thereby endangering population persistence (Lenormand 2002; Garant et al. 2007; Bridle et al. 2010). Second, the rate of loss at quantitative loci can be compensated by new mutations. However, this requires quite large population sizes, and minimal effective sizes needed are estimated to range from 500 (Frankham et al. 2002) to low thousands (Willi et al. 2006) up to 5000 (Lande 1995). As effective population sizes are generally much smaller than the census size, this will require even higher census sizes, up to one order of magnitude (Frankham 1995). However, traits governed by a single gene or a few genes, for example heavy metal and disease resistance, lose genetic variation much faster than quantitative traits, with the rate of change being proportional to the number of underlying loci (Malcom 2011 and references therein), and the frequency of beneficial mutations is generally much lower. Given the small population sizes of fragmented populations and the low mutation rates, new mutations will rarely play an important role and populations have to rely on the standing genetic variation to adapt (Lynch and Lande 1993; Blows and Hoffmann 2005; Orr and Unckless 2008; Teotónio et al. 2009).
Population size and adaptability
If levels of adaptive variation decrease with decreasing population size and the potential to respond to selection depends on the standing level of genetic variation, small populations that have been subject to genetic erosion are expected to show reduced adaptive potential. Several authors have addressed the consequences of bottlenecks and inbreeding for the selection response of quantitative traits. For many traits, a decrease in genetic variance was observed, consistent with the expectations for additive variation (Wade et al. 1996; Whitlock and Fowler 1999; Sacheri et al. 2001; Day et al. 2003; Kristensen et al. 2005; Swindell and Bouzat 2005). Swindell and Bouzat (2005) investigated the selection response of sternopleural bristles in D. melanogaster at regular intervals during consecutive generation of inbreeding. They showed that the response continuously declined over the generations, concluding that the longer populations have been subject to genetic erosion, the lower their adaptive potential. There are also indications that populations that are slowly inbred retain a higher evolutionary potential than rapidly inbred populations despite the same level of inbreeding, probably because balancing selection does retard the loss of genetic variation (Day et al. 2003; Kristensen et al. 2005). This might be of importance when assessing the tolerance to inbreeding and the evolutionary potential of fragmented populations.
Bakker et al. (2010) used an experimental approach to investigate the adaptive potential of fragmented populations of D. melanogaster. They compared the adaptive potential of populations that had been subdivided (six small subpopulations with on average 50 individuals each) with undivided populations of nearly the same total size as the divided populations (on average 220 individuals). For the divided population, each generation between 0.5 and 1.3 individuals was exchanged between the subpopulations, mimicking natural metapopulations (see Bakker et al. 2010 for details). All these populations were maintained for 40 generations (reaching an inbreeding coefficient of around 0.25 for the fragmented populations) whereafter their adaptive response to three stresses (temperature, ethanol, and salt stress) was tested. Figure 5 shows the adaptive response after six generations of selection in each of the stress environments. It shows that the adaptive response was larger for the undivided populations than for the divided populations, even though there was large variation both among subpopulations within metapopulations and among populations. This study demonstrated that the history of fragmentation does impede adaptive responses. Frankham et al. (1999) observed that inbred populations of D. melanogaster also showed a lower adaptive response to salt stress than noninbred population resulting in higher extinction rates of inbred populations toward this stress.
Figure 5.
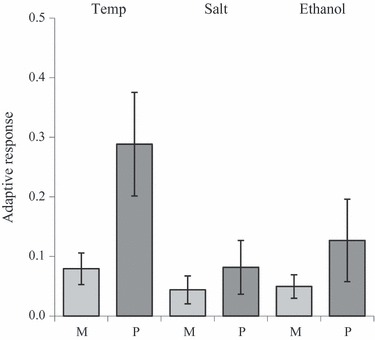
Mean adaptive response (±SE) after six generations of adaptation at three stress environments, temperature stress (Temp), salt stress (Salt) and ethanol stress (Ethanol) for fragmented (M) and nonfragmented (P) populations of Drosophila melanogaster. The adaptive response for each population was calculated as the difference in viability of adapted flies minus the viability of nonadapted flies for each population at each stress (from Bakker et al. 2010).
Discussion
Inbreeding affects evolutionary responses
We explored the consequences of small population size and the concomitant process of genetic erosion of populations to address two fundamental questions: (i) How do the effects of genetic drift and inbreeding affect the stress perception of populations? (ii) To what extent does genetic erosion impede evolutionary adaptation to stressful environments of such populations? These questions are important as many fragmented populations currently suffer both from genetic erosion and changing and deteriorating environmental conditions (e.g. climate change, chemical pollution), which endanger their persistence.
The evidence presented here shows that generally the fitness reduction because of inbreeding increases significantly under stress and that this effect becomes amplified as inbreeding coefficients increase (Bijlsma et al. 2000; Armbuster and Reed 2005; Fox and Reed 2011). This phenomenon causes populations to be much more sensitive to environmental stress, making it still harder to cope with the rapid deteriorating environmental conditions. This increases the extinction risk of inbred populations greatly (Bijlsma et al. 2000). The increased sensitivity to environmental stress of genetically eroded populations signifies that effects of human-induced stress cannot properly be evaluated without taking this phenomenon into account. Given its importance, further investigations are called for to understand the causation and consequences of the synergistic interaction between inbreeding and stress.
Small populations are also subject to loss of genetic variation due to genetic drift. The available data show that this, in general, also holds for adaptive variation. The decrease in standing genetic variation in small populations could potentially decrease evolutionary responses. This is well supported by the experimental evidence for traits that are not or only marginally related to fitness for which genetically eroded populations showed significantly reduced selection responses. There is also increasing evidence that this may also hold for fitness-related traits (Frankham et al. 1999; Willi and Hoffmann 2009; Bakker et al. 2010). This indicates that fragmentation and the accompanying genetic erosion will limit the evolutionary responses to stressful environmental conditions.
However, the effects of inbreeding depression and loss of adaptive variation are not independent. When a population is subjected to a novel environmental stress, the selection intensity will increase and the growth rate of the population will decrease and will often become negative initially leading to a decrease in population size that even may reach a critical low level because of selective deaths, the cost of selection (Haldane 1957; Gomulkiewicz and Holt 1995; Orr and Unckless 2008; Bell and Gonzalez 2009 and references therein). Only after adapted individuals have reached a sufficient high frequency in the stressed population, this trend will be reversed and the population will show a positive growth rate again. As such, persistence/extinction can be regarded as a race between adaptation and demographic decay (Maynard Smith 1989; Bell and Gonzalez 2009). In the presence of increased inbreeding depression under stress, the extra reduction in individual fitness is expected to reduce population numbers and growth rates much further, making adaptive recovery a great deal more difficult. Thus, the increased cost of inbreeding under stress coupled with the cost of adaptation is expected to limit adaptation and severely increase the extinction probability of small populations. In this process, inbreeding plays a pivotal role. The interaction between inbreeding depression and reduced levels of genetic variation will critically limit evolutionary responses. One has to realize that the dynamics of the adaptive responses may differ considerably depending on whether the environmental changes occur gradually or abruptly.
Chevin et al. (2010) recently published a stimulating paper in which they expanded the evolutionary model by Lynch and Lande (1993) by including phenotypic plasticity. What they did not (yet) include were, among others, genetic drift and inbreeding. As inbreeding increases the sensitivity to stress and decreases fitness, it is expected to increase the environmental sensitivity to selection considerably. Moreover, inbreeding can significantly impair plastic responses (Auld and Relyea 2010). Based on the model of Chevin et al. (2010), it is expected that the joint effects of inbreeding would be that the critical maximum rate of environmental change that allows long-term persistence would become decreased. This effect will even be strengthened if inbreeding makes plasticity more costly. We advocate that future modeling of the persistence of biodiversity in changing environments preferably should include the effects of genetic erosion.
Some general remarks
Our inferences are restricted to species that are liable to inbreeding depression. Clearly not all species will suffer in the same way: normally selfing species will show little loss in fitness upon inbreeding, while species that normally outcross will show much higher levels of inbreeding depression. We have focused solely on the effects of genetic erosion and have not discussed nongenetic factors, nor did we discuss possible positive effects of fragmentation and geographic isolation in the context of the buildup of local races and possible speciation events (Howard 1993). These issues go beyond the framework of this paper.
The evidence presented here mostly comes from laboratory experiments, and the situation undoubtedly will be more complex in nature. However, our findings suggest that inbreeding depression is a key factor in the adaptive process, and the magnitude of inbreeding depression generally increases in the wild. Therefore, we are confident that inbreeding depression also plays a pivotal role in the adaptive process in nature. Moreover, population sizes in nature fluctuate considerably in time. This will on average deflate the effective population size (Ne) and strengthen the effects of genetic drift.
Future prospects and practical approaches
Since the 1980s, conservation genetics has recognized the need to avert the negative effects of genetic erosion as it does increase the extinction probability of species (Frankham et al. 2002). Here, we showed that genetic erosion, and particularly inbreeding depression, will also significantly impair the adaptive potential to (future) stressful challenges, like climate change and pollution. This leads to two questions that we will shortly address: (i) How can we identify populations that are threatened by genetic erosion? (ii) How can we alleviate the negative effects of genetic erosion and decrease extinction probabilities?
The first question implies that we need methods that reveal when populations are genetically eroded and suffer from inbreeding depression. Although individual inbreeding can be detected in nature using pedigrees, estimates of the inbreeding coefficients and the related fitness effects are difficult to determine at the population level. In many investigations, Wright's fixation index FIS is used as a measure of the level of inbreeding. However, this is incorrect as in small randomly mating populations, FIS will be always zero, even though biparental inbreeding increases the inbreeding coefficient (f) continuously (Keller and Waller 2002; Biebach and Keller 2010). In this situation, population-specific FST can be used to infer the level of inbreeding and offers a convenient way to estimate the average level of inbreeding in a population (Vitalis et al. 2001; Biebach and Keller 2010; and references therein). Both the rate of loss of variation and the rate of inbreeding critically depend on the genetically effective population size (Ne). Neutral molecular markers, such of microsatellite loci, are currently the markers of choice to study the genetics of populations and allow estimation of important parameters that contribute to loss of genetic variation and the rate of inbreeding. In combination with constantly improving estimation procedures, it is now feasible to estimate these parameters, like effective population size, inbreeding coefficient and migration rate, with increasing accuracy. This makes it possible to evaluate the genetic risks of the population under investigation and to device effective management decisions (Palstra and Ruzzante 2008; Biebach and Keller 2010; Luikart et al. 2010).
Comparing the performance of small versus large populations for several fitness traits, known to be affected by inbreeding, does facilitate the detection of populations that suffer from inbreeding depression. This approach has been successfully applied to several species, leading to management measures to alleviate the genetic problems (Westemeier et al. 1998; Madsen et al. 1999; Vilà et al. 2003; Hedrick and Fredrickson 2010). However, the performance of a population also depends on ecological and environmental factors, which may confound the genetic effects. Recent developments in landscape genetics may help to entangle the different factors contributing to the endangerment of populations (Segelbacher et al. 2010).
Although neutral molecular markers provide crucial information about the dynamics of neutral genetic variation and the level of inbreeding, we are also highly interested in the dynamics and loss of adaptive variation. Unfortunately, there is only a weak correlation between molecular genetic diversity and quantitative genetic variation (Hedrick 2001; Gilligan et al. 2005), and thus the levels of neutral variation are not predictive for the levels of adaptive variation. Also, heterozygosity at these markers shows little correlation with the inbreeding coefficient (Pemberton 2004). However, the rapid advances in genomic techniques promise to solve these problems. In the near future, molecular tools will allow us to obtain the complete sequence of any species. It is expected that this will enable us to successfully address hot topics like the genetic basis of inbreeding depression, the structure and amount of adaptive variation, the level of local adaptation and the causes of G×E interactions (Allendorf et al. 2010; Kristensen et al. 2010; Ouborg et al. 2010). This would greatly advance our understanding of genetic processes in small populations and the dynamics of biodiversity in a changing world.
The second question, in essence, involves (i) alleviating inbreeding depression and restoring adaptive genetic variation and (ii) increasing population sizes to a level at which genetic erosion will be minimal. These issues have been regularly discussed in the field of conservation genetics. We will discuss these issues here briefly, while further ideas can be found elsewhere (e.g. Frankham et al. 2002; Sgrò et al. 2011). Table 1 presents the outline of a three-step program of interventions that will alleviate the problems caused by genetic erosion. Improving gene flow and connectivity between the population fragments that previously formed a single large population is the first step. It is now well documented that influx of migrants in genetically eroded populations rapidly decreases inbreeding depression, increases genetic diversity and positively affects population growth (Westemeier et al. 1998; Madsen et al. 1999; Vilà et al. 2003; Hedrick and Fredrickson 2010). This process of genetic rescue will be only successful in the long term if gene flow levels stay high as otherwise genetic erosion will arise again (Liberg et al. 2005; Biebach and Keller 2010). Genetic rescue often raises concerns about outbreeding depression (Edmands 2007). Unless the environment has changed drastically since a population became fragmented and the population fragments have become locally adapted, outbreeding depression is not expected to be a problem at this scale as increased connectivity only restores the previous situation. This step is expected to decrease both the genetic and demographic risks of populations.
Table 1.
Management measures to improve the genetic constitution of fragmented and genetically eroded populations in three steps
| Measure | Expected result |
|---|---|
| 1. Increase gene flow between fragments and/or increase connectivity | As populations in habitat fragments are expected to be fixed for different (mildly) deleterious alleles, this measure will immediately decrease inbreeding depression levels; it will increase genetic variation levels; local adaptation in general will not be a problem as it restores former undivided conditions and establishes a metapopulation with sufficient gene flow levels. |
| 2. Increase habitat and/or population size | This measure will decrease the impact of genetic drift and inbreeding; it will buffer the genetic erosion of populations that will occur in the future; it will mitigate the cost of selection upon adaptation. |
| 3. Facilitate genetic exchange with more distant populations and populations from different habitats | This measure will mitigate inbreeding depression even more; it will boost the level of genetic diversity; it will supplement adaptive genetic variants not yet present in the population; this will facilitate evolutionary responses in the future. Dangers: this measure may disrupt local adaptation and cause outbreeding depression; if the total population size is large enough, recombination and selection may nullify this loss of fitness over the generations. |
Even so promoting gene flow will cancel the negative effects of genetic erosion, the total (meta)population size often is still limited, and genetic erosion might arise again. Therefore, to ensure long-term viability, in a second phase of the rescue process, it would be advisable to increase population size to a level at which genetic erosion is expected to be minimized. This could be achieved by increasing the habitat size or by improving the quality of the habitat (e.g., by removing edge effects) so it can sustain larger numbers of individuals. This action would decrease genetic risks even further and, in addition, make the population more resistant against environmental stochasticity (Lande 1993).
If required, gene flow between more distant and (slightly) different habitats could be promoted as a third step. This step will increase genetic diversity even further by an influx of genetic variants that may not have been present in the target population. Such an intervention is expected to facilitate adaptive response in the (near) future. For instance, importing immigrants from the warmer regions (e.g., lower latitudes) of a species distribution into the cooler regions may improve the ability of the receiving population to cope with ongoing climate change. A drawback of this action is that the immigrants might be maladapted and disrupt local adaptation, thereby causing outbreeding depression (Edmands 2007; Frankham et al. 2011). However, Frankham et al. (2011) showed that when a careful decision procedure is followed, the probability of outbreeding depression to occur might be considerably lower than generally anticipated. Moreover, after a hybridization event, recombination and natural selection can overcome the initial decrease in fitness and improve fitness to levels higher than before hybridization within a few generations (Edmands et al. 2005; Erickson and Fenster 2006). This suggests that outbreeding depression might only be a temporal problem. If the receiving population is healthy and sufficiently large, it is expected to be able to cope with this short-term problem. Nevertheless, great care should be taken before implementing this third step.
Conclusion
Fragmentation of habitats leads to small isolated populations that become subject to genetic erosion. Such populations of normally outcrossing species will usually show decreased levels of genetic variation and a decrease in fitness because of inbreeding depression. More importantly, the magnitude of inbreeding depression generally increases considerably under stressful environmental conditions, like extreme temperatures. This makes inbred populations more vulnerable to stressful environments. At the same time, loss of genetic variation has been found to decrease the selective response of genetically eroded populations. The combined action of the decrease in tolerance because of inbreeding and loss of adaptive potential clearly impede adaptive responses and significantly increase extinction risks under stressful environmental conditions. We argue that fragmented populations are much more vulnerable to changes in environmental conditions than large nonfragmented populations. Models developed to predict the persistence of biodiversity under changing and deteriorating conditions, like climate change, should, therefore, include the negative effects of genetic erosion.
Acknowledgments
We thank Torsten N. Kristensen, and three anonymous reviewers for critical comments on the manuscript. We thank Joke Bakker, Désiree Joubert, and Marielle van Rijswijk for use of their original data. This study was in part supported by the Netherlands Organisation for Scientific Research (NWO) by grants from the sections Earth and Life Sciences (ALW, grant 805.33.362P) and Science for Global Development (WOTRO, CPG-NWO-895.100.013). RB thanks the section of Ecology and Genetics of the Department of Biosciences at Aarhus University for their hospitality and the Aarhus Stress Network for financial support while writing this manuscript.
Literature cited
- Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nature Reviews Genetics. 2010;11:697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- Armbuster P, Reed DH. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. [DOI] [PubMed] [Google Scholar]
- Auld JR, Relyea RA. Inbreeding depression in adaptive plasticity under predation risk in a freshwater snail. Biology Letters. 2010;6:222–224. doi: 10.1098/rsbl.2009.0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld JR, Agrawal AA, Relyea RA. Re-evaluating the costs and limits of phenotypic plasticity. Proceedings of the Royal Society London B. 2010;227:503–511. doi: 10.1098/rspb.2009.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker J, van Rijswijk MEC, Weissing FJ, Bijlsma R. Consequences of fragmentation for the ability to adapt to novel environments in experimental Drosophila metapopulations. Conservation Genetics. 2010;11:449–462. [Google Scholar]
- Barton NH, Turelli M. Effects of genetic drift on variance components under a general model of epistasis. Evolution. 2004;28:2111–2132. doi: 10.1111/j.0014-3820.2004.tb01591.x. [DOI] [PubMed] [Google Scholar]
- Beldade P, Mateus AA, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Molecular Ecology. 2011;20:1347–1363. doi: 10.1111/j.1365-294X.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- Bell G, Gonzalez A. Evolutionary rescue can prevent extinction following environmental change. Ecology Letters. 2009;12:942–948. doi: 10.1111/j.1461-0248.2009.01350.x. [DOI] [PubMed] [Google Scholar]
- Bell G, Gonzalez A. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science. 2011;332:1327–1330. doi: 10.1126/science.1203105. [DOI] [PubMed] [Google Scholar]
- Biebach I, Keller LF. Inbreeding in reintroduced populations: the effects of early reintroduction history and contemporary processes. Conservation Genetics. 2010;11:527–538. [Google Scholar]
- Bijlsma R, Bundgaard J, van Putten WF. Environmental dependence of inbreeding depression and purging in Drosophila melanogaster. Journal of Evolutionary Biology. 1999;12:1125–1137. [Google Scholar]
- Bijlsma R, Bundgaard J, Boerema AC. Does inbreeding affect the extinction risk of small populations?: predictions from Drosophila. Journal of Evolutionary Biology. 2000;13:502–514. [Google Scholar]
- Bijlsma R, Westerhof MDD, Roekx LP, Pen I. Dynamics of genetic rescue in inbred Drosophila melanogaster populations. Conservation Genetics. 2010;11:435–448. [Google Scholar]
- Bishop J, Cook L. Genetic Consequences of Man-Made Change. London: Academic Press; 1981. [Google Scholar]
- Blows MW, Hoffmann AA. A reassessment of genetic limits to evolutionary change. Ecology. 2005;86:1371–1384. [Google Scholar]
- Bradshaw AD. Evolutionary significance of phenotypic plasticity in plants. Advances in Genetics. 1965;13:115–155. [Google Scholar]
- Bradshaw WE, Holzapfel CM. Climate change - Evolutionary response to rapid climate change. Science. 2006;312:1477–1478. doi: 10.1126/science.1127000. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD, McNeilly T. Evolutionary response to global climate change. Annals of Botany. 1991;67(Suppl):5–14. [Google Scholar]
- Bridle JR, Polechova J, Kawata M, Butler RK. Why is adaptation prevented at ecological margins? New insights from individual-based simulations. Ecology Letters. 2010;13:485–494. doi: 10.1111/j.1461-0248.2010.01442.x. [DOI] [PubMed] [Google Scholar]
- Bryant EH, McCommas SA, Combs LM. The effect of an experimental bottleneck upon quantitative genetic variance in the housefly. Genetics. 1986;114:1191–1211. doi: 10.1093/genetics/114.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R, Lynch M. Evolution and extinction in a changing environment: a quantitative genetic analysis. Evolution. 1995;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Carson RL. Silent Spring. Boston: Houghton Mifflin; 1962. [Google Scholar]
- Charlesworth B, Charlesworth D. Inbreeding depression and its evolutionary consequences. Annual Review of Ecology and Systematics. 1987;18:237–268. [Google Scholar]
- Charlesworth D, Willis JH. The genetics of inbreeding depression. Nature Reviews Genetics. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- Cheptou P-O, Donohue K. Environment-depend inbreeding depression: its ecological and evolutionary significance. New Phytologyst. 2011;189:395–407. doi: 10.1111/j.1469-8137.2010.03541.x. [DOI] [PubMed] [Google Scholar]
- Cheptou P-O, Imbert E, Lepart J, Escarre J. Effects of competition on lifetime estimates of inbreeding depression in the outcrossing plant Crepis sancta (Asteraceae) Journal of Evolutionary Biology. 2000;13:522–531. [Google Scholar]
- Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. Plos Biology. 2010;8:e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL, Hoffmann AA, Kristensen TN, Angilletta MJ, Jr, Stenseth NC, Pertoldi C. Adapting to climate change: a perspective from evolutionary physiology. Climate Research. 2010;43:3–15. [Google Scholar]
- Crnokrak P, Roff DA. Inbreeding depression in the wild. Heredity. 1999;83:260–270. doi: 10.1038/sj.hdy.6885530. [DOI] [PubMed] [Google Scholar]
- Dahlgaard J, Hoffmann AA. Stress resistance and environmental dependency of inbreeding depression in Drosophila melanogaster. Conservation Biology. 2000;14:1187–1192. [Google Scholar]
- Dahlgaard J, Hasson E, Loeschcke V. Behavioral differentiation in oviposition activity in Drosophila buzzatii from highland and lowland populations in Argentina: plasticity or thermal adaptation? Evolution. 2001;55:738–747. doi: 10.1554/0014-3820(2001)055[0738:bdioai]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Day SB, Bryant EH, Meffert LM. The influence of variable rates of inbreeding on fitness, environmental responsiveness, and evolutionary potential. Evolution. 2003;57:1314–1324. doi: 10.1111/j.0014-3820.2003.tb00339.x. [DOI] [PubMed] [Google Scholar]
- DeRose MA, Roff DA. A comparison of inbreeding depression in life-history and morphological traits in animals. Evolution. 1999;53:1288–1929. doi: 10.1111/j.1558-5646.1999.tb04541.x. [DOI] [PubMed] [Google Scholar]
- DiBattista JD. Patterns of genetic variation in anthropogenically impacted populations. Conservation Genetics. 2008;9:141–156. [Google Scholar]
- Dobzhansky Th, Pavlovsky O, Spassky B, Spassky N. Genetics of natural populations. XXIII. Biological role of deleterious recessives in populations of Drosophila. Genetics. 1955;40:781–796. doi: 10.1093/genetics/40.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Molecular Ecology. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- Edmands S, Feaman HV, Harrison JS, Timmerman CC. Genetic consequences of many generations of hybridization between divergent copepod populations. Journal of Heredity. 2005;96:114–123. doi: 10.1093/jhered/esi014. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Fenster CB. Intraspecific hybridization and the recovery of fitness in the native legume Chamaecrista fasiculata. Evolution. 2006;60:225–233. [PubMed] [Google Scholar]
- Fischer M, van Kleunen M, Schmid B. Genetic Allee effects on performance, plasticity and developmental stability in a clonal plant. Ecology Letters. 2000;3:530–539. [Google Scholar]
- Fox CW, Reed DH. Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution. 2011;65:246–258. doi: 10.1111/j.1558-5646.2010.01108.x. [DOI] [PubMed] [Google Scholar]
- Frankham R. Effective population size – adult population size ratio's in the wild. Genetical Research. 1995;66:95–107. [Google Scholar]
- Frankham R, Lees K, Mongomery ME, England PR, Lowe EH, Brisco DA. Do population size bottlenecks reduce evolutionary potential? Animal Conservation. 1999;2:255–260. [Google Scholar]
- Frankham R, Ballou JD, Brisco DA. Introduction to Conservation Genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Frankham R, Ballou JD, Eldridge MBD, Lacy RC, Ralls K, Dudash MR, Fenster CB. Predicting the probability of outbreeding depression. Conservation Biology. 2011;25:465–475. doi: 10.1111/j.1523-1739.2011.01662.x. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to climate fluctuations. Proceedings of the National Academy of Sciences of the United states of America. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant D, Forde SE, Hendry AP. The multifarious effects of dispersal and gene flow on contemporary adaptation. Functional Ecology. 2007;21:434–443. [Google Scholar]
- Ghalambor CK, McKay JK, Carrol SP, Reznick DN. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptations in new environments. Functional Ecology. 2007;21:394–407. [Google Scholar]
- Gienapp P, Teplitsky C, Alho JS, Mills JA, Merilä J. Climate change and Evolution: disentangling environmental and genetic responses. Molecular Ecology. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- Gilligan DM, Briscoe DA, Frankham R. Comparative losses of quantitative and molecular variation in finite populations of Drosophila melanogaster. Genetical Research. 2005;85:47–55. doi: 10.1017/s0016672305007342. [DOI] [PubMed] [Google Scholar]
- Gomulkiewicz R, Holt RD. When does evolution by natural selection prevent extinction? Evolution. 1995;49:201–207. doi: 10.1111/j.1558-5646.1995.tb05971.x. [DOI] [PubMed] [Google Scholar]
- Grether GF. Environmental change, phenotypic plasticity, and genetic compensation. American Naturalist. 2005;166:E115–E123. doi: 10.1086/432023. [DOI] [PubMed] [Google Scholar]
- Haag CR, Hottinger JW, Riek M, Ebert D. Strong inbreeding depression in a Daphnia metapopulation. Evolution. 2002;56:518–526. [PubMed] [Google Scholar]
- Haldane JBS. The cost of natural selection. Journal of Genetics. 1957;55:511–524. [Google Scholar]
- Hedrick PW. Purging inbreeding depression and the probability of extinction – full-sib mating. Heredity. 1994;73:363–372. doi: 10.1038/hdy.1994.183. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Conservation genetics: where are we now? Trends in Ecology and Evolution. 2001;16:629–636. [Google Scholar]
- Hedrick PW, Fredrickson R. Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conservation Genetics. 2010;11:615–625. [Google Scholar]
- Hedrick PW, Kalinowski ST. Inbreeding depression in conservation biology. Annual Review of Ecology and Systematics. 2000;31:139–162. [Google Scholar]
- Hitch AT, Leberg PL. Breeding distributions of North American bird species moving north as a result of climate change. Conservation Biology. 2006;21:534–539. doi: 10.1111/j.1523-1739.2006.00609.x. [DOI] [PubMed] [Google Scholar]
- Hoeck PA, Bollmer JL, Parker PG, Keller LF. Differentiation with drift: a spatio-temporal genetic analysis of Galápagos mocking bird populations (Mimus ssp.) Philosophical Transaction of the Royal Society B. 2010;365:1127–1138. doi: 10.1098/rstb.2009.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DJ. Small populations, inbreeding, and speciation. In: Thornhill NW, editor. The Natural History of Inbreeding and Outbreeding. Chicago: University of Chicago Press; 1993. pp. 118–142. [Google Scholar]
- Huey RB, Berrigan D, Gilchrist GW, Herron JC. Testing the adaptive significance of acclimation: a strong inference approach. American Zoologist. 1999;39:323–336. [Google Scholar]
- Jiménez JA, Hughes KA, Alaks G, Graham L, Lacy RC. An experimental study of inbreeding depression in a natural habitat. Science. 1994;266:271–273. doi: 10.1126/science.7939661. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Bellinger MR, Toepfer JE, Dunn P. Temporal changes in allele frequencies and low effective population size in greater prairie-chickens. Molecular Ecology. 2004;13:2617–2630. doi: 10.1111/j.1365-294X.2004.02264.x. [DOI] [PubMed] [Google Scholar]
- Joubert D, Bijlsma R. Interplay between habitat fragmentation and climate change: inbreeding affects the response to thermal stress in Drosophila melanogaster. Climate Research. 2010;43:57–70. [Google Scholar]
- Keller LF. Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia. Evolution. 1998;52:240–250. doi: 10.1111/j.1558-5646.1998.tb05157.x. [DOI] [PubMed] [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology and Evolution. 2002;17:230–241. [Google Scholar]
- Keller LF, Grant PR, Grant BR, Petren K. Environmental conditions affect the magnitude of inbreeding depression in survival of Darwin's finches. Evolution. 2002;55:937–942. doi: 10.1111/j.0014-3820.2002.tb01434.x. [DOI] [PubMed] [Google Scholar]
- Kellerman VK, van Heerwaarden B, Hoffmann AA, Sgró C. Very low additive genetic variance and evolutionary potential in multiple populations of two rainforest Drosophila species. Evolution. 2006;60:1104–1108. doi: 10.1554/05-710.1. [DOI] [PubMed] [Google Scholar]
- Ketola T, Kotiaho JS. Inbreeding, energy use and condition. Journal of Evolutionary Biology. 2009;22:770–781. doi: 10.1111/j.1420-9101.2009.01689.x. [DOI] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Koelewijn HP. Effects of different levels of inbreeding on progeny fitness in Plantago coronopus. Evolution. 1998;52:692–702. doi: 10.1111/j.1558-5646.1998.tb03694.x. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Sørensen AC. Inbreeding – lessons from animal breeding, evolutionary biology and conservation genetics. Animal Science. 2005;80:121–133. [Google Scholar]
- Kristensen TN, Sørensen AC, Sorensen D, Pedersen KS, Sørensen JG, Loeschcke V. A test of quantitative genetics theory using Drosophila– effects of inbreeding and the rate of inbreeding on heritabilities and variance components. Journal of Evolutionary Biology. 2005;18:736–770. doi: 10.1111/j.1420-9101.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Barker JSF, Pedersen KS, Loeschcke V. Extreme temperatures increase the deleterious consequences of inbreeding under both laboratory and semi-natural conditions. Proceedings of the Royal Society London B. 2008;275:2055–2061. doi: 10.1098/rspb.2008.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen TN, Pedersen KS, Vermeulen CJ, Loeschcke V. Research on inbreeding in the “omics” era. Trends in Ecology and Evolution. 2010;25:44–52. doi: 10.1016/j.tree.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Loeschcke V, Bilde T, Hoffmann AA, Sgró C, Noreikienė K, Ondrésik M, et al. No inbreeding depression for developmental temperature acclimation across multiple Drosophila species. Evolution. 2011;65:3195–3201. doi: 10.1111/j.1558-5646.2011.01359.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Risk of population extinction from demographic and environmental stochasticity and random catastrophes. American Naturalist. 1993;142:911–927. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- Lande R. Mutation and conservation. Conservation Biology. 1995;9:782–791. [Google Scholar]
- Leimu R, Vergeer P, Angeloni F, Ouborg NJ. Habitat fragmentation, climate change, and inbreeding in plants. Annals of the New York Academy of Sciences. 2010;1195:84–98. doi: 10.1111/j.1749-6632.2010.05450.x. [DOI] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits of natural selection. Trends in ecology and Evolution. 2002;17:183–189. [Google Scholar]
- Liberg O, Andrén H, Pederson H-C, Sejberg D, Wabakken P, Åkesson M, Bensch S. Severe inbreeding depression in a wild wolf (Canis lupus) population. Biology Letters. 2005;1:17–20. doi: 10.1098/rsbl.2004.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conservation Genetics. 2010;11:355–373. [Google Scholar]
- Luquet E, Léna J-P, David P, Joly P, Lengagne T, Perrin N, Plénet S. Consequences of genetic erosion on fitness and phenotypic plasticity in European tree frog populations (Hyla aborea. Journal of Evolutionary Biology. 2011;24:99–110. doi: 10.1111/j.1420-9101.2010.02138.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Lande R. Evolution and extinction in response to environmental change. In: Kareiva PM, Kingsolver JG, Huey RB, editors. Biotic Interactions and Global Change. Sunderland, MA: Sinauer; 1993. pp. 234–250. [Google Scholar]
- MacNair MR. The evolution of plants in metal-contaminated environments. In: Bijlsma R, Loeschcke V, editors. Environmental Stress, Adaptation, and Evolution. Basel: Birkhauser Verlag; 1997. pp. 3–24. [Google Scholar]
- Madsen T, Shine R, Olsson M, Wittzell H. Restoration of an inbred adder population. Nature. 1999;402:34–35. [Google Scholar]
- Malcom JW. Smaller, scale-free gene networks increase quantitative trait heritability and result in faster population recovery. PLoS ONE. 2011;6:14645. doi: 10.1371/journal.pone.0014645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. The causes of extinction. Philosophical Transaction of the Royal Society B. 1989;325:241–252. doi: 10.1098/rstb.1989.0086. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Clarke JM, Hollingsworth MJ. The expression of hybrid vigour in Drosophila subobscura. Proceedings of the Royal Society London B. 1955;144:159–171. doi: 10.1098/rspb.1955.0042. [DOI] [PubMed] [Google Scholar]
- McKenzie JA, Batterham P. Predicting insecticide resistance: mutagenesis, selection and response. Philosophical Transactions Royal Society B. 1998;353:1729–1734. doi: 10.1098/rstb.1998.0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey DH, Wilson AJ, Brommer JF. The evolutionary ecology of individual phenotypic plasticity in wild populations. Journal of Evolutionary Biology. 2007;20:831–844. doi: 10.1111/j.1420-9101.2007.01300.x. [DOI] [PubMed] [Google Scholar]
- Orr HA, Unckless RL. Population extinction and the genetics of adaptation. American Naturalist. 2008;172:160–169. doi: 10.1086/589460. [DOI] [PubMed] [Google Scholar]
- Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma R, Hedrick PW. Conservation genetics in transition to conservation genomics. Trends in Genetics. 2010;26:177–187. doi: 10.1016/j.tig.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Palstra FP, Ruzzante DE. Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Molecular Ecology. 2008;17:3428–3447. doi: 10.1111/j.1365-294x.2008.03842.x. [DOI] [PubMed] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology and Systematics. 2006;37:637–669. [Google Scholar]
- Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- Paschke M, Bernasconi G, Schmid B. Population size and identity influence the reaction norm of the rare, endemic plant Cochlearia bavaria across a gradient of environmental stress. Evolution. 2003;57:496–508. doi: 10.1111/j.0014-3820.2003.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Pedersen LD, Pedersen AR, Bijlsma R, Bundgaard J. The effects of inbreeding and heat stress on male sterility in Drosophila melanogaster. Biological Journal of the Linnean Society. 2011;104:432–442. [Google Scholar]
- Pemberton J. Measuring inbreeding depression in the wild: the old ways are the best. Trends in Ecology and Evolution. 2004;19:613–615. doi: 10.1016/j.tree.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Pertoldi C, Bijlsma R, Loeschcke V. Conservation genetics in a globally changing environment: present problems, paradoxes and future challenges. Biodiversity Conservation. 2007;16:4147–4163. [Google Scholar]
- Pigliucci M. Evolution of phenotypic plasticity: where are we going now? Trends in Ecology and Evolution. 2005;20:482–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pluess AR, Stöcklin J. Genetic diversity and fitness in Scabiosa columbaria in the Swiss Jura in relation to population size. Conservation Genetics. 2004;5:145–156. [Google Scholar]
- Ralls K, Ballou JD, Templeton AR. Estimates of lethal equivalents and the cost of inbreeding in mammals. Conservation Biology. 1988;2:185–193. [Google Scholar]
- Rattner BA. History of wildlife toxicology. Ecotoxicology. 2009;18:773–783. doi: 10.1007/s10646-009-0354-x. [DOI] [PubMed] [Google Scholar]
- Reed DH. Relationship between population size and fitness. Conservation Biology. 2005;19:563–568. [Google Scholar]
- Reusch T, Wood T. Molecular ecology of global change. Molecular Ecology. 2007;16:3973–3992. doi: 10.1111/j.1365-294X.2007.03454.x. [DOI] [PubMed] [Google Scholar]
- Roberts DR, Andre RG. Insecticide resistance issues in vector-borne disease control. American Journal of Tropical Medicine and Hygiene. 1994;50(6 Suppl):21–36. doi: 10.4269/ajtmh.1994.50.21. [DOI] [PubMed] [Google Scholar]
- Ross-Gillespie A, O'Riain MJ, Keller LF. Viral epizootic reveals inbreeding depression in a habitually inbreeding mammal. Evolution. 2007;61:2268–2273. doi: 10.1111/j.1558-5646.2007.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacheri IJ, Nichols RA, Brakefield PM. Effects of bottlenecks on quantitative genetic variation in the butterfly Bicyclus anynana. Genetical Research Cambridge. 2001;77:167–181. doi: 10.1017/s0016672301004906. [DOI] [PubMed] [Google Scholar]
- Schiegg K, Pasinelli G, Walters JR, Daniels SJ. Inbreeding and experience affect response to climate change by endangered woodpeckers. Proceedings of the Royal Society London B. 2002;269:1153–1159. doi: 10.1098/rspb.2002.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting CD. The evolution of phenotypic plasticity in plants. Annual Review of Ecology and Systematics. 1986;17:667–693. [Google Scholar]
- Schlichting CD, Levin DA. Effects of inbreeding on phenotypic plasticity in cultivated Phlox. Theoretical and Applied Genetics. 1986;72:114–119. doi: 10.1007/BF00261465. [DOI] [PubMed] [Google Scholar]
- Segelbacher G, Cushman SA, Epperson BK, Fortin M-J, Francois O, Hardy OJ, Holderegger R, et al. Applications of landscape genetics in conservation biology: concepts and challenges. Conservation Genetics. 2010;11:375–385. [Google Scholar]
- Sgrò CM, Lowe AJ, Hoffmann AA. Building evolutionary resilience for conserving biodiversity under climate change. Evolutionary Applications. 2011;4:326–337. doi: 10.1111/j.1752-4571.2010.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Bouzat JL. Modelling the adaptive potential of isolated populations: experimental simulations using Drosophila. Evolution. 2005;59:2159–2169. [PubMed] [Google Scholar]
- Teotónio H, Chelo IM, Bradić M, Rose MR, Long AD. Experimental evolution reveals natural selection on standing genetic variation. Nature Genetics. 2009;41:251–257. doi: 10.1038/ng.289. [DOI] [PubMed] [Google Scholar]
- Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collinham YC, Erasmus BFN, et al. Extinction risks from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- Van Buskirk J, Willi Y. The change in quantitative genetic variation with inbreeding. Evolution. 2006;60:2428–2434. [PubMed] [Google Scholar]
- Vermeulen CJ, Bijlsma R. Changes in mortality patterns and temperature-dependence of life span in Drosophila melanogaster caused by inbreeding. Heredity. 2004a;92:275–281. doi: 10.1038/sj.hdy.6800412. [DOI] [PubMed] [Google Scholar]
- Vermeulen CJ, Bijlsma R. Characterization of conditionally expressed mutants effecting age-specific survival in inbred lines of Drosophila melanogaster: lethal conditions and temperature sensitive period. Genetics. 2004b;167:1241–1248. doi: 10.1534/genetics.103.023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà C, Sundqvist A-K, Flagstad Ø, Seddon J, Bjørnerfeldt S, Kojola I, Casulli A, et al. Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proceedings of the Royal Society London B. 2003;270:91–97. doi: 10.1098/rspb.2002.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis R, Dawson K, Boursot P. Interpretation of variation across marker loci as evidence of selection. Genetics. 2001;158:1811–1823. doi: 10.1093/genetics/158.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ, Schuster SM, Stevens L. Inbreeding: its effects on the response to selection for pupal weight and the heritable variance in fitness in the flour beetle, Tribolium castaneum. Theoretical Population Biology. 1996;25:138–139. doi: 10.1111/j.1558-5646.1996.tb03882.x. [DOI] [PubMed] [Google Scholar]
- Waller DM, Dole J, Bersch AJ. Effects of stress and phenotypic variation on inbreeding depression in Brassica rapa. Evolution. 2008;62:917–931. doi: 10.1111/j.1558-5646.2008.00325.x. [DOI] [PubMed] [Google Scholar]
- Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershmer EL, et al. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, Fowler K. The changes in genetic and environmental variance with inbreeding in Drosophila melanogaster. Genetics. 1999;152:345–353. doi: 10.1093/genetics/152.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. Is local provenance important in habitat creation? Journal of Applied Ecology. 2001;38:1371–1373. [Google Scholar]
- Willi Y, Hoffmann AA. Demographic factors and genetic variation influence population persistence under environmental change. Journal of Evolutionary Biology. 2009;22:124–133. doi: 10.1111/j.1420-9101.2008.01631.x. [DOI] [PubMed] [Google Scholar]
- Willi Y, Van Buskirk J, Hoffmann AA. Limits to the adaptive potential of small populations. Annual Review of Ecology and Systematics. 2006;37:433–458. [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright LI, Tregenza T, Hosken DJ. Inbreeding, inbreeding depression and extinction. Conservation Genetics. 2008;9:833–843. [Google Scholar]


