Abstract
Environmental heterogeneity influences coevolution and local adaptation in host–parasite systems. This also concerns applied issues, because the geographic range of parasites may depend on their capacity to adapt to abiotic conditions. We studied temperature-specific adaptation in the wheat yellow/stripe rust pathogen, Puccinia striiformis f.sp. tritici (PST). Using laboratory experiments, PST isolates from northern and southern France were studied for their ability to germinate and to infect bread and durum wheat cultivars over a temperature gradient. Pathogen origin × temperature interactions for infectivity and germination rate suggest local adaptation to high- versus low-temperature regimes in south and north. Competition experiments in southern and northern field sites showed a general competitive advantage of southern over northern isolates. This advantage was particularly pronounced in the southern ‘home’ site, consistent with a model integrating laboratory infectivity and field temperature variation. The stable PST population structure in France likely reflects adaptation to ecological and genetic factors: persistence of southern PST may be due to adaptation to the warmer Mediterranean climate; and persistence of northern PST can be explained by adaptation to commonly used cultivars, for which southern isolates are lacking the relevant virulence genes. Thus, understanding the role of temperature-specific adaptations may help to improve forecast models or breeding programmes.
Keywords: climate change, genotype × environment interaction, local adaptation, plant pathogen, Puccinia striiformis f.sp. tritici, temperature adaptation, wheat, yellow/stripe rust
Introduction
Adaptation to heterogeneous environments is considered a main driver of the evolution and maintenance of genetic diversity in natural populations (Levins 1968; Hedrick 1986; Ravigné et al. 2009). This argument hinges on the idea that adaptation to one environment reduces fitness in others, resulting in genetic trade-offs between environments (genotype × environment interactions, G × E) and concomitant patterns of local adaptation (Kawecki and Ebert 2004).
Environmental heterogeneity may also influence coevolution and patterns of local adaptation in host–parasite interactions (Thompson 2006; Nuismer and Gandon 2008; Gandon and Nuismer 2009). In these systems, the ‘environment’ takes different forms. For example, the parasite is confronted with the genetic environment of the host (resistance genes, etc.) and the abiotic environment, either directly or indirectly via the host. Traditionally, empirical and theoretical work have focused on parasite adaptation to the genetic host environment (and vice versa) and on the role of host genotype × parasite genotype interactions (GH × GP) in maintaining genetic polymorphism and producing geographic patterns of local adaptation (Kaltz and Shykoff 1998; Gandon 2002; Dybdahl and Storfer 2003). However, the abiotic environment can strongly affect disease development and transmission, and recent studies have demonstrated G × E, or even GH × GP × E, interactions for these traits (Thomas and Blanford 2003; Vale et al. 2008; Wolinska and King 2009). This may add substantial complexity to the traditional view of host–parasite coevolution. How does environmental variation modulate the strength or specificity of reciprocal selection (Hochberg and van Baalen 1998; Gomulkiewicz et al. 2007; Laine 2008; Lopez-Pascua and Buckling 2008)? Can patterns of parasite local adaptation be decomposed into adaptation to the local environment and adaptation to the genetic structure of the local host population (Gandon and Nuismer 2009)? These questions are also highly relevant in an applied context, especially in the light of global climate change (Harvell et al. 2002; Lafferty 2009). Indeed, understanding how present environmental conditions shape the evolution of medically or agronomically relevant parasites and pathogens may help predict future adaptation and changes in their geographic range.
Agrosystems are convenient for the study of (co) evolutionary processes. The physiology and genetics underlying the interaction between crop and pathogen are generally well characterised, and breeder-mediated coevolution is sometimes well documented (Thrall et al. 2011). Typically, this process follows boom-and-bust cycles (Bonman et al. 1992; Wolfe and McDermott 1994; McDonald and Linde 2002), whereby new resistance genes introduced by breeders are countered by the recurrent emergence of corresponding virulence mutations in the pathogen population.
However, the role of the environment in pathogen adaptation is largely unclear. Many studies in plant pathology have described environmental effects on disease development and transmission (Garrett et al. 2006; Hau and de Vallavieille-Pope 2006), but often use only few host or pathogen genotypes (e.g. Pfister et al. 2004; Nordskog et al. 2007). Therefore, still relatively little is known about the strength of G × E interactions and their adaptive significance. This may explain why environmental variables are rarely considered when discussing resistance durability (McDonald and Linde 2002; Niks and Rubiales 2002; Burdon and Thrall 2008).
We explored the influence of the abiotic environment on local adaptation of the fungal pathogen Puccinia striiformis Westend. f.sp. tritici (PST), the worldwide causal agent of wheat yellow (stripe) rust. Typically, PST populations are clonal and adapt through a stepwise mutation process and selective sweeps (Wellings and McIntosh 1990; Hovmøller and Justesen 2007). Thus, within only few years, populations acquire virulence factors overcoming major qualitative resistance genes used in cultivars (Wellings and McIntosh 1990; Wan et al. 2004).
In France, the situation is particular: a stable genetic differentiation exists between northern and southern PST populations, with two clonal lineages diverging for virulence factors and molecular markers (Enjalbert et al. 2005). As no geographic barriers prevent migration and because PST has long-distance wind dispersal (1500 km: Brown and Hovmøller 2002), the north/south genetic structure may be explained by selection. In part, the pattern reflects adaptation of PST to the wheat resistance gene landscape. Over the past 30 years, intense selection against yellow rust in north-western Europe involved the successive introduction of new resistance genes by plant breeders and rapid emergence of new pathotypes, resulting in a large spectrum of virulence genes in the northern PST population (de Vallavieille-Pope et al. 1990, 2011). In contrast, PST from the Mediterranean region has a smaller virulence spectrum and generally lack the virulences necessary to overcome the Yr resistance genes present in most widely used cultivars in the north (de Vallavieille-Pope et al. 1990; Bayles et al. 2000).
These genetic differences would explain the absence of southern strains in northern areas, but not the absence of northern strains in the south. The northern PST population harbours all the virulence genes necessary to infect wheat cultivars used in southern France (de Vallavieille-Pope et al. 2011. We, therefore, hypothesised that the geographic differentiation also involves adaptation to the local climate, namely to a north–south cline in temperature. Temperature strongly influences PST development (de Vallavieille-Pope et al. 1995; Chen 2005; Hau and de Vallavieille-Pope 2006), and a recent theoretical study detailed how high temperature is a major limiting factor for PST epidemic spread (Papastamati and van den Bosch 2007). Thus, local adaptation to higher temperatures in the Mediterranean region may protect southern PST against invasion by northern PST.
To test for temperature-specific adaptation, we first measured infectivity and spore germination rate of northern and southern PST isolates over a range of temperatures on bread and durum wheat cultivars under controlled climate chamber conditions. Second, competition experiments between northern and southern isolates were carried out in experimental field sites in the north and south of France. Third, we integrated climate chamber data and meteorological data into a simple model and compared predicted competitive success with realised outcomes in the field.
Materials and methods
Origin of fungal isolates and host cultivars
We tested 10 PST isolates collected from northern France and seven from southern France (Table S1), representing predominant genotypes in the two geographic areas over a 20-year period. Molecular markers and virulence profiles indicate that southern isolates belong to a North-African population, whereas northern isolates belong to the northwestern-Europe PST population (Enjalbert et al. 2005; Bahri et al. 2009b). Low within-group genetic diversity indicates a narrow origin for each clonal lineage. Southern isolates (6E16 pathotype) exhibit a simple virulence spectrum (Table S1) and cannot infect most of the northern France cultivars, which carry one or several of the resistance genes Yr1, Yr3, Yr4, Yr9, Yr17, Yr25, Yr32 (de Vallavieille-Pope et al. 1990; Bayles et al. 2000). Epidemics in the south (1996–2001), therefore, typically occurred on non-Yr bread and durum wheat cultivars, such as Victo and Acalou (Enjalbert et al. 2005; de Vallavieille-Pope et al. 2011). In contrast, northern isolates belong to different pathotypes with a more complex virulence spectrum (Table S1), evolved in response to various cultivars introduced by breeders (de Vallavieille-Pope et al. 2000, 2011). For example, over the past 15 years, epidemics in the north involved the successive breakdown of Yr17, Yr17 + Yr6 and Yr32 resistance genes (Bayles et al. 2000; de Vallavieille-Pope et al. 2011). In principle, the genetic composition of northern PST allowed infection of cultivars used in the south (e.g. Victo and Acalou), but nonetheless, only 11.7% of the isolates collected in the south during the 1996–2001 period were typical northern pathotypes. Conversely, only 4% of the isolates collected in northern France were 6E16 and mainly found on the Victo cultivar, which is rarely grown in the north.
Bread wheat (Triticum aestivum) is predominantly used in the north of France and durum wheat (Triticum turgidum) in the south; the main experiments were performed on one bread wheat cultivar (Victo) and one durum wheat cultivar (Acalou), both highly susceptible to PST. In supplementary experiments, using a subset of six northern and southern isolates, additional cultivars were tested (see Supporting Information).
Preparation of fungal spores
To minimise maternal or environmental effects, urediniospores of the 17 isolates were simultaneously inoculated on 10-day-old wheat seedlings of the cultivar Michigan Amber. Infected plants were kept in a climate chamber, under a 8-h 14°C dark period and 16-h 17°C light period regime (light intensity: 300 μmol quanta m−2 s−1). After 15 days, harvested urediniospores were placed in a glycerol-filled desiccator for 4 days at 4°C, and then stored in liquid nitrogen. Before inoculation, frozen spores were heat shocked for 10 min at 40°C to facilitate germination.
Inoculation protocol for climate chamber experiments
In independent experimental tests, we measured infectivity of the 17 northern and southern isolates at five temperatures: 7, 10, 15, 20 and 25°C. The range of temperatures chosen here is representative of spring and early summer night/dawn, where spore germination and subsequent leaf infection occur subsequently to dew formation.
Plants were inoculated as described in Bahri et al. (2009a). Replicates were grouped in 25 cm2 pots, in which 10 seeds of a given host cultivar were sown. After 10 days (two-leaf stage), we chose six similarly developed seedlings in each replicate pot and removed their second leaf; the remaining seedlings were discarded. Replicate pots were inoculated in a settling tower using 0.5 mg of urediniospores. The density of deposited spores (per mm2) was estimated on glass microscopy slides, placed in the settling tower. Inoculated plants were placed for 24 h in a dew chamber in the dark to permit penetration of the fungus (de Vallavieille-Pope et al. 1995; Hall et al. 2006). During this 24-h period, replicate pots were exposed to one of the five temperature treatments. Then, plants from all treatments were kept for 1 week in the greenhouse, under optimal growth conditions (8-h 14°C dark, 16-h 17°C light).
After 1 week of incubation, chloroses (i.e. early symptoms because of spores successfully penetrating the leaf) were counted on each leaf. We calculated infectivity as the number of chloroses divided by the leaf surface (length × width, mm2) and the density of deposited spores (spores per mm2). In total, we established five replicate pots for each combination of fungal isolate, host cultivar (Victo or Acalou) and temperature, with a total of 5100 inoculated seedlings (17 isolates × five temperatures × two cultivars × five pots × six seedlings). For a subset of isolates, these tests were repeated at 10 and 20°C, with additional host cultivars (see Supporting Information).
Germination rate
To measure the effect of temperature on spore germination rate, urediniospores were deposited on glass microscopy slides in the settling tower, as described earlier. The spores were then incubated for 24 h at 7, 10, 15 and 20°C in a dew chamber. For each PST isolate and temperature, germination was assessed on two slides, by examining 100 spores per slide under a microscope at 10× magnification. Spores were considered as germinated when the germ tube length was at least equal to the diameter of the spore. We repeated this experiment at 10 and 20°C (see Supporting Information).
Field competition experiments
In 2005 and 2006, we carried out pairwise competition experiments between southern and northern isolates in experimental field plots in southern France (Mauguio, 43°37′N 4°0′E) and northern France (Grignon, 48°50′N 1°55′E). Four pairs of southern/northern isolates (Table S1) were tested in each location, on host cultivars Acalou and Victo. For each competing pair, 50:50 spore mixtures were established and inoculated on Victo seedlings, as described earlier. In late March, two pots of sporulating seedlings (20 seedlings per pot) were planted in the centre of a 10 m × 1.5 m plot of a given cultivar (approximately 1300 plants). The Victo plots were separated from the Acalou plots by a 7-m band of the cultivar Caphorn, which is fully resistant to all our isolates as well as to naturally occurring isolates. In total, 32 field plots were established (2 years × two locations × two cultivars × four pairs of competing isolates).
At the end of the epidemics (early June), spores from the centre of each infection focus in a plot were harvested with a vacuum collector. The initially introduced infected plants, in most cases already dead by the end of the experiment, were excluded from spore harvest. The relative proportions of the two competing isolates were determined by inoculating 0.1 mg of the collected urediniospores on five pots, each containing 15 seedlings of the Compair (Yr8) cultivar. This selective cultivar is resistant to the northern isolates and susceptible to the southern isolates (carrying V8, Table S1). We then determined the relative proportion of southern and northern isolates in two steps. First, 7 days after inoculation, we counted the number of chlorotic spots on the seedlings and marked each spot with an ink marker. These chlorotic spots correspond to both successful infections (by the southern isolates) and hypersensitive reactions (by the northern isolates). Second, on day 14, we recorded the number of sporulating chloroses and could clearly distinguish the successful infections from the necrotic spots (=failed infection by northern isolates). Thus, the proportion of southern isolates among the harvested spores was taken as the ratio of the number of sporulating choroses/total chlorotic spots (necrotic + sporulating). Additional experiments, using different ratios of isolates in the spore mixtures, confirmed the capacity of this protocol to detect variation in isolate frequency (Supporting Information).
Yellow rust sporulation is characterised by long-lasting lesions and short-distance spore dispersal (de Vallavieille-Pope et al. 2000), resulting in local spread of infection in our experimental plots. Therefore, the spatial separation of inoculation spots minimised the risk of cross-contamination between experimental plots. In all plots, we always observed circular development of secondary infection foci around the planted pots with the infected seedlings, indicating highly localised spread of infection. Furthermore, molecular analysis of spore samples from four secondary infection foci (100 spores per focus) showed no evidence for contamination with pathotypes used in other experiments in neighbour plots.
Statistical analyses
First, for the climate chamber data, effects of host cultivar, pathogen origin and temperature were investigated in factorial analyses of variance (anova). Isolate identity was nested as a random factor within pathogen origin. To meet assumptions of homoscedasticity and normality, we used square-root transformation for infectivity and arcsine transformation for germination rate. For infectivity, we took averages over seedlings and replicate pots to obtain the mean infectivity per combination of isolate, temperature and cultivar. Similarly, germination rate was averaged over microscope replicate slides.
Second, for the field competition data, anova was used to test for effects of year, host cultivar and identity of pairs of southern/northern isolates on the (arcsine-transformed) frequency of the southern genotype at the end of the epidemics. Year and pair identity were considered random factors.
Third, we tested whether outcomes in the field experiment could be predicted from a combination of climate chamber data and meteorological data. To this end, we calculated daily predicted values of infectivity for each individual isolate, based on the daily minimum temperature recorded in each field location and the infectivity estimated at that temperature from the climate chamber data. From these predicted daily infectivity values, we calculated a cumulated predicted growth difference (i.e. a difference in spore production) between competing isolates, and this for each host cultivar, field location and year (for details, see Figures S8 and S9). Analyses of covariance were tested for a relationship between predicted growth difference (southern minus northern isolate) and the observed change in southern isolate frequency. These analyses included pair identity, field location, host cultivar and year as cofactors.
We used the JMP statistical package (SAS 2009). Where appropriate, initial full models were simplified by sequential removal of nonsignificant factors, starting with higher-order interactions.
Results
Infectivity
Infectivity, that is the fraction of spores able to infect host tissue and cause symptom development, was strongly dependent on temperature (Fig. 1, Table 1). We observed hump-shaped reaction norms, with high levels of infection at 10 and 15°C and low levels of infection at 7 and 20°C. Dew incubation at 25°C induced a hypersensitive reaction, rendering hosts almost entirely resistant to infection at this temperature.
Figure 1.
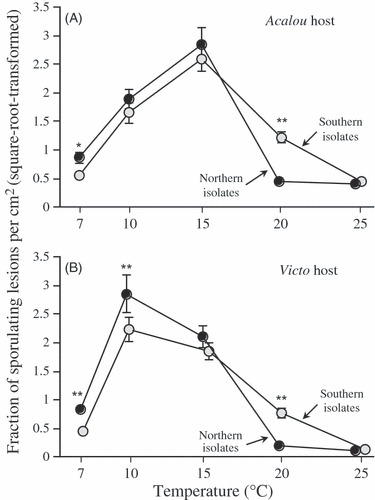
Mean infectivity of northern and southern Puccinia striiformis f.sp. tritici isolates for five incubation temperatures, on the (A) Acalou and (B) Victo host cultivar. Means and standard errors based on the averages of 10 northern and seven southern isolates. *Significant differences between northern and southern isolates at a given temperature, as revealed by Student–Newman–Keuls a posteriori tests. *P < 0.05; **P < 0.01.
Table 1.
Analyses of variance of infectivity, as a function of pathogen origin (north versus south), host cultivar (Victo versus Acalou) and pathogen isolate, at five incubation temperatures
| Source | df | Denominator | Denominator df | Mean square (×10−2) | F | P |
|---|---|---|---|---|---|---|
| Origin | 1 | (1) | 15 | 0.123 | 0.28 | 0.6075 |
| Cultivar | 1 | (2) | 15 | 0.817 | 8.48 | 0.0110 |
| Temperature | 4 | (3) | 60 | 32.870 | 82.52 | <0.0001 |
| Isolate[origin] (1) | 15 | (2)+(3)−(4) | 52 | 0.446 | 1.07 | 0.4063 |
| Isolate*cultivar[origin] (2) | 15 | (4) | 60 | 0.097 | 1.25 | 0.2653 |
| Isolate*temperature[origin] (3) | 60 | (4) | 60 | 0.398 | 5.11 | <0.0001 |
| Origin*temperature | 4 | (3) | 60 | 1.580 | 3.97 | 0.0064 |
| Cultivar*temperature | 4 | (4) | 60 | 2.651 | 34.03 | <0.0001 |
| Origin*cultivar | 1 | (2) | 15 | 0.170 | 1.76 | 0.2053 |
| Origin*cultivar*temperature | 4 | (4) | 60 | 0.033 | 0.43 | 0.7888 |
| Residual (4) | 60 | 0.078 |
The denominator column indicates the terms used as denominator for F-tests. Isolate was taken as a random factor.
Statistical analysis revealed significant interactions of temperature with pathogen and cultivar identity (Table 1). First, the temperature × pathogen origin interaction indicated differential performance of northern and southern isolates at different temperatures. At lower temperatures (≤15°C), southern isolates were less infectious than northern isolates, in particular at 7 and 10°C (significant pairwise a posteriori comparisons reported in Fig. 1). In contrast, at high temperature (20°C), the pattern was inverted, with southern isolates showing up to 10-fold higher infection success than the northern isolates (Fig. 1). These temperature-dependent differences were very similar on both host cultivars (nonsignificant temperature × pathogen origin × cultivar interaction; Table 1).
Second, the temperature × cultivar interaction indicated an approximately 5°C difference in the peak levels of infection between the two cultivars. The bread wheat cultivar Victo was most susceptible to infection at 10°C, whereas the durum wheat cultivar Acalou became most infected at 15°C.
For a subset of isolates, we repeated the experiment at 10 and 20°C on additional wheat cultivars (see Supporting Information). These experiments showed, first, a high level of repeatability of the relative performance of individual isolates at a given temperature (Figure S1, Table S2). Second, the temperature × pathogen origin interaction was statistically robust against variation between replicate experiments (Table S3). Thus, in independent replicate experiments, there was an advantage in infection success of southern isolates at 20°C and an advantage of northern isolates at 10°C (Figure S2). Third, this trade-off was consistent over several host cultivars, albeit more pronounced on bread wheat than on durum wheat (Figure S3, Table S4).
Germination rate
Germination rate peaked at 10 and 15°C (Fig. 2), but precise responses varied with pathogen origin (temperature × pathogen origin interaction, Table 2). As for infectivity, southern isolates had a clear advantage over northern isolates at high temperature (20°C); conversely, northern isolates tended to have higher germination rates at lower temperatures of 10 and 15°C, but not at 7°C (Fig. 2, with results from pairwise contrasts). A complementary experiment showed that the relative performance of a subset of isolates was repeatable at 10 and 20°C (Table S5, Figure S4), confirming the higher germination rate of southern isolates at 20°C (Figure S5).
Figure 2.
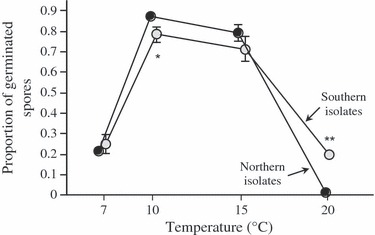
Mean germination rate of northern and southern Puccinia striiformis f.sp. tritici isolates for four incubation temperatures. Means and standard errors based on the averages of 10 northern and seven southern isolates. *Significant differences between northern and southern isolates at a given temperature, as revealed by Student–Newman–Keuls a posteriori tests. *P < 0.05; **P < 0.01.
Table 2.
Analyses of variance of germination rate, as a function of pathogen origin (north versus south) and pathogen isolate, at four incubation temperatures. Isolate was taken as a random factor
| Source | df | Mean square | F | P |
|---|---|---|---|---|
| Origin | 1 | 0.029 | 1.57 | 0.2301 |
| Temperature | 3 | 2.970 | 251.34 | <0.0001 |
| Origin*temperature | 3 | 0.183 | 15.51 | <0.0001 |
| Isolate[origin] | 15 | 0.019 | 1.57 | 0.1229 |
| Residual | 45 | 0.012 |
Across isolates, there was a significant positive relationship between germination rate and infectivity (ancova of infectivity, with temperature and isolate origin as cofactors and germination rate as covariate: F1,62 = 8.40, P = 0.0052). Thus, germination rate was a good predictor of infection success. Genotypic correlations, calculated separately per isolate origin and temperature treatment, were almost all positive (seven of eight), with a mean effect size of 0.35 (±0.13 SE).
Field competition experiment
In the southern location, a hypersensitive resistance reaction of one of the two cultivars (Acalou) precluded the collection of spores for analysis in both 2005 and 2006. In the remaining 24 experimental plots, the southern isolates generally increased in frequency both in the northern and southern location (Fig. 3); in only one plot, their final frequency fell below the initial 50% (overall mean: 78 ± 3%; test for difference from 50:50 ratio: t23 = 9.65, P < 0.0001).
Figure 3.
Mean frequency of southern isolates after pairwise competition with northern isolates in experimental field plots in northern France (left panel) and southern France (right panel). Independent replicate experiments were carried out in 2005 and 2006, on two host cultivars (Victo, Acalou), with an initial 1:1 ratio of the southern and the northern isolate. Means and standard errors taken over four different southern/northern pairs of isolates. In the south, a high-temperature-induced resistance reaction rendered the Acalou cultivar resistant to fungal development, leading to missing data.
To accommodate for the unbalanced data set, we first performed an analysis for the northern location only, where data for both the Acalou and Victo cultivars were available; a second analysis was restricted to the Victo cultivar, but included both locations. In the first analysis, the outcome of competition in the north did not significantly vary among the different pairs of isolates (F3,9 = 1.28, P = 0.3385), with southern isolates often reaching frequencies of over 80%. In 2005, the increase of southern isolates was only marginal on the Victo cultivar (significant year × cultivar interaction: F1,12 = 69.1, P < 0.0001; Fig. 3). However, additional sampling 1 month later revealed that southern isolates had reached frequencies of >90%, indicating a delayed competitive advantage in 2005.
The second analysis, considering competition on the Victo cultivar, showed that the mean competitive success of southern isolates was significantly higher in the southern than in the northern location (F1,13 = 7.66, P = 0.016; Fig. 3). This pronounced southern advantage was consistent over the different competing isolates and between years (interactions with location: P > 0.08): in seven of eight cases (four pairs × 2 years), competitive success of southern isolates was higher in the south than in the north (Wilcoxon signed-rank test: W = 17.0, P = 0.0156; Fig. 4).
Figure 4.
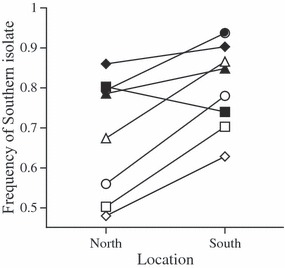
Final frequencies of southern isolates in the pairwise competition experiment on the Victo cultivar in southern and northern field locations. Experiments started from an initial 50:50 ratio of southern/northern isolates. Different symbols correspond to the four different pairs of competing isolates (i.e. different southern and northern isolates); open symbols = 2005, filled symbols = 2006.
Predicting field competition outcome from climate chamber data
By combining climate chamber data and meteorological data, we calculated predicted growth differences between southern and northern isolates in the field experiment. First, analysis of this predicted difference revealed a significant effect of assay location (F1,17 = 15.44, P = 0.0011, in anova controlling for cultivar, year and pair identity): southern isolates were predicted to be relatively more competitive in the southern than in the northern location (Fig. 5A). This is consistent with their observed higher competitive success in the south (Fig. 4). Second, there also was a quantitative match between predicted and observed values: across all replicates, we found a significant positive correlation between predicted and observed competitive success of southern isolates (r = 0.43, n = 24, P = 0.0356; Fig. 5B). Thus, within certain limits, it was possible to predict competition outcomes for a given isolate in a given field plot.
Figure 5.
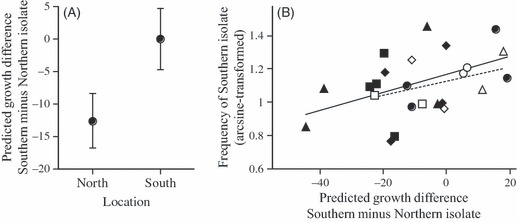
(A) Mean predicted cumulated growth differences between competing southern and northern isolates in the southern and northern location. Means and standard errors calculated over competing pairs of isolates, years and host cultivars. (B) Relationship between predicted growth differences and the observed final frequency of southern isolates in the field experiment. Open symbols denote the field location in the south of France and filled symbols the location in the north. Different symbols refer to the four different pairs of competing isolates, pooled over the two host cultivars and field seasons. The two regression lines illustrate the relationship in the southern location (dotted line) and in the northern location (solid line).
Discussion
We found strong effects of temperature on pathogen fitness traits as well as genotype × temperature interactions for these traits in the laboratory experiment. The field experiment revealed competitive superiority of southern over northern PST isolates, especially reinforced in the southern assay location. We will discuss implications of these results regarding climate-specific (local) adaptation of PST and whether such specificity explains the stable spatial genetic structure of PST in France. We will also consider the agronomic relevance of our findings.
Temperature × host or pathogen genotype interactions
The climate chamber experiments revealed optimal infectivity and spore germination at intermediate temperatures, a pattern typical of PST and other rust species (de Vallavieille-Pope et al. 1995). However, the shape of these curves varied with host and pathogen identity, resulting in genotype × temperature interactions. First, the PST origin × temperature interaction was caused by higher infectivity of northern isolates at lower temperatures and higher infectivity of southern isolates at higher temperature. A high-temperature advantage of southern isolates was also observed for germination, and additional experiments revealed shorter latency and higher spore production of southern isolates (Figure S6), indicating a generally higher fitness at high temperature. Positive genetic correlations between these traits suggest that directional selection can act on them simultaneously, allowing fast and efficient adaptation to a high-temperature regime. Genetic variation in germination and aggressiveness at high temperature is also known for PST isolates from other countries (Elahinia 2000; Milus et al. 2006).
Second, there was a 5-degree difference in the infection optima between the bread wheat (Victo) and durum wheat (Acalou) cultivar. This is a clear demonstration of a G × E interaction quantitatively affecting susceptibility to infection in wheat. While air temperature was controlled in these experiments, leaf surface temperature is the relevant parameter for infection success. Surface temperature is modulated by leaf colour and orientation or stomata reactivity (Claus et al. 1995), and these traits may differ between cultivars, thereby producing the observed G × E interaction, in addition to cultivar-specific thermal responses of metabolism or pathogen defences.
Local adaptation
High temperature strongly limits PST infection success and epidemic spread. We had, therefore, hypothesised that the geographic distribution of PST isolates in France reflects adaptation to different temperature regimes, namely the higher temperatures in the southern Mediterranean region. This is consistent with the superiority of southern isolates at high temperature in the laboratory experiments, and together with the higher performance of northern isolates at lower temperatures, these results suggest a pattern of temperature-dependent local adaptation. Unlike in a study on temperature and local adaptation in a natural plant–pathogen system (Laine 2008), this temperature specificity varied little with host cultivar or species identity. This shows that maximal complexity, that is strong Host × Parasite × Environment interactions, is not necessarily the rule in host–parasite systems (Vale et al. 2008; Vale and Little 2009).
While the laboratory results indicate local adaptation, the field data are less conclusive. Southern isolates were overall more competitive than northern isolates both in the northern and southern field location. Thus, one criterion of local adaptation, the ‘resident-versus-foreign advantage’ (Gandon and Van Zandt 1998; Kawecki and Ebert 2004), is not met for both locations.
Nonetheless, there were other signatures of local adaptation. First, the advantage of southern isolates was significantly more pronounced in the southern location. This fulfils the ‘home-versus-away advantage’ criterion of local adaptation (Gandon and Van Zandt 1998; Kawecki and Ebert 2004), with both southern and northern isolates being relatively more successful in their respective local ‘home’ location than in the foreign ‘away’ location (Fig. 3). Second, observed competitive outcomes were positively correlated with outcomes predicted from combined climate chamber and meteorological data. Predicted competitive ability of southern isolates was also higher in the southern location, as was observed in the experiment. These correlations indicate that the temperature trade-off found under controlled conditions is meaningful and has a significant influence on competition outcomes in the field.
The field experiment investigated competition during spring–summer epidemics, and perhaps, a full pattern of local adaptation would emerge over the entire season. Predicted values suggest a ‘resident’ advantage of northern isolates in the north in early spring and autumn, because of the lower infectivity of southern isolates at low temperature (see also Figure S9). Furthermore, PST survives the winter as dormant mycelium in leaves of volunteers and autumn-sown wheat, and it is known that cold winter periods can decrease the occurrence of PST (Hovmøller 2001; Gladders et al. 2007). Thus, it would be interesting to compare northern and southern isolates for their survival at below-zero temperatures, frequent in the northern winter.
Finally, it is possible that the performance of northern isolates was influenced by the carriage of costly virulence genes (Van der Plank 1968). Virulence genes confer nonrecognition by the host defence, but may come at the cost of reduced pathogen development. Indeed, Thrall and Burdon (2003) observed a trade-off between the number of virulence genes and spore production in the rust pathogen Melampsora lini, and in PST, already one additional virulence gene can reduce competitive ability of PST strains (Bahri et al. 2009a). Here, northern isolates carried 2–4 more virulences than their southern counterparts (Table S1), and this may have lowered their overall competitive success. However, virulence costs alone may not explain the observed temperature specificity: while costs for northern isolates may be reinforced under suboptimal conditions at high temperature (see Quance and Travisano 2009, Brown et al. 2006 for examples of condition-dependent costs in other organisms), it is less conceivable that additional virulence genes would become beneficial at lower temperatures. Clearly, to assess the precise contribution of virulence costs to patterns of temperature adaptation, it would be necessary to recombine these virulence genes into identical genetic backgrounds, a difficult task with this predominantly clonal pathogen.
A geographic mosaic of local adaptation?
A central point of our study is that the rare occurrence of northern pathotypes in the south can be explained by the competitive superiority of southern isolates in that region. As discussed earlier, this result likely reflects temperature-specific adaptation to the warmer southern climate, although a general fitness advantage on susceptible cultivars cannot be ruled out when considering our field data. Conversely, a selective advantage of northern isolates in the north may not be exclusively driven by differential temperature adaptation. In fact, the absence of southern isolates in the north can be explained by the geographic structure of host resistance genes, because of regional use of cultivars. While the northern PST population harbours the virulences necessary to overcome the Yr resistances in the most commonly used cultivars in the north (and in the south), the southern isolates are lacking these virulences.
Altogether, our results suggest a geographic mosaic of local adaptation in this system, with ecological factors (temperature) and genetic factors (resistance/virulence structure) playing different roles in different regions, a scenario explored by recent theoretical work (Nuismer and Gandon 2008). These results also partly resemble patterns in a natural system (Barrett et al. 2008). Two lineages of M. lini, a rust pathogen of wild flax (Linum marginale), are confined to inland and coastal habitats, respectively. The coastal lineage, showing a resident advantage in the cooler coastal habitat, has the genes necessary to infect plants in the inland habitat, but is lacking adaptation to the higher inland temperatures. Thus, unlike in our case, adaptation to ecological factors seems to be the main driver of the maintenance of genetic polymorphism in both regions.
Implications for disease management
Predicting climate impact on the geographic range of pests and pathogens is a major challenge for fundamental and applied research (Harvell et al. 2002; Lafferty 2009). Our experiments confirm correlational studies and theoretical work, identifying temperature as an important determinant of PST epidemics (Rapilly 1979; Hau and de Vallavieille-Pope 2006). Moreover, we have demonstrated a direct link between temperature, infectivity and competitive success, and it was even possible to predict the relative success of individual PST isolates under variable environmental field conditions. This suggests that already simple and rapid experimental tests (germination rates) and baseline information on climatic parameters may help to generate forecasting models for different production areas.
Our study also points to the limits of forecasting efforts if the capacity of pathogens to adapt to the environment is ignored. For PST, temperature-specific adaptation may not only explain the regional persistence of the Mediterranean population but also likely influence the spread into new habitats. Indeed, the last decade has seen the worldwide expansion of two PST strains that are very aggressive at high temperatures (Hovmøller et al. 2008; Milus et al. 2009). These strains have caused damaging epidemics in regions usually considered too hot for PST (Boshoff et al. 2002; Wellings et al. 2003; Yahyaoui et al. 2004; Milus et al. 2006).
The observed genotype × environment interactions have practical implications for resistance management. For adequate characterisation of resistance and disease phenology, it is advisable to test isolates or cultivars under different environmental conditions. We found a substantial difference between the two host cultivars for the temperatures at which they exhibit maximum susceptibility. Such differences can guide plant breeders to design new cultivars for particular climatic regions. For example, resistance genes may be crossed into genetic backgrounds that are already more resistant under the climatic conditions in the region of interest. Thus, the spread of the high-temperature-adapted PST strains may be countered by a double-protection strategy, recombining specific resistance genes with unspecific resistance genes, activated at high temperature (Uauy et al. 2005). This unspecific resistance, also observed in our study at 25°C, is a very efficient defence and a potential source of durable resistance (Qayoum and Line 1985; Milus and Line 1986).
Conclusions
In line with findings in natural systems (Wolinska and King 2009), we observed substantial pathogen and host G × E interactions for infection success. The G × E interactions in the pathogen illustrate how natural selection may shape evolutionary trajectories over the geographic range of the interacting species (Thompson 2006). In particular, our results highlight the importance of disentangling the relative contributions of environmental and genetic factors to the coevolutionary process (Nuismer and Gandon 2008). In our case, adaptation of PST seems to have a strong environmental component, contributing to the spatial structure of genetic diversity in this pathogen. The same conceptual framework may help to understand the determinants of the geographic range of pathogens and also guide strategies of resistance management, be it through forecasting models or through climate-specific plant breeding efforts.
Acknowledgments
We thank Nathalie Galet and Laurent Gerard for their help during the experimentations. We are grateful to Tatiana Giraud, AurélienTellier, Maud Tenaillon, Thierry Langin, Angus Buckling, Simon Fellous, Samuel Soubeyrand Alison Duncan and three anonymous reviewers for their advice and comments on early versions of the manuscript. This work was supported by the European Integrate Project Bioexploit, FOOD-CT-2005-513959 and EMERFUNDIS, ANR 07-BDIV-003.
Data archiving statement
The data used to generate the results presented can be found in the INRA server. The link is available here: http://moulon.inra.fr/deap/Mboup_2011_Evol_App_data.xls.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Relationship between infectivity in the main experiment and in the additional experiment, shown for individual PST isolates at 10 and 20°C.
Figure S2. Mean difference (±SE) in infectivity between Southern and Northern isolates in the additional experiment, shown for two temperatures (10 and 20°C).
Figure S3. Mean difference (±SE) in infectivity between Southern and Northern isolates in the additional experiment, shown for two temperatures (10 and 20°C) and for different durum wheat and bread wheat cultivars.
Figure S4. Relationship between germination rates in the main experiment and in the additional experiment, shown for individual PST isolates at 10 and 20°C.
Figure S5. Mean (±SE) difference ingermination rate between Southern and Northern isolates at 10 and 20°C, as measured in the main experiment (dark grey circles) and additional experiment (light grey circles).
Figure S6. Time to sporulation (latency time)and spore production of Northern and Southern PST isolates,on the Victo host cultivar.
Figure S7. Correlation between mean proportion of vir8 isolates in initial vir8/Avir8 spore mixture and estimated vir8 frequency, as measured on the cultivarCompare (Yr8).
Figure S8. Infectivity curves of the eight isolates used in field competitions, fitted to the observed values (A: Acalou cultivar, B: Victo cultivar), using a beta function.
Figure S9. Time course of predicted daily differences in infectivity (black lines) between competing Southern and Northern isolates in the field experiment in each year (2005, 2006) and location (North, South), as illustrated for one selected pair of isolates in each panel.
Table S1. Characteristics of 17 isolates ofPuccinia striiformis f.sp. tritici used in this study.
Table S2. Analysis of covariance of infectivity in the additional replicate experiment, testing effects of pathogen isolate, temperature (10 vs 20°C) and infectivity in the main experiment (covariate); all other interactions (P > 0.8) removed from model.
Table S3. Analysis of variance of infectivity, testing effects of temperature (10 vs 20°C), pathogen origin (North versus South), identity of pathogen isolate and replicate experiment.
Table S4. Analyses of variance of infectivity in additional replicate experiments, testing effects of pathogenorigin (North versus South), identity of pathogen isolate and host cultivar, at two incubation temperatures (10, 20°C).
Table S5. ancova of germination ratein additional replicate experiments, as a function of pathogenisolate, temperature (10 vs 20°C) and germination rate in main experiments (=covariate).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Bahri B, Kaltz O, Leconte M, de Vallavieille-Pope C, Enjalbert J. Tracking costs of virulence in natural populations of the wheat pathogen. Puccinia striiformis f.sp. tritici. BMC Evolutionary Biology. 2009a;9:1–12. doi: 10.1186/1471-2148-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahri B, Leconte M, Ouffroukh A, de Vallavieille-Pope C, Enjalbert J. Geographic limits of a clonal population of wheat yellow rust in the Mediterranean region. Molecular Ecology. 2009b;18:4165–4179. doi: 10.1111/j.1365-294X.2009.04267.x. [DOI] [PubMed] [Google Scholar]
- Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host–parasite interactions. Trends in Ecology & Evolution. 2008;23:678–685. doi: 10.1016/j.tree.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles RA, Flath K, Hovmøller MS, deVallavieille-Pope C. Breakdown of the Yr17 resistance to yellow rust of wheat in northern Europe. Agronomie. 2000;20:805–811. [Google Scholar]
- Bonman JM, Khush GS, Nelson RJ. Breeding rice for resistance to pests. Annual Review of Phytopathology. 1992;30:507–528. [Google Scholar]
- Boshoff WHP, Pretorius ZA, Van Niekerk BD. Establishment, distribution, and pathogenicity of Puccinia striiformis f.sp. tritici in South Africa. Plant Disease. 2002;86:485–492. doi: 10.1094/PDIS.2002.86.5.485. [DOI] [PubMed] [Google Scholar]
- Brown JKM, Hovmøller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- Brown JKM. 2006. Being more costly under conditions that are suboptimal for the fungus, as shown for B. graminis and M. graminicola. 13th Conference of the Spanish Society for Phytopathology, Murcia: 15.
- Burdon JJ, Thrall PH. Pathogen evolution across the agro-ecological interface: implications for disease management. Evolutionary Applications. 2008;1:57–65. doi: 10.1111/j.1752-4571.2007.00005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM. Epidemiology and control of stripe rust Puccinia striiformis f. sp. tritici on wheat. Canadian Journal of Plant Pathology. 2005;27:314–337. [Google Scholar]
- Claus S, Wernecke P, Pigla U, Dubsky G. A dynamic model describing leaf temperature and transpiration of wheat plants. Ecological Modelling. 1995;81:31–40. [Google Scholar]
- Dybdahl MF, Storfer A. Parasite local adaptation: red queen versus suicide king. Trends in Ecology and Evolution. 2003;18:523–530. [Google Scholar]
- Elahinia SA. Assessment of urediniospore germination of Puccinia striiformis at various temperatures on agar and detached leaves of wheat. Journal of Agricultural Science and Technology. 2000;1:41–47. [Google Scholar]
- Enjalbert J, Duan X, Leconte M, Hovmøller MS, de Vallavieille-Pope C. Genetic evidence of local adaptation of wheat yellow rust (Puccinia striiformis f. sp. tritici) within France. Molecular Ecology. 2005;14:2065–2073. doi: 10.1111/j.1365-294X.2005.02566.x. [DOI] [PubMed] [Google Scholar]
- Gandon S. Local adaptation and the geometry of host-parasite coevolution. Ecology Letters. 2002;5:246–256. [Google Scholar]
- Gandon S, Nuismer SL. Interactions between genetic drift, gene flow, and selection mosaics drive parasite local adaptation. The American Naturalist. 2009;173:212–224. doi: 10.1086/593706. [DOI] [PubMed] [Google Scholar]
- Gandon S, Van Zandt PA. Local adaptation and host-parasite interactions. Trends in Ecology & Evolution. 1998;13:214–216. doi: 10.1016/s0169-5347(98)01358-5. [DOI] [PubMed] [Google Scholar]
- Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE. Climate change effects on plant disease: genomes to ecosystems. Annual Review of Phytopathology. 2006;44:489–509. doi: 10.1146/annurev.phyto.44.070505.143420. [DOI] [PubMed] [Google Scholar]
- Gladders P, Langton SD, Barrie IA, Hardwick NV, Taylor MC, Paveley ND. The importance of weather and agronomic factors for the overwinter survival of yellow rust (Puccinia striiformis) and subsequent disease risk in commercial wheat crops in England. Annals of Applied Biology. 2007;150:371–382. [Google Scholar]
- Gomulkiewicz R, Drown DM, Dybdahl MF, Godsoe W, Nuismer SL, Pepin KM, Ridenhour BJ, et al. Dos and don’ts of testing the geographic mosaic theory of coevolution. Heredity. 2007;98:249–258. doi: 10.1038/sj.hdy.6800949. [DOI] [PubMed] [Google Scholar]
- Hall SR, Tessier AJ, Duffy MA, Huebner M, Caceres CE. Warmer does not have to mean sicker: temperature and predators can jointly drive timing of epidemics. Ecology. 2006;87:1684–1695. doi: 10.1890/0012-9658(2006)87[1684:wdnhtm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, Samuel MD. Climate warming and disease risks for terrestrial and marine biota. Science. 2002;296:2158–2162. doi: 10.1126/science.1063699. [DOI] [PubMed] [Google Scholar]
- Hau B, de Vallavieille-Pope C. Wind-dispersed diseases. In: Cooke BM, Gareth Jones D, Kaye B, editors. The Epidemiology of Plant Diseases. 2nd edn. Dordrecht: Springer; 2006. pp. 387–416. [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: a decade later. Annual Review of Ecology and Systematics. 1986;17:535–566. [Google Scholar]
- Hochberg ME, van Baalen M. Antagonistic coevolution over productivity gradients. The American Naturalist. 1998;152:620–634. doi: 10.1086/286194. [DOI] [PubMed] [Google Scholar]
- Hovmøller MS. Disease severity and pathotype dynamics of Puccinia striiformis f.sp tritici in Denmark. Plant Pathology. 2001;50:181–189. [Google Scholar]
- Hovmøller MS, Justesen AF. Rates of evolution of avirulence phenotypes and DNA markers in a northwest European population of Puccinia striiformis f. sp. tritici. Molecular Ecology. 2007;16:4637–4647. doi: 10.1111/j.1365-294X.2007.03513.x. [DOI] [PubMed] [Google Scholar]
- Hovmøller MS, Yahyaoui AH, Milus EA, Justesen AF. Rapid global spread of two aggressive strains of a wheat rust fungus. Molecular Ecology. 2008;17:3818–3826. doi: 10.1111/j.1365-294X.2008.03886.x. [DOI] [PubMed] [Google Scholar]
- Kaltz O, Shykoff JA. Local adaptation in host-parasite systems. Heredity. 1998;81:361–370. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Lafferty KD. The ecology of climate change and infectious diseases. Ecology. 2009;90:888–900. doi: 10.1890/08-0079.1. [DOI] [PubMed] [Google Scholar]
- Laine AL. Temperature-mediated patterns of local adaptation in a natural plant-pathogen metapopulation. Ecological Letters. 2008;11:327–337. doi: 10.1111/j.1461-0248.2007.01146.x. [DOI] [PubMed] [Google Scholar]
- Levins R. Evolution in Changing Environments. Princeton, NJ: Princeton University Press; 1968. [Google Scholar]
- Lopez-Pascua LC, Buckling A. Increasing productivity accelerates host-parasite coevolution. Journal of Evolutionary Biology. 2008;21:853–860. doi: 10.1111/j.1420-9101.2008.01501.x. [DOI] [PubMed] [Google Scholar]
- McDonald BA, Linde C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica. 2002;124:163–180. [Google Scholar]
- Milus EA, Line RF. Gene action for inheritance of durable, high-temperature, adult-plant resistance to stripe rust in wheat. Phytopathology. 1986;76:435–441. [Google Scholar]
- Milus EA, Seyran E, McNew R. Aggressiveness of Puccinia striiformis f. sp tritici isolates in the South-Central United States. Plant Disease. 2006;90:847–852. doi: 10.1094/PD-90-0847. [DOI] [PubMed] [Google Scholar]
- Milus EA, Kristensen K, Hovmøller MS. Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp tritici causing stripe rust of wheat. Phytopathology. 2009;99:89–94. doi: 10.1094/PHYTO-99-1-0089. [DOI] [PubMed] [Google Scholar]
- Niks RE, Rubiales D. Potentially durable resistance mechanisms in plants to specialised fungal pathogens. Euphytica. 2002;124:201–216. [Google Scholar]
- Nordskog B, Gadoury DM, Seem RC, Hermansen A. Impact of diurnal periodicity, temperature, and light on sporulation of Bremia lactucae. Phytopathology. 2007;97:979–986. doi: 10.1094/PHYTO-97-8-0979. [DOI] [PubMed] [Google Scholar]
- Nuismer SL, Gandon S. Moving beyond common-garden and transplant designs: insight into the causes of local adaptation in species interactions. The American Naturalist. 2008;171:658–668. doi: 10.1086/587077. [DOI] [PubMed] [Google Scholar]
- Papastamati K, van den Bosch F. The sensitivity of the epidemic growth rate to weather variables, with an application to yellow rust on wheat. Phytopathology. 2007;97:202–210. doi: 10.1094/PHYTO-97-2-0202. [DOI] [PubMed] [Google Scholar]
- Pfister SE, Halik S, Bergdahl DR. Effect of temperature on Thekopsora minima urediniospores and uredinia. Plant Disease. 2004;88:359–362. doi: 10.1094/PDIS.2004.88.4.359. [DOI] [PubMed] [Google Scholar]
- Qayoum A, Line RF. High-temperature, adult-plant resistance to stripe rust of wheat. Phytopathology. 1985;75:1121–1125. [Google Scholar]
- Quance MA, Travisano M. Effects of temperature on the fitness cost of resistance to bacteriophage T4 in Escherichia coli. Evolution. 2009;63:1406–1416. doi: 10.1111/j.1558-5646.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Rapilly F. Yellow rust epidemiology. Annual Review of Phytopathology. 1979;17:59–73. [Google Scholar]
- Ravigné V, Dieckmann U, Olivieri I. Live where you thrive: joint evolution of habitat choice and local adaptation facilitates specialization and promotes diversity. The American Naturalist. 2009;174:E141–E169. doi: 10.1086/605369. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. SAS® 9.2 Macro Language: Reference. Cary, NC: SAS Institute Inc; 2009. [Google Scholar]
- Thomas MB, Blanford S. Thermal biology in insect-parasite interactions. Trends in Ecology and Evolution. 2003;18:344–350. [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution. Chicago, IL: University of Chicago Press; 2006. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Oakeshott JG, Fitt G, Southerton S, Burdon JJ, Sheppard A, Russell RJ, et al. Evolution in agriculture: the application of evolutionary approaches to the management of biotic interactions in agroecosystems. Evolutionary Applications. 2011;4:200–215. doi: 10.1111/j.1752-4571.2010.00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Brevis JC, Chen XM, Khan I, Jackson L, Chicaiza O, Distelfeld A, et al. High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theoretical and Applied Genetics. 2005;112:97–105. doi: 10.1007/s00122-005-0109-x. [DOI] [PubMed] [Google Scholar]
- Vale PF, Little TJ. Measuring parasite fitness under genetic and thermal variation. Heredity. 2009;103:102–109. doi: 10.1038/hdy.2009.54. [DOI] [PubMed] [Google Scholar]
- Vale PF, Kaltz O, Salvaudon L, Fellous S. The role of the environment in the evolutionary ecology of host parasite interactions. Infection, Genetics and Evolution. 2008;8:302–305. doi: 10.1016/j.meegid.2008.01.011. Meeting report. Paris, 5th December 2007. [DOI] [PubMed] [Google Scholar]
- de Vallavieille-Pope C, Picard-Formery H, Radulovic S, Johnson R. Specific resistance factors to yellow rust in seedlings of some French wheat varieties and races of Puccinia striiformis Westend in France. Agronomie (Paris) 1990;10:103–113. [Google Scholar]
- de Vallavieille-Pope C, Huber L, Leconte M, Goyeau H. Comparative effects of temperature and interrupted wet periods on germination, penetration, and infection of Puccinia recondita f.sp. tritici and P. striiformis on wheat seedlings. Phytopathology. 1995;85:409–415. [Google Scholar]
- de Vallavieille-Pope C, Giosue S, Munk L, Newton AC, Niks RE, Ostergard H, Pons-Kuhnemann J, et al. Assessment of epidemiological parameters and their use in epidemiological and forecasting models of cereal airborne diseases. Agronomie. 2000;20:715–727. [Google Scholar]
- de Vallavieille-Pope C, Ali S, Leconte M, Enjalbert J, Delos M, Rouzet J. Virulence dynamics and regional structuring of Puccinia striiformis f. sp. tritici in France between 1984 and 2009. Plant Disease. 2011;96:131–140. doi: 10.1094/PDIS-02-11-0078. [DOI] [PubMed] [Google Scholar]
- Van der Plank JE. Disease Resistance in Plants. New York, NY and London: Academic Press; 1968. p. 206. [Google Scholar]
- Wan A, Zhao Z, Chen XM, He Z, Jin S, Jia Q, Yao G, et al. Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Disease. 2004;88:896–904. doi: 10.1094/PDIS.2004.88.8.896. [DOI] [PubMed] [Google Scholar]
- Wellings CR, McIntosh RA. Puccinia striiformis f.sp. tritici in Australia: pathogenic changes during the first 10 years. Plant Pathology. 1990;39:316–325. [Google Scholar]
- Wellings CR, Wright DG, Keiper F, Loughman R. First detection of wheat stripe rust in Western Australia: evidence for a foreign incursion. Australasian Plant Pathology. 2003;32:321–322. [Google Scholar]
- Wolfe MS, McDermott JM. Population-genetics of plant pathogen interactions – the example of Erysiphe-graminis hordeum-vulgare pathosystem. Annual Review of Phytopathology. 1994;32:89–113. [Google Scholar]
- Wolinska J, King K. Environment can alter selection in host-parasite interactions. Trends in Parasitology. 2009;25:236–244. doi: 10.1016/j.pt.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Yahyaoui AH, Hovmøller MS, Ezzahiri B, Jahoor A, Maatougui MH, Wolday A. Survey of barley and wheat diseases in the central highlands of Eritrea. Phytopathologia Mediterranea. 2004;43:39–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to generate the results presented can be found in the INRA server. The link is available here: http://moulon.inra.fr/deap/Mboup_2011_Evol_App_data.xls.


