Abstract
Identifying natural barriers to movements of hosts associated with infectious diseases is essential for developing effective control strategies. Raccoon rabies variant (RRV) is a zoonosis of concern for humans because its main vector, the raccoon (Procyon lotor), is found near residential areas. In Québec, Canada, all cases of RRV found in raccoons since 2006 were detected on the eastern side of the Richelieu River, suggesting that this river acts as a barrier to gene flow and thus the potential for RRV to spread. The objectives of this study were to characterize the genetic structure of raccoon populations and assess the effect of the Richelieu River on the population structure in southern Québec, Canada. We also evaluated whether RRV spread potential differed between sex and at a larger spatial scale. Our analyses revealed a weak signal of genetic differentiation among individuals located on each side of the Richelieu River. At a larger spatial scale, genetic structuring was weak. Our results suggest that rivers might not always efficiently restrain raccoon movements and spread of RRV. We suggest that the difference in genetic structure found between sexes can be partly explained by male movements during the breeding season in winter, when ice bridges allow passage over most rivers in Québec.
Keywords: genetic differentiation, isolation by distance, landscape genetics, microsatellites, Procyon lotor, raccoon rabies variant
Introduction
Understanding how environmental variables contribute to the emergence, distribution and persistence of infectious diseases is crucial to effectively predict and control their spread (Ostfeld et al. 2005). As several infectious diseases affecting wild animals are transmittable to humans (e.g. H5N1, rabies), predicting ‘why’, ‘where’ and ‘how far’ hosts disperse are key questions for wildlife managers and health agencies (Anderson and May 1991; Cullingham et al. 2009). Typically, the effect of environmental variables on animal population structure is assessed using landscape genetics methods, which provide information on the relationships between landscape features and potential for gene flow (Manel et al. 2003; Holderegger and Wagner 2006). As infectious diseases depend on their hosts for dispersal (Grenfell and Dobson 1995), several authors suggested that approaches from landscape genetics could be used to understand the potential for disease spread (Biek et al. 2007; Blanchong et al. 2008; Archie et al. 2009; Biek and Real 2010). This approach also presumes that diseased and healthy animals have similar probabilities of dispersal and average distance moved. Recent studies that applied landscape genetic methods suggest that the variability in the disease progression can often be explained by the presence of rivers, mountains and roads (Childs et al. 2000; Russel et al. 2006; Blanchong et al. 2008; Neaves et al. 2009). For example, research on the prevalence of chronic wasting disease (CWD) in white-tailed deer (Odocoileus virginianus) found that the presence of rivers and highways significantly reduced the extent of gene flow and limited the spread of CWD (Blanchong et al. 2008). Patterns of sex-specific dispersal can also influence the spatial genetic structure of a host population (Lawson Handley and Perrin 2007). In mammals, for example, most species have polygamous or promiscuous mating systems, where juvenile and adult males have a higher probability of dispersal compared to adult females (Greenwood 1980). Yet, few studies of landscape genetic have incorporated landscape variables and sex-specific patterns of dispersal to predict host population structure and risks of disease spread.
In this article, we applied a landscape genetic approach to evaluate the effects of landscape barriers and sex-biased dispersal on the potential for raccoon rabies variant (RRV) spread in southern Québec. In the case of RRV, the assumption that healthy animals provide a representative picture of dispersal patterns of infected specimens is realistic because behaviour of rabid animals is different from healthy ones only during the period of morbidity (between 3 and 8 days in raccoon, see McLean 1975). The RRV has a long incubation period [an average of 39–79 days for animals with natural exposure (McLean 1975)], and therefore, for most of the period during which raccoon are carrying rabies, the behaviour of infected animals is similar to healthy individuals (Rosatte et al. 2006). Similarity in behaviour between healthy and rabid animals was also found in striped skunks (Mephitis mephitis) by Greenwood et al. (1997). Thus, describing the way healthy hosts are organized in space can help providing insights on the potential for RRV spread.
Rabies is a zoonotic disease with a significant impact, as more than 55 000 human deaths occur every year as a result of it (World Health Organization 2012). While most of these cases are found in developing countries, rabies is a great concern for public health agencies worldwide. For example, in the USA, the costs for the control of rabies and postexposition treatment are estimated between 230 million and 1 billion US dollars annually (Rupprecht et al. 1995). In particular, the RRV has been in constant northward progression in North America over the last four decades (Curtis 1999). RRV is problematic because the distribution range of its main host, the raccoon (Procyon lotor), is linked to human presence, which increases the risk of spread to domestic animals and humans (Cullingham et al. 2009). In Canada, the first cases of the RRV were found in 1999 in Ontario (Rosatte et al. 2001), in 2000 in New Brunswick and in 2006 in Québec (Fig. 1). Government agencies in North America are thus aiming to develop cost-efficient methods to control the spread of this disease in wild populations and limit its negative impact on public health and finances. One of the avenues to explore is whether landscape elements (rivers, mountain ranges, etc.) have the potential to be used in concert with other control methods by restraining animal movement.
Figure 1.
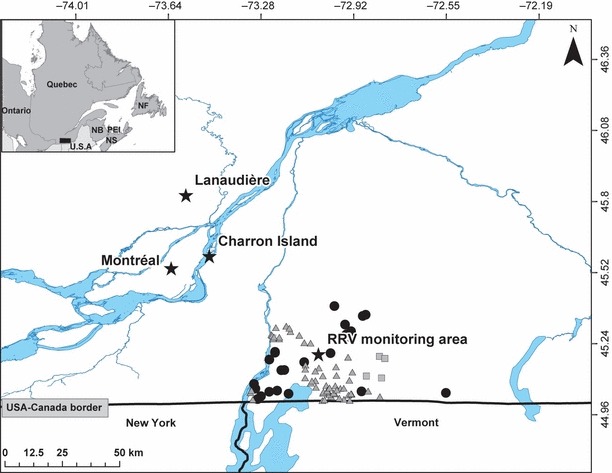
Map of the study area in southern Québec (indicated by a black rectangle on the map in the top left corner), Canada, showing the locations of the raccoon rabies variant (RRV) cases documented in raccoons by the enhanced provincial surveillance programme and the Canadian Food Inspection Agency’s passive surveillance programme (i.e. following human and pet postexposure investigations) in 2006 (filled squares, n = 4), 2007 (filled triangles, n = 59) and 2008 (filled circles, n = 26). The ‘stars’ indicate the centre of sampling sites.
Recent molecular evidence suggests that rivers are natural barriers to effective dispersal of terrestrial hosts potentially carrying rabies. Indeed, Cullingham et al. (2009) reported reduced gene flow across the Niagara River between Ontario (Canada) and New York (USA), implying that raccoons’ ability to disperse could be associated with the permeability of the river to animal movement. The degree to which a river is permeable to animal movement may vary with many factors, such as the width of the river, the rate of flow or human activities related to the river. For example, in Ontario, authorities mechanically prevent ice formation on the Niagara River during the winter, keeping water flowing heavily throughout the year (Cullingham et al. 2009). Thus, the extent to which rivers can limit raccoon movements and be effectively reinforced with oral vaccination to be used as immunological barriers to control rabies remains unclear. This is especially true in temperate regions where ice formation occurs in winter and facilitates animal movement. In southern Québec, where rabies surveillance has been conducted since 2007 (Rees et al. 2011; hereafter referred to as the RRV monitoring area), all cases of RRV found in raccoons since 2006 were detected on the eastern side of the Richelieu River (see Fig. 1). Here, we applied landscape genetic methods to investigate the potential of RRV spread in raccoons in this area. Specifically, we wanted to test whether the distribution of RRV cases on the east side of the Richelieu River was most likely explain by a barrier effect of this river or instead because of other factors, including the control activities that were deployed following the detection of the first cases in 2006. While these activities have clearly interfered with RRV spread in this area, cases were still detected until 2008 despite a large control effort. Yet, the absence of the disease from the west side of the Richelieu River during this period thus suggests that this river acts as a potential barrier to movement for this species (Fig. 1). To examine this possibility, we first used microsatellite markers to assess the population genetic structure of raccoons in the RRV monitoring area. We then evaluated whether disease spread potential was different depending on sex (because of differences in movements between genders) by assessing patterns of isolation by distance and the effect of the Richelieu River on the structure of relatedness detected for males and females. Then, we explored the genetic structure of raccoons at a larger spatial scale to get a better understanding of animal dispersal around the RRV monitoring area and to evaluate the potential for rabies spread in the province of Québec. To do so, we quantified the extent of genetic diversity and population structuring found in sites located in peripheral areas compared to that of the RRV monitoring area.
Materials and methods
Study area and raccoon DNA sampling
Our main study area is located in southern Québec, Canada, and covers approximately 21 300 km2. It spans North up to Lanaudière and is bordered West by the St. Lawrence River and to the South and East by the USA border (states of New York, Vermont, New Hampshire and Maine) (Fig 2). We randomly selected a subsample of raccoons for genetic analysis from all samples collected by the Ministère des Ressources naturelles et de la Faune of Québec (MRNFQ) and its partners during monitoring and control activities against RRV in raccoons, such as the recovery of road-killed animals (Rees et al. 2011) and field tests of oral vaccination efficiency in 2008 and 2009. Location of each animal collected was obtained using hand-held global positioning system (GPS), and a skin biopsy was collected from the ear with a 2-mm punch for subsequent genetic analyses. Samples were stored in 95% ethanol until DNA extraction. The sex of each animal collected was assessed both during field manipulations and using molecular techniques as described in Shaw et al. (2003) (Table 1). We used 273 animals genotyped from the RRV monitoring zone (Fig. 2) to conduct our so-called RRV monitoring area analyses. We also genotyped an additional 149 samples that were collected from areas peripheral to the zone were RRV cases were found to explore the genetic structure at a larger spatial scale. Samples collected in the peripheral areas yield from three sites on islands in the St. Lawrence River [Montréal (n = 20), Charron (n = 29) and Orléans (n = 52)] and one site on the North shore of the St. Lawrence River (Lanaudière, n = 48).
Figure 2.
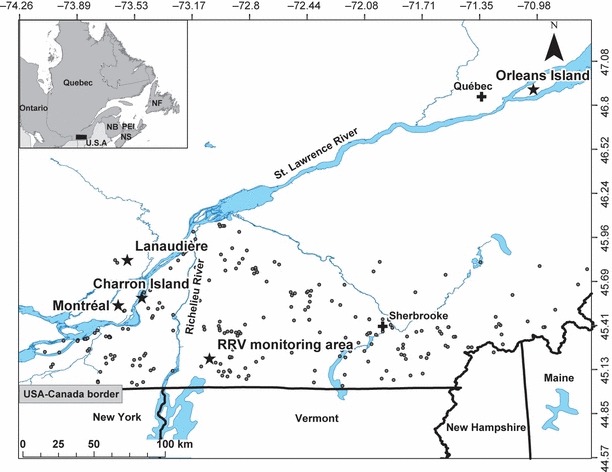
Location of the samples (grey circles) obtained from the raccoon rabies variant (RRV) monitoring area in southern Québec, Canada. Sites for the comparison between RRV monitoring and peripheral areas are also represented by stars, which are located in the centre of sampling locations (from West to East): Montréal (45.54°, −73.63°), Charron Island (45.58°, −73.48°), Lanaudière (45.80°, −73.47°), Orléans Island (46.90°, −71.01°). The ‘cross’ indicates major cities. Sample sizes for these sites are reported in the Materials and methods section.
Table 1.
Sample sizes of genotyped individuals with observed (HO) and expected (HE) heterozygosity, allelic richness (AR– using Montréal as the common sample size) and inbreeding coefficient (FIS) for sites sampled in southern Québec, Canada
| Lanaudière | Orléans Island | Montréal | Charron Island | RRV monitoring area | ||
|---|---|---|---|---|---|---|
| Locus | Sample size | 48 (27, 21) | 52 (25, 27) | 20 (11, 9) | 29 (13, 16) | 273 (121, 152) |
| PLOM15 | HO | 0.88 | 0.73 | 0.90 | 0.71 | 0.87 |
| HE | 0.88 | 0.73 | 0.79 | 0.71 | 0.86 | |
| AR | 10.62 | 5.94 | 9.00 | 8.56 | 11.32 | |
| FIS | 0.001 | 0.009 | −0.101 | 0.011 | −0.012 | |
| PLO2-117 | HO | 0.94 | 0.90 | 1.00 | 0.90 | 0.86 |
| HE | 0.91 | 0.83 | 0.90 | 0.88 | 0.92 | |
| AR | 14.48 | 10.04 | 13.00 | 12.23 | 15.79 | |
| FIS | −0.025 | −0.074 | −0.092 | −0.007 | 0.059 | |
| PLOM3 | HO | 0.71 | 0.67 | 0.65 | 0.65 | 0.78 |
| HE | 0.78 | 0.75 | 0.75 | 0.64 | 0.80 | |
| AR | 5.60 | 4.77 | 6.00 | 4.90 | 5.65 | |
| FIS | 0.087 | 0.118 | 0.158 | −0.002 | 0.010 | |
| PLOM 20 | HO | 0.88 | 0.71 | 0.90 | 0.90 | 0.83 |
| HE | 0.88 | 0.72 | 0.83 | 0.86 | 0.89 | |
| AR | 11.98 | 7.70 | 11.00 | 12.01 | 12.29 | |
| FIS | 0.01 | 0.024 | −0.054 | −0.022 | 0.054 | |
| PLO2-14 | HO | 0.83 | 0.75 | 0.90 | 0.72 | 0.88 |
| HE | 0.84 | 0.75 | 0.88 | 0.83 | 0.89 | |
| AR | 11.09 | 9.73 | 13.00 | 11.54 | 13.64 | |
| FIS | 0.017 | 0.013 | −0.003 | 0.161 | 0.002 | |
| PLOM2 | HO | 0.81 | 0.90 | 0.80 | 0.82 | 0.89 |
| HE | 0.81 | 0.78 | 0.75 | 0.83 | 0.88 | |
| AR | 9.74 | 6.36 | 9.00 | 7.91 | 10.40 | |
| FIS | −0.006 | −0.143 | −0.036 | 0.035 | 0.002 | |
| PLOM17 | HO | 0.73 | 0.58 | 0.75 | 0.79 | 0.77 |
| HE | 0.74 | 0.69 | 0.67 | 0.78 | 0.81 | |
| AR | 5.32 | 5.71 | 5.00 | 5.99 | 6.28 | |
| FIS | 0.013 | 0.177 | −0.100 | 0.003 | 0.052 | |
| PLM06 | HO | 0.55 | 0.91 | 0.80 | 0.59 | 0.68 |
| HE | 0.69 | 0.73 | 0.68 | 0.67 | 0.74 | |
| AR | 3.85 | 4.62 | 4.00 | 3.69 | 5.08 | |
| FIS | 0.195 | −0.225 | −0.156 | 0.142 | 0.096 | |
| PLM10 | HO | 0.88 | 0.81 | 0.85 | 0.75 | 0.83 |
| HE | 0.85 | 0.78 | 0.82 | 0.71 | 0.86 | |
| AR | 7.88 | 5.76 | 8.00 | 7.32 | 9.02 | |
| FIS | −0.025 | −0.027 | −0.013 | −0.034 | 0.029 | |
| PLM20 | HO | 0.69 | 0.73 | 0.70 | 0.48 | 0.73 |
| HE | 0.74 | 0.72 | 0.74 | 0.65 | 0.76 | |
| AR | 7.70 | 5.79 | 9.00 | 7.27 | 7.81 | |
| FIS | 0.066 | −0.011 | 0.081 | 0.279 | 0.065 | |
| Overall mean | HO | 0.79 | 0.77 | 0.83 | 0.73 | 0.81 |
| HE | 0.81 | 0.75 | 0.78 | 0.76 | 0.84 | |
| AR | 8.83 | 6.64 | 8.70 | 8.14 | 9.73 | |
| FIS | 0.033 | −0.014 | −0.032 | 0.057 | 0.036 |
RRV, raccoon rabies variant.
The numbers of females and males genotyped for each site are presented in parentheses (F, M). Lanaudière is located on the North shore of the St. Lawrence River (see Fig. 2).
DNA extraction and microsatellite analyses
DNA was extracted using a salting out protocol as described in Chambers and Garant (2010). The following ten microsatellite loci, developed specifically for raccoons, were selected based on their size and polymorphism (see Table S1 for details about loci): PLO-M2, PLO-M3, PLO-M15, PLO-M17, PLO-M20, PLO2-14, PLO2-117, PLM06, PLM10 and PLM20 (Cullingham et al. 2006; Siripunkaw et al. 2008).
Microsatellite loci amplification was performed using GeneAmp 9700 thermocyclers (Applied Biosystems, Foster City, CA, USA). The amplification conditions are described in Tables S2 and S3. PCR products were visualized using an AB 3130 capillary DNA sequencer (Applied Biosystems). A volume of 1 μL of PCR product was added to 8.9 μL of Hi-Di Formamide and 0.1 μL of GeneScan Liz 600 (Applied Biosystems). Allele size was scored using the software genemapper version 4.0 (Applied Biosystems).
Data analysis
We tested all loci for departure from Hardy–Weinberg equilibrium and linkage disequilibrium and obtained observed and expected heterozygosity for each locus and site using the software GenePop version 4.0 (Raymond and Rousset 1995). We used the software fstat version 2.9.3.2 (Goudet 1995) to calculate allelic richness (AR). Finally, we used cervus version 3.0.3 (Marshall et al. 1998; Kalinowski et al. 2007) to assess the presence of null alleles.
RRV monitoring area population genetic structure
We first quantified the extent of genetic differentiation among groups located on each side of the Richelieu River by calculating FST among them using fstat (Goudet 1995). We also assessed the maximum level of genetic differentiation among these two groups using G’ST (as in Hedrick 2005) as well as Jost’s D (Jost 2008). We then used the bayesian clustering software structure version 2.3.3 to estimate the most likely number of genetic clusters in the RRV monitoring area (Pritchard et al. 2000). We performed the analyses using a model with admixture, separate α for each population, allele frequencies correlated (using λ = 1.0) among genetic clusters and without using prior information on sampling location. For each value of K (number of clusters: from one to four), we ran five independent models with 250 000 iterations, plus a burn in period of 100 000 iterations. Means of the ln-probabilities of all independent runs for a given K were then calculated. We also used the software geneland version 3.3 (Guillot et al. 2005) and baps version 5.2 (Corander et al. 2004) to assess the number of genetic clusters within the RRV monitoring area, but because results were similar to those obtained with structure (results not shown), we only reported the latter. We also specifically ran structure with K = 2 and then assessed the proportion of individuals from each side of the Richelieu that were assigned to each cluster (i.e. the number of individuals genetically assigned to the western side of the river among those that were actually sampled on that side, and the same proportion was calculated for the eastern side). We then compared these proportions to the overall proportion of individuals assigned to each given cluster using binomial proportion tests.
To further explore the relative importance of the landscape in shaping the spatial distribution of genetic structure, we assessed the effect of linear geographic distance and that of the Richelieu River. We calculated the geographic distance (km) between pairs of individuals using the extension Hawth’s tool in ArcGIS version 3.2.2. We used Genepop to provide a first assessment of the isolation by distance (IBD) between pairs of individuals using the method described in Rousset (2000). We then used SPAGeDI version 1.3 (Hardy and Vekemans 2002) to estimate pairwise genetic relatedness (Rxy) between all sampled individuals with Wang’s estimator (Wang 2002) and used 1-Rxy as a measure of genetic distance between individuals. The presence of IBD among individuals was then further assessed using Mantel tests (Mantel 1967) implemented in fstat. We conducted analyses over all individuals and then separately for each sex to assess whether potential for RRV spread differs according to gender. Finally, we used Partial Mantel tests implemented in fstat to establish the strength and significance of the partial correlations between the presence of the Richelieu River, the geographic distance and the genetic distance among individuals. Again, these tests were performed over all individuals and separately for each sex to assess whether spread potential and effect of barriers were different depending on gender.
Comparisons between RRV monitoring areas and peripheral areas
We compared allelic richness (AR), observed heterozygosity and the inbreeding coefficient (FIS) among sites using analyses of variance (anovas) because none of the parameters significantly deviated from normality assumptions (Shapiro Wilk tests all P > 0.05. We performed these analyses using the software r version 2.0 (R Core Development Team, 2008). We also calculated pairwise genetic differentiation among sites using FST. Again, we tested for IBD, but this time assessing the relationships between the genetic differentiation (FST) and geographic distance (km) among sites. As individuals from each group were distributed over a large territory, we used the central sampling point of each group to calculate the geographic distances between them.
Results
Microsatellite polymorphism
A total of 422 raccoons were genotyped at ten microsatellites (Table 1). All loci were highly polymorphic, exhibiting from 7 to 37 alleles per locus with an average of 17.1 (±10.3 SD). Observed heterozygosity per locus ranged from 0.66 to 0.92 (Table 1). Expected heterozygosity ranged from 0.72 for PLM-06 to 0.93 for PLO2-117. Overall FIS within each sampling site ranged from −0.032 (Montréal) to 0.057 (Charron Island) (Table 1). FIS were significantly higher than expected in both Charron Island (FIS = 0.057, P = 0.035) and in the RRV area samples (FIS = 0.036, P = 0.001). No significant deviation from Hardy–Weinberg equilibrium or linkage disequilibrium was detected after Bonferroni correction. Null alleles were found at low frequencies within our data (<5%).
Genetic differentiation analyses
RRV monitoring area
The level of genetic differentiation estimated between the individuals sampled on the East and West sides of the Richelieu River was low but highly significant (FST = 0.009, P < 0.001). The maximum level of differentiation expected (G’ST) among these two groups was 0.053 and Jost’s D was 0.049. structure analyses, however, indicated the presence of a single genetic cluster within the RRV monitoring area (K = 1, estimated likelihood = −11413.2; Fig. 3). We found similar results when the analyses were conducted independently for each sex, with K = 1 for either males or females. Yet, when running structure with K = 2, we found that individuals from each side of the Richelieu River were grouped in separate clusters in proportions always higher than expected given the overall proportions of individuals assigned to each of the clusters (all P < 0.02, results not shown).
Figure 3.
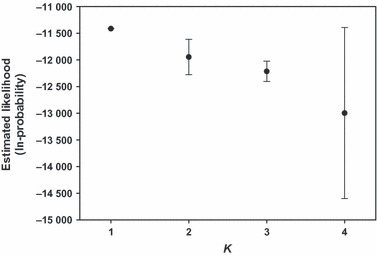
Mean estimated likelihood (ln-probability) of genetic clusters (K = 1–4) obtained from structure analyses within the raccoon rabies variant (RRV) monitoring area in southern Québec, Canada. Mean of five replicated analyses per cluster are presented with their standard deviations.
Analyses of IBD using Rousset’s (2000) genetic distance among individuals showed that within the RRV monitoring area, raccoon pairs that were more genetically related were, on average, also geographically closer to each other (slope, β = 0.0020, P < 0.017). This relationship was, however, only significant in females (males: β = 0.0015, P = 0.13; females: β = 0.0029, P = 0.009). Mantel tests conducted in fstat using pairwise genetic relatedness among individuals also showed a positive pattern of IBD over all individuals, although this relationship disappeared when the analyses were conducted on both sexes separately (Table 2). The results of the Partial Mantel tests suggested that animals located on the same side of the Richelieu River were more genetically related than individuals found on opposite sides. IBD disappeared after accounting for the presence of the river (Table 2). Analyses conducted in each sex separately suggested that the presence of the Richelieu River affects males and females differently. Females located on the same side of the river were significantly more related than females located on opposite sides, whereas no such differences were found in males (Table 2).
Table 2.
Mantel and Partial Mantel test results for the correlations between the geographic distance (km) and the presence of the Richelieu River with the pairwise estimated genetic distance among individual raccoons sampled in the raccoon rabies variant (RRV) monitoring zone in southern Québec, Canada
| Test | r2 | Explanatory variable | β | r | P-value |
|---|---|---|---|---|---|
| Mantel tests | Overall (r2 = 0.0001) | Geographic distance | 0.00003 | 0.0119 | 0.02 |
| Females (r2 = 0.0003) | Geographic distance | 0.00005 | 0.0170 | 0.14 | |
| Males (r2 = 0.0001) | Geographic distance | 0.00002 | 0.0066 | 0.44 | |
| Partial Mantel tests | Overall (r2 = 0.0005) | Geographic distance | 0.000001 | 0.0119 | 0.94 |
| Presence of Richelieu River | 0.00611 | 0.0189 | 0.001 | ||
| Females (r2 = 0.0015) | Geographic distance | −0.00002 | −0.0170 | 0.72 | |
| Presence of Richelieu River | 0.01143 | 0.0343 | 0.007 | ||
| Males (r2 = 0.0003) | Geographic distance | −0.000004 | −0.0066 | 0.90 | |
| Presence of Richelieu River | 0.00479 | 0.0151 | 0.10 |
Pairwise genetic distances were defined as 1 – Rxy and using the Rxy estimator of Wang (2002). The percentage of variation explained by the models (r2), the slopes (β), the partial correlation coefficients (r) and their associated P-values are reported.
Comparison between RRV monitoring areas and peripheral areas
The mean distance between sites was 125.5 km (range between 16 and 254 km), and the average FST value between them was 0.044 ± 0.006 (Table 3). The highest level of genetic differentiation was detected between Orléans and Charron Islands (FST = 0.099), and the lowest value was found between the Lanaudière region (North shore of the St. Lawrence River) and the Island of Montréal (FST = 0.021, Table 3). We found a signal of IBD among all our sites, as revealed by a significant positive correlation between the level of genetic differentiation and the geographic distance (r = 0.64, slope = 0.00021, P = 0.037).
Table 3.
Genetic differentiation (pairwise FST values) estimated with FSTAT between all sampled sites in southern Québec, Canada (above the diagonal) and geographic distance (in km) among sites (below the diagonal)
| Lanaudière | Orléans Island | Montréal | Charron Island | RRV monitoring area | |
|---|---|---|---|---|---|
| Lanaudière | – | 0.084 | 0.021 | 0.079 | 0.027 |
| Orléans Island | 226 | – | 0.094 | 0.099 | 0.054 |
| Montréal | 33 | 254 | – | 0.061 | 0.022 |
| Charron Island | 33 | 243 | 16 | – | 0.034 |
| RRV monitoring area | 86 | 200 | 90 | 74 | – |
RRV, raccoon rabies variant.
All pairwise FST values are significant (P < 0.05).
Analyses comparing genetic diversity revealed that allelic richness (AR) was significantly different among sites (F = 13.8, df = 4, P < 0.001). This difference was because of a lower allelic richness on Orléans Island (mean AR = 6.64 alleles) than in other sites (all P < 0.01). On the other hand, the RRV monitoring area harboured a higher AR compared to every other site (mean AR = 9.73 alleles, all P < 0.05). We found no difference in FIS (F = 1.88, df = 4, P = 0.14) or observed heterozygosity values (F = 2.15, df = 4, P = 0.09) among sites.
Discussion
The objectives of this study were to use landscape genetic approaches to investigate the potential of the Richelieu River to act as a natural barrier to RRV host movements that may be used to increase the efficiency of RRV control methods in the future and to evaluate the potential for RRV spread outside the current infected area. All RRV cases documented in raccoons in Québec from 2006 to 2008 were found on the East side of the Richelieu River suggesting that this river might restrict raccoon movements (see Rees et al. 2011). Accordingly, we found that there was low but significant genetic differentiation among groups located on either side of this river. However, the river appears to be a relatively weak barrier to gene flow, as we identified a single genetic cluster within the RRV monitoring area and found a signal of IBD among individuals over this area. Also, our larger scale investigation suggests that the genetic differentiation observed in the RRV monitoring area is relatively small. Similarly, the genetic differentiation of raccoons among islands and on the North shore of the St. Lawrence River was significant but relatively small, suggesting that in the absence of control operations, RRV could potentially spread into northern areas by crossing the St. Lawrence River, which is larger than the Richelieu River (Fig. 1).
RRV monitoring area population genetic structure
Previous studies based on spatial dynamics of terrestrial rabies hosts suggested that rivers could act as natural barriers slowing the spread of this disease (Smith et al. 2002; Russel et al. 2006; Arjo et al. 2008) and recent molecular evidence partly supported this contention (Cullingham et al. 2009). Our results indicate that rivers may not always act as impermeable barriers to gene flow, as we detected only a weak genetic substructuring in relation to the Richelieu River. However, the presence of some substructuring does suggest that the Richelieu River contributed to restricting RRV spread in that area, as all RRV cases in southern Québec were found on the East side of the Richelieu River (Fig. 1). Although comparisons between FST values stemming from different studies must be made cautiously (Meirmans and Hedrick 2011), it is interesting to note that the low level of genetic differentiation found in this study area is even weaker than the one reported in a study in the Niagara region conducted by Cullingham et al. (2009). A potential explanation for these low levels of genetic differentiation is that the presence of human infrastructure (such as bridges) could allow animals to cross-natural barriers. Cullingham et al. (2009), however, argued that walking or ‘hitch-hiking’ across bridges was unlikely to explain this low differentiation because the FST values were not correlated with the number of bridges present. In our study area, there are five bridges and two railways that could allow raccoons to cross the Richelieu River. Moreover, raccoons are known as pest animals, often relying on domestic garbage as food sources and habitations as shelters (Rosatte et al. 2010). Thus, humans can also promote raccoon movements via translocation of undesirable animals, a practice that should not be encouraged in the context of disease spread.
The weak level of genetic structure observed in our study suggests that over a longer time period, RRV would have been likely to spread west of the Richelieu River. There are a few hypotheses that can explain why over 3 years, the RRV in raccoons was not found on the west side of the Richelieu River, despite genetic evidences of apparent movements between these sites. First, rabies might induce costs (physiological, immunological, etc.) that could influence the capacity of dispersal and ultimately prevent the spread of infected raccoons across the Richelieu River. Obviously, rabid animals do exhibit drastic changes in their behaviour once they develop the disease symptoms (Rosatte et al. 2006). Anecdotal data on movements of five rabid raccoons, however, suggest that the distances moved by rabid raccoon are not different from those of nonrabid (see Rosatte et al. 2006). As the costs of the disease on animal movement would need to be extremely high to explain the observed RRV pattern in our study area, this hypothesis seems unlikely to explain our results. More studies focusing specifically on the behaviour of infected animals are needed to draw more general conclusions about their capacity for dispersal and the resulting impact on population structure. However, as rabid individuals are typically eliminated as quickly as possible because of obvious risks of disease spread, such studies might be difficult to conduct in the wild. A second hypothesis to explain the lack of RRV cases in raccoons on the west side of the Richelieu River is that the environmental conditions in this area may be unfavourable or suboptimal for the disease to spread (e.g. Arjo et al. 2008). Although possible, this also seems unlikely as landscape characteristics and host population densities appear fairly uniform across the region (J. Mainguy, unpublished data). A more likely explanation is that the observed spatial distribution of infected animals might only be transient and that without any control operations, the disease front would have eventually progressed at the same rate that it did historically across eastern North America (30–50 km per year, Childs et al. 2000), where it crossed several rivers of different widths. Indeed, as the Québec government was very active in preventing the disease from spreading in that region from 2006 and onward through population reduction, trap–vaccinate–release and oral rabies vaccination (Canac-Marquis et al. 2007; Guérin et al. 2008), it seems likely that both these control interventions and the presence of the river explained the observed distribution of RRV cases in raccoons between 2006 and 2008 in that region.
We also detected the presence of IBD among individuals within our study area. Indeed, we found that, on average, animals that were spatially close were also genetically more related to each other. Such a pattern has been reported for other carnivorous species. For example, Frantz et al. (2010) detected a strong pattern of IBD among European badgers (Meles meles) in western England. In our study area, however, further analyses revealed that the global pattern was weak, as IBD was only detected in females when analyses were conducted on each sex separately. Female genetic structure also seemed to be shaped by the presence of the Richelieu River. Indeed, a Partial Mantel test indicated that, when accounting for the presence of the Richelieu River, females located on the same side of the river were more related to each other than females located on different sides. While the use of Partial Mantel tests in such contexts has been questioned (see Raufaste and Rousset 2001; but see also Castellano and Balletto 2002; Legendre and Fortin 2010), our results are still concordant with previous empirical studies suggesting that female raccoons are often philopatric, while males are more likely to disperse (Gehrt and Fritzell 1998; Cullingham et al. 2009; Rosatte et al. 2010). The absence of a barrier effect of the Richelieu River for males could also be explained by the difference in mating strategies between the sexes. Indeed, as our study area is located at the northern edge of the distribution of this species (Gehrt 2003), the mating season occurs in winter (February–March) when most major rivers are covered with ice and snow. Although raccoons are mostly inactive during winter (Pitt et al. 2008), male raccoons are more likely to move during that period when searching for mating partners (F. Pelletier, unpublished results). Breeding dispersal (and therefore gene flow) of male raccoons is, therefore, less likely to be limited by rivers at this time of the year (Cullingham et al. 2009). Other studies on raccoons over a heterogeneous landscape in north-central Indiana, USA, found evidence of genetic structure over a relatively small scale and of male-biased dispersal (Dharmarajan et al. 2009). Altogether, these results emphasize the importance of considering landscape heterogeneity and behavioural differences in dispersal patterns between males and females when trying to understand patterns of genetic differentiation in mammals, as well as their respective potential impact on disease spread.
Comparison between the RRV monitoring area and peripheral areas
While our analyses conducted within the RRV monitoring area provided detailed information on gene flow within the region where RRV cases have been detected in Québec, our characterization of population genetic differentiation in relation to the peripheral area provides insights on the main potential paths for rabies propagation in Québec. At this larger scale, we found evidence of IBD thus suggesting that long distance migrants are not common enough to prevent genetic structuring within our system. Raccoons are medium-sized omnivorous mammals, and although some specimens have already been observed more than 200 km from their point of origin (Rosatte et al. 2007), it appears that the majority of raccoons from both sexes move on considerably shorter distances (<5 km, Rosatte et al. 2010), possibly explaining the rather homogeneous genetic structure observed within the bounds of the Québec RRV monitoring area. Dharmarajan et al. (2009) reported that typically, without the presence of major barriers to dispersal, approximately 10% of raccoons dispersed more than 20 km. A previous study, conducted in Pennsylvania, also found a strong signature of IBD among groups of raccoons using a landscape genetics approach (Root et al. 2009).
The greatest average level of genetic differentiation was found between Orléans Island and the other sites (mean FST = 0.083). Part of this differentiation is because of the distance separating Orléans Island from those other sites (mean = 231 km) and more importantly, to the fact that it is an island, which further isolates the raccoons inhabiting it from those living on the mainland. Orléans Island also harboured a low allelic richness, potentially suggesting a founder effect on this island, which might have further increased the genetic differentiation with the other sites examined in this study. Further detailed investigation with additional sampling sites located on each side of the St. Lawrence River is warranted before reaching a general conclusion on the strength of the isolation among populations and on the potential for RRV spread. Yet, our findings of significant levels of genetic differentiation among populations from each side of the St. Lawrence River contrast with the previous study conducted in Ontario by Cullingham et al. (2009), in which there was no such effect for this particular river. Such potentially conflicting results could likely be explained by the variation in the extent of river flow between the two study sites. In Québec, for example, the flow of the St. Lawrence River is stronger than the stretch of the river where the study was conducted in Ontario (see Cullingham et al. 2009). In winter, the flow of the St. Lawrence River in Québec remains important, and ice is generally carried away by the current, preventing both shores from being joined by an ice bridge. Additionally, the Canadian Coast Guard maintains maritime traffic in the waters of the St. Lawrence River through the winter. Such measures may have an important impact on the movements of raccoons in winter and spring. However, other parameters, such as differences in raccoon density, sex-specific dispersal patterns of animals within the available habitats of a given landscape, as well as shape differences in the landscapes among study sites, are important to consider in order to better assess the risks of disease spread (Cullingham et al. 2009; Rees et al. 2009).
Potential applications to the control of rabies spread in Québec
Identification of barriers to rabies host movements is important as this information can be used to improve disease containment strategies. Planning interventions, such as rabies vaccination (through capture or distribution of oral baits) in conjunction with natural barriers, would create a better buffer zone for limiting subsequent rabies epidemics (Rosatte et al. 2001). Our research only partially supports the suggestion that large rivers can be used for this purpose, at least in northern locations. Indeed, even a large river like the Richelieu River had a relatively weak effect on the patterns of relatedness and genetic substructuring within the RRV monitoring area, suggesting little impact on raccoon movement. Our results and those of Cullingham et al. (2009) also suggest that the degree of permeability of a river is likely to be affected by the presence of ice in winter. This result has two main consequences for modelling and controlling the propagation of infectious diseases such as RRV. First, the presence of rivers needs to be included in the investigation of potential barriers against disease spread, but especially in combination with information on the likelihood of ice-bridge formation during winter. The presence of ice bridges is likely to affect gene flow between populations and rivers with long periods of ice presence should consequently have a high permeability to raccoon movements. Second, if ice bridges affect river permeability, then one should expect that the use of rivers to target control efforts should be more efficient in regions where ice is unlikely to form during winter. In the Québec RRV monitoring area, the long winter period likely allows male raccoons to move from one shore to the other during the mating season, therefore, increasing the risk of RRV spread, although it has not occurred so far with the help of large-scale control programmes. Finally, as the RRV is also transmitted to striped skunks (Mephitis mephitis) in our study area and elsewhere (Guerra et al. 2003), future comparative studies should aim at quantifying the population structure of this species in the same region to better assess the potential spread of this multi-host disease and improve control activities against this zoonotic disease.
Acknowledgments
We are grateful to trappers, veterinaries and technicians from the Ministère des Ressources Naturelle et de la Faune du Québec (MRNFQ), the ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec (MAPAQ) and the Centre Québécois sur la Santé des Animaux Sauvages (CQSAS) for collecting and providing samples. In particular, we thank Frédérick Lelièvre, Pierre Canac-Marquis, Isabelle Rémillard and Denis Fournier for providing samples and data from sites located in the peripheral area of the RRV monitoring area. We are also grateful to Melody Porlier, Jennifer Chambers and Hélène Presseault-Gauvin for their support and help with genetic analyses. We acknowledge the financial contribution of the MRNFQ to this research. D.G and F.P. are funded by Natural Sciences and Engineering Research Council of Canada (NSERC) discovery grants and the Canada Foundation for Innovation. F.P. holds the Canada Research Chair in Evolutionary Demography and Conservation. D.G. FP. and J.M are funded by a research team grant provided by the Fonds Québécois de la Recherche sur la Nature et les Technologies (FQRNT).
Data archiving statement
Data for this study are available at: Dryad: doi: 10.5061/dryad.fp659276.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. GenBank accession number, 5′ and 3′ primer sequences [Forward (F) and Reverse (R)], repetition pattern, dye, allele size (in base pairs – bp), and annealing temperature (Ta) for microsatellite loci used in this study.
Table S2. Polymerase chain reaction reagent volume and concentrations (final volume of 10 µL per sample with 10 ng of DNA) for the ten microsatellite loci used in this study.
Table S3. Amplification conditions for the ten microsatellite loci used in this study.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford, UK: Oxford University Press; 1991. [Google Scholar]
- Archie EA, Luikart G, Ezenwa V. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends in Ecology and Evolution. 2009;24:21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Arjo WM, Fisher CE, Armstrong J, Boyd F, Slate D. Effect of natural barriers and habitat on the western spread of raccoon rabies in Alabama. Journal of Wildlife Management. 2008;72:1725–1735. [Google Scholar]
- Biek R, Real L. The landscape genetics of infectious disease emergence and spread. Molecular Ecology. 2010;19:3515–3531. doi: 10.1111/j.1365-294X.2010.04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceedings of the National Academy of Sciences, USA. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchong JA, Samuel MD, Scribner KT, Weckworth BV, Langenberg JA, Filcek KB. Landscape genetics and the spatial distribution of chronic wasting disease. Biology Letters. 2008;4:130–133. doi: 10.1098/rsbl.2007.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canac-Marquis P, Rioux R, Dicaire A, Rajotte D, Sirois C, Huot M, Guérin D, et al. 2007. p. 140. Le contrôle de la rage du raton laveur en Montérégie en 2006: rapport des opérations de terrain. Ministère des Ressources naturelles et de la Faune, Direction du développement de la faune, Direction de la protection de la faune, Direction de l’aménagement de la faune de Montérégie, Département de Santé publique de la Montérégie.
- Castellano S, Balletto E. Is the partial Mantel test inadequate? Evolution. 2002;56:1871–1873. doi: 10.1111/j.0014-3820.2002.tb00203.x. [DOI] [PubMed] [Google Scholar]
- Chambers J, Garant D. Determinants of population genetic structure in eastern chipmunks (Tamias striatus): the role of landscape barriers and sex-biased dispersal. Journal of Heredity. 2010;101:413–422. doi: 10.1093/jhered/esq029. [DOI] [PubMed] [Google Scholar]
- Childs JE, Curns AT, Dey ME, Real LA, Feinstein L, Bjornstad ON, Krebs JW. Predicting the local dynamics of epizootic rabies among raccoons in the United States. Proceedings of the National Academy of Sciences. 2000;97:13666–13671. doi: 10.1073/pnas.240326697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpää M. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004;20:2363–2369. doi: 10.1093/bioinformatics/bth250. [DOI] [PubMed] [Google Scholar]
- Cullingham CI, Kyle CJ, White BN. Isolation, characterization and multiplex genotyping of raccoon tetranucleotide microsatellite loci. Molecular Ecology Notes. 2006;6:1030–1032. [Google Scholar]
- Cullingham CI, Kyle CJ, Pond BA, Rees EE, White B. Differential permeability of rivers to raccoon gene flow corresponds to rabies incidence in Ontario, Canada. Molecular Ecology. 2009;18:43–53. doi: 10.1111/j.1365-294X.2008.03989.x. [DOI] [PubMed] [Google Scholar]
- Curtis A. Using a spatial filter and a geographic information system to improve rabies surveillance data. Emerging Infection Diseases. 1999;5:603–606. doi: 10.3201/eid0505.990501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmarajan G, Beasley JC, Fike JA, Rhodes OE. Population genetic structure of raccoons (Procyon lotor) inhabiting a highly fragmented landscape. Canadian Journal of Zoology. 2009;87:814–824. [Google Scholar]
- Frantz AC, Pope LC, Etherington TR, Wilson GJ, Burke T. Using isolation-by-distance-based approaches to assess the barrier effect of linear landscape elements on badger (Meles meles) dispersal. Molecular Ecology. 2010;19:1663–1674. doi: 10.1111/j.1365-294X.2010.04605.x. [DOI] [PubMed] [Google Scholar]
- Gehrt SD. Raccoons and allies. In: Feldhamer GA, Chapman JA, Thompson BC, editors. Wild Mammals of North America. 2nd edn. Baltimore, Maryland: Johns Hopkins University Press; 2003. pp. 611–634. [Google Scholar]
- Gehrt SD, Fritzell EK. Duration of familial bonds and dispersal patterns for raccoons in south Texas. Journal of Mammalogy. 1998;79:859–872. [Google Scholar]
- Goudet J. FSTAT version 1.2: a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Greenwood PJ. Mating systems, philopatry and dispersal in birds and mammals. Animal Behaviour. 1980;28:1140–1162. [Google Scholar]
- Greenwood RJ, Newton WE, Pearson GL, Schamber GJ. Population and movement characteristics of radio-collared striped skunks in North Dakota during an epizootic of rabies. Journal of Wildlife Disease. 1997;33:226–241. doi: 10.7589/0090-3558-33.2.226. [DOI] [PubMed] [Google Scholar]
- Grenfell BT, Dobson AP. Ecology of Infectious Diseases in Natural Populations. New York: Cambridge University press; 1995. [Google Scholar]
- Guérin D, Jolicoeur H, Canac-Marquis P, Landry F, Gagnier M. 2008. p. 148. Le contrôle de la rage du raton laveur en Montérégie en 2007: rapport des interventions terrestres et aérienne. Ministère des Ressources naturelles et de la Faune, Direction générale de l’expertise sur la faune et ses habitats.
- Guerra MA, Curns AT, Rupprecht CE, Hanlon CA, Krebs JW, Childs JE. Skunk and raccoon rabies in the eastern United States: temporal and spatial analysis. Emerging Infectious Diseases. 2003;9:1143–1150. doi: 10.3201/eid0909.020608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G, Mortier F, Estoup A. Geneland: a computer package for landscape genetics. Molecular Ecology Notes. 2005;5:712–715. [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Holderegger R, Wagner H. A brief guide to landscape genetics. Landscape Ecology. 2006;21:793–796. [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Lawson Handley LJ, Perrin N. Advances in our understanding of mammalian sex-biased dispersal. Molecular Ecology. 2007;16:1559–1578. doi: 10.1111/j.1365-294X.2006.03152.x. [DOI] [PubMed] [Google Scholar]
- Legendre P, Fortin M-J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Molecular Ecology Resources. 2010;10:831–844. doi: 10.1111/j.1755-0998.2010.02866.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology and Evolution. 2003;18:189–197. [Google Scholar]
- Mantel N. The detection of disease clustering and generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- McLean RG. Raccoon rabies. In: Baer GM, editor. The Natural History of Rabies. London, UK: Academic Press, Inc; 1975. pp. 53–77. [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: Fst and related measures. Molecular Ecology Resources. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Neaves LE, Zenger KR, Prince RIT, Eldridge MDB, Cooper DW. Landscape discontinuities influence gene flow and genetic structure in a large, vagile Australian mammal, Macropus fuliginosus. Molecular Ecology. 2009;18:3363–3378. doi: 10.1111/j.1365-294X.2009.04293.x. [DOI] [PubMed] [Google Scholar]
- Ostfeld RS, Glass GE, Keesing F. Spatial epidemiology: an emerging (or re-emerging) discipline. Trends in Ecology and Evolution. 2005;20:328–336. doi: 10.1016/j.tree.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Pitt JA, Larivière S, Messier F. Social organization and group formation of raccoons at the edge of their distribution. Journal of Mammalogy. 2008;89:646–653. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly PJ. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Raufaste N, Rousset F. Are partial Mantel tests adequate? Evolution. 2001;55:1703–1705. doi: 10.1111/j.0014-3820.2001.tb00689.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GenePop (version1.2): population-genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rees EE, Pond BA, Cullingham CI, Tinline RR, Ball D, Kyle CJ, White BN. Landscape modeling spatial bottlenecks: implications for raccoon rabies disease spread. Biology Letters. 2009;5:387–390. doi: 10.1098/rsbl.2009.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees EE, Bélanger D, Lelièvre F, Côté N, Lambert L. Targeted surveillance of raccoon rabies in Québec, Canada. Journal of Wildlife Management. 2011;75:1406–1416. [Google Scholar]
- Root JJ, Puskas RB, Fisher JW, Swope CB, Neubaum MA, Reeder SA, Piaggio AJ. Landscape genetics of raccoons (Procyon lotor) associated with ridges and valleys of Pennsylvania: implications for oral rabies vaccination programs. Vector-Borne and Zoonotic Diseases. 2009;9:583–588. doi: 10.1089/vbz.2008.0110. [DOI] [PubMed] [Google Scholar]
- Rosatte R, Donovan D, Allan M, Howes L-A, Silver A, Bennett K, Macinnes C, et al. Emergency response to raccoon rabies introduction into Ontario. Journal of Wildlife Management. 2001;37:265–279. doi: 10.7589/0090-3558-37.2.265. [DOI] [PubMed] [Google Scholar]
- Rosatte R, Sobey K, Donovan D, Bruce L, Allan M, Silver A, Bennett K, et al. Behavior, movements, and demographics of rabid raccoons in Ontario, Canada: management implications. Journal of Wildlife Diseases. 2006;43:589–605. doi: 10.7589/0090-3558-42.3.589. [DOI] [PubMed] [Google Scholar]
- Rosatte R, MacDonals E, Sobey K, Donovan D, Bruce L, Allan M, Silver A, et al. The elimination of raccoon rabies from Wolfe Island, Ontario: animal density and movements. Journal of Wildlife Diseases. 2007;43:242–250. doi: 10.7589/0090-3558-43.2.242. [DOI] [PubMed] [Google Scholar]
- Rosatte R, Ryckman M, Ing K, Proceviat S, Allan M, Bruce L, Donovan D, et al. Density, movements, and survival of raccoons in Ontario, Canada: implications for disease spread and management. Journal of Mammalogy. 2010;91:122–135. [Google Scholar]
- Rousset F. Genetic differentiation between individuals. Journal of Evolutionary Biology. 2000;13:58–62. [Google Scholar]
- Rupprecht CE, Smith JS, Fekadu M, Childs JE. The ascension of wildlife rabies: a cause for public health concern or intervention? Emerging Infection Diseases. 1995;1:107–114. doi: 10.3201/eid0104.950401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel CA, Real LA, Smith DL. Spatial control of rabies on heterogeneous landscapes. PLoS ONE. 2006;1:e27. doi: 10.1371/journal.pone.0000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CN, Wilson PJ, White B. A reliable molecular method of gender determination for mammals. Journal of Mammalogy. 2003;84:123–128. [Google Scholar]
- Siripunkaw C, Kongrit C, Faries KM, Monello RJ, Gompper ME, Eggert LS. Isolation and characterization of polymorphic microsatellite loci in the raccoon (Procyon lotor. Molecular Ecology Resources. 2008;8:199–201. doi: 10.1111/j.1471-8286.2007.01922.x. [DOI] [PubMed] [Google Scholar]
- Smith DL, Lucey B, Waller LA, Childs JE, Real LA. Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proceedings of the National Academy of Sciences. 2002;99:3668–3672. doi: 10.1073/pnas.042400799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2012. Human and animal rabies http://www.who.int/rabies/en/ (accessed on 4 January 2012)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this study are available at: Dryad: doi: 10.5061/dryad.fp659276.


