Abstract
Aleutian mink disease (AMD) is a prominent infectious disease in mink farms. The AMD virus (AMDV) has been well characterized in Europe where American mink (Neovison vison) are an introduced species; however, in North America, where American mink are native and the disease is thought to have originated, the virus’ molecular epidemiology is unknown. As such, we characterized viral isolates from Ontario free-ranging mink of domestic, hybrid, and wild origin at two proteins: NS1, a nonstructural protein, and VP2, a capsid protein. AMDV DNA was detected in 25% of free-ranging mink (45 of 183), indicating prevalent active infection. Median-joining networks showed that Ontario AMDV isolates formed two subgroups in the NS1 region and three in the VP2 region, which were somewhat separate from, but closely related to, AMDVs circulating in domestic mink worldwide. Molecular analyses showed evidence of AMDV crossing from domestic to wild mink. Our results suggest that AMDV isolate grouping is linked to both wild endogenous reservoirs and the long-term global trade in domestic mink, and that AMD spills back and forth between domestic and wild mink. As such, biosecurity on mink farms is warranted to prevent transmission of the disease between mink farms and the wild.
Keywords: Aleutian disease, American mink, disease transmission, domestic, epidemiology, molecular ecology, Neovison vison
Introduction
Aleutian mink disease (AMD) is a persistent and progressive parvovirus infection primarily affecting American mink (Neovison vison). Several strains of AMD, ranging from nonvirulent to highly virulent, have been identified, with several known to be highly virulent (Utah-1, Danish-K and Ontario isolates) (Bloom et al. 1980; Hadlow et al. 1983; Gottschalck et al. 1994). Highly virulent strains cause disease in all mink, whereas low-virulence isolates typically cause disease only in mink with the Aleutian coat color (Bloom et al., 1994; Schuierer et al. 1997). Clinical signs of progressive AMD infection in adult mink include excess plasma and gammaglobulins in the blood, inflammation of the glomeruli of the kidneys, decreased fertility, spontaneous abortion, and severe chronic immune dysfunction (Bloom et al., 1994; Henson et al. 1976). In newborn kits, the virus causes acute, rapidly progressing interstitial pneumonia, with extremely high mortality rates (Alexandersen et al. 1994). Aleutian disease transmission can be horizontal, via blood, feces, urine, saliva, and insect vectors, or vertical, through infection of kits by seropositive dams during the perinatal period (Alexandersen et al. 1985; Shen et al. 1973).
The AMD virus (AMDV) is the only member of the genus Amdovirus, subfamily Parvovirinae, family Parvoviridae (Murphy et al. 1995). In other parvoviruses, infection typically elicits a protective host response; however, in AMD infection, the host response actually contributes to the progression of the disease symptoms (Castelruiz et al. 2005; McKenna et al. 1999). Infection with AMDV elicits a strong immune response, resulting in high levels of non-neutralizing antibodies, which are unable to inhibit virus replication (Castelruiz et al. 2005). These non-neutralizing antibodies build up and form immune complexes that are deposited in tissue, leading to inflammation and subsequent disease progression (McKenna et al. 1999). As such, there is presently no effective treatment or vaccine, as vaccines developed thus far have caused mink to suffer from hyperacute disease (Aasted et al. 1998). On mink farms, the only effective means of eradicating the virus involves repeated anti-AMD antibody testing using counterimmunoelectrophoresis followed by the culling of seropositive individuals (Cho and Greenfield 1978).
The 5-kb linear single-stranded AMDV genome codes for three nonstructural proteins (NS1, NS2, and the putative NS3) and two structural (VP1, VP2) proteins (Bloom et al., 1994; Qiu et al. 2006). The major nonstructural protein of parvoviruses, NS1, is responsible for viral DNA replication, and regulation of transcription and some enzymatic activities (Best et al. 2003; Christensen et al. 1995). VP2 capsid proteins are responsible for capsid assembly and DNA packaging (Parrish and Kawaoka 2005). A role in pathogenicity and viral host range has been suggested for both the NS1 and VP2 proteins, as AMD isolates display a high degree of variability in both of these regions; however, these functions have yet to be conclusively demonstrated (Gottschalck et al. 1994; Oie et al. 1996; Olofsson et al. 1999). The AMDV genome is highly variable, and previous analyses of the virus in Europe suggest that strains can be divided into three main lineages, which do not appear to be based on virulence, geographic origin, or year of isolation (Knuuttila et al. 2009; Olofsson et al. 1999; Schuierer et al. 1997). Thus, it is unclear what governs the phylogenetic grouping of AMDV isolates, and whether North American isolates fit into the previously identified subtypes.
Since the domestication of American mink in the late 1800s for the fur industry, feral mink populations have become established across Europe and South America as a result of deliberate releases by animal rights activists and accidental escapes on farms (Gerell 1967; Joergensen 1985; Medina 1997; Kruska and Sidorovich 2003; Wildhagen 1956). These escaped domestic mink have had considerable negative effects on native biodiversity in these regions, both through predation and competition (e.g. Bonesi and Palazon, 2007; Ferreras and Macdonald 1999; Halliwell and Macdonald 1996). In North America, wild mink populations are declining, and escaped domestic mink may pose a serious threat to native mink through hybridization, competition, and disease transmission (Bowman et al. 2007; Kidd et al. 2009).
Aleutian mink disease is the most commercially important infectious disease affecting farmed mink worldwide, causing considerable economic losses to mink farmers through both reduced reproductive output and decreased pelt value (Hunter 2008). The prevalence of Aleutian disease on mink farms in Canada has not been accurately assessed; however, several voluntary surveys have reported that a ‘considerable number’ of mink farms in some parts of the country are AMD positive (Newman and Reed 2006; OARSC, 2004; OMAFRA, unpublished data). Voluntary testing in Ontario has found that the percentage of farms with AMD-positive reactors has been as low as 14% and as high as 60% between 1986 and 2006, although farm participation tends to be low (OMAFRA, unpublished data).
Aleutian disease exposure has been reported in free-ranging mink populations in Europe, where domesticated American mink are invasive (UK: Yamaguchi and Macdonald 2001; France: Fournier-Chambrillon et al. 2004; and Spain: Mañas et al. 2001), and the disease may be contributing to population declines of the native European mustelid, Mustela lutreola (Fournier-Chambrillon et al. 2004; Knuuttila et al. 2009). Aleutian disease is proposed to have been introduced to fur farms in Europe and Asia via importation of domestic American mink from North America for the fur farming industry (Mañas et al. 2001). Surprisingly, the prevalence of AMD in native North American mink populations has been largely ignored; however, we recently surveyed the free-ranging mink population in Ontario, Canada, and found high Aleutian disease seroprevalence in wild, escaped domestic, and wild-domestic hybrid mink (Nituch et al. 2011). We also found an association between AMD seroprevalence and the distribution of mink farms at both large- and small-spatial scales, suggesting that the farms are sources of disease spillover to wild populations (Nituch et al. 2011). Aleutian disease may have a negative impact on wild mink populations because of its effects on immune function, reproduction, and survival.
There have been few genetic analyses of AMDV; only a small number of isolates have been examined, sampled predominantly from regions where American mink are an introduced species (Gottschalck et al. 1994; Knuuttila et al. 2009; Schuierer et al. 1997; Olofsson et al. 1999; Shackelton et al. 2007). In North America, where American mink are native and the disease is thought to have originated, the prevalence of both active AMD infection and the AMD viral isolates present in the free-ranging population is unknown. To obtain a clearer picture of the molecular epidemiology and genetic diversity of AMDV in North American mink, we have characterized viral isolates obtained from free-ranging mink of both domestic and wild origin in Ontario. Our study had several objectives. First, we wished to identify viral strains in the free-ranging mink population. Second, we sought to compare the geographic distribution of these strains to potential sources and transmission routes, including mink farms and the commercial mink trade. We hypothesized that AMDV is spread between free-ranging domestic and wild mink, and between mink farms and wild mink. The first hypothesis predicts that free-ranging domestic and wild mink should be infected with similar AMDV strains. The second hypothesis predicts that if there are strains of AMDV common to the commercial mink trade, then we should also observe these strains in wild mink. Finally, we explored the potential origins of the virus. All previous AMDV studies have used either the NS1 gene or the VP2 gene. Thus, we included both NS1 and VP2 proteins in our analyses to allow for direct comparison with all previously characterized AMDV isolates.
Methods
Sample collection
Free-ranging mink (n = 206) were collected, primarily by fur trappers, within 19 counties across Ontario during winters 2005–2009. Counties varied in their mink farm density from 0 to 0.35 farms/100 km2. During necropsy, we collected hair or muscle tissue for DNA analyses and a spleen sample for AMDV genetic sequencing. We chose to use spleen samples for AMDV analyses as high concentrations of replicative forms of AMDV DNA have been previously found in spleen of AMD-infected mink (Bloom et al., 1985; Haas et al., 1988). During necropsy and sample collection, precautions were taken to avoid cross-contamination of samples.
AMDV sequencing and mink genotyping
We extracted DNA from an approximately 10 mg piece of tissue from each spleen sample using a QIAGEN DNeasy kit (Qiagen Inc., Mississauga, ON, Canada). Samples were extracted according to the manufacturer’s protocol, except for the last step when the samples were eluted with 70°C TE0.1 (1.0 m Tris–HCl, 0.5 m EDTA) rather than buffer AE (Qiagen Inc.).
We amplified nucleotide fragments of the hypervariable regions of both the AMDV NS1 and VP2 genes from the isolated DNA by polymerase chain reaction (PCR) using previously described primers (Oie et al. 1996; Olofsson et al. 1999). PCR was performed in 20-μL reaction mixtures containing 5 μL template DNA, 1× PCR buffer with (NH4)2SO4, 0.2 mm of each dNTP, 1.5 mm MgCl2, 0.5 μm of each primer, and 1 unit of Taq polymerase (Fermentas, Burlington, ON, Canada). We amplified all samples under the following conditions: 94°C for 5 min, 35 cycles of 94°C for 30 s, 65°C for 1 min (NS1) or 60°C for 1 min (VP2), and 72°C for 1 min, with a final 45 min elongation step at 60°C. Negative controls were used in each set of amplifications, and included normal mink spleen and reactions from which template was omitted. As a positive control, we used AMDV previously isolated from a known AMD-positive mink. All DNA and reagents were processed and manipulated in a remote laboratory suite where no AMDV molecular biology had been performed.
We subjected PCR products (10 μL) to agarose gel electrophoresis and visualized them under UV light following ethidium bromide staining to determine presence and relative quality of extracted viral DNA. A positive result was indicated by the presence of a DNA fragment of the expected size (approximately 500 bp). The AMDV DNA product from positive PCRs was purified with ExoSAP and precipitated in ethanol to remove excess salts. Sequencing reactions were performed using BigDye® Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Samples then were resuspended in 15 μL of Hi-Di Formamide, and both strands of the labeled DNA samples were run on an ABI 3730 automated sequencer (Applied Biosystems) at the Ontario Ministry of Natural Resources genetics laboratory at Trent University.
Mink were genotyped and assigned to population clusters (domestic, wild or hybrid) according to previously published methods (Kidd et al. 2009). Individuals were assigned to populations with a minimum membership of q ≥ 0.80, whereas all animals with q < 0.80 for both groups were considered hybrids.
Sequence analysis
We compared our AMDV sequences with AMDV NS1 and VP2 sequences available on GenBank (http://www.ncbi.nlm.nih.gov/entrez). The majority of the GenBank sequences were isolated from domestic mink on fur farms. Although we were unable to directly sample AMDV on mink farms in Ontario for our study, the GenBank samples were indicative of AMDV strains circulating in the commercial mink trade. The GenBank sequences originated from seven different countries, including many of the world’s largest producers of domestic mink pelts (Supporting Information). Hereafter, we refer to these GenBank sequences as global AMDV isolates. We edited all nucleotide sequences in BioEdit 7.0.5.3 (Hall 1999) and conducted multiple sequence alignment using MEGA 4.0 (Tamura et al. 2007). Percent nucleotide divergence values were also calculated using MEGA. Median-joining networks for both the NS1 and the VP2 regions were created in NETWORK 4.6 (http://www.fluxus-engineering.com). Phylogenetic trees were constructed using the maximum likelihood method, using bootstrap analyses of 1000 replicates in PhyML (Guindon et al. 2010).
Results
PCR
Aleutian mink disease viral DNA was detected using PCR in 25% (45 of 183) of free-ranging mink. The acquired NS1 sequences were edited to a length of 322 bp corresponding to nucleotide positions 601–922 of the complete sequence of the nonpathogenic cell-culture-adapted ADV-G genome. The acquired VP2 sequences were edited to a length of 531 bp corresponding to nucleotide positions 2725–3255 of ADV-G.
Sequence analysis
The NS1 gene nucleotide sequences of AMDV isolates from free-ranging Ontario mink had 82–100% identity to each other, and 83–89% identity to the cell-culture-adapted ADV-G genome. AMDV sequences from two mink sampled in Niagara were identical to one another at NS1, as were two mink from Essex and three mink from Timiskaming. One NS1 isolate from Essex (isolate 8) appeared to be highly divergent from other Ontario mink isolates. Isolate 8 was most closely related to another isolate from Essex, with 88% sequence identity, and shared as little as 82% sequence similarity with other Ontario isolates. Comparative sequence analyses of our AMDV isolates from free-ranging Ontario mink with previously characterized global AMDV isolates showed high sequence divergence, with up to 20% difference between nucleotide sequences in the NS1 region. The two most distantly related samples in our comparison of 127 AMDV isolates were from Wellington County, Ontario (isolate 29), and from China (CH1).
The VP2 gene nucleotide sequences of the newly sequenced strains had 89–100% identity to each other and were 90–96% homologous with ADV-G. Sequences from two mink sampled in York were identical to one another at VP2, as were two mink from Niagara and three mink from Timiskaming. As was the case with NS1, isolate 8 (from Essex) was also divergent from other Ontario isolates in the VP2 region. Isolate 8 was most closely related to an isolate from Niagara (isolate 22), with 93% sequence identity, and least related to a mink from Leeds and Grenville (isolate 37), with which it shared only 89% sequence identity. Comparison of our acquired AMDV sequences with published AMDV VP2 isolates found up to 11% difference between nucleotide sequences, with the two most distantly related samples being isolate 8 from Essex and a mink from China (CH4).
Phylogenetic analysis
NS1
The AMDV NS1 median-joining network (Fig. 1) demonstrated that Ontario AMDV isolates formed two main haplogroups, closely related to, but somewhat separate from global AMDV isolates. Isolates from wild, hybrid and free-ranging domestic mink sampled in Ontario appeared in both of these haplogroups, which exhibited some geographic clustering (Fig. 2). Ontario haplogroup I contained AMDV isolates from mink originating in southwestern Ontario (Essex, Niagara, and Wellington counties), whereas haplogroup II consisted of AMDV isolates from across the sampling region. Overall, including the global isolates, there appeared to be some clustering of haplotypes within countries; however, several cases of crossover between countries were present, such as between Denmark, Finland and Sweden, and between Sweden and the Netherlands. The AMDV NS1 maximum likelihood phylogeny (Fig. 3) also formed two main Ontario haplogroups, with haplogroup I again consisting of isolates from mink originating in southwestern Ontario (Essex, Niagara, and Wellington counties) and haplogroup II consisting of isolates from across the sampling region. Mink isolates 5 and 7, sampled in Ontario from free-ranging domestic mink, did not fit into either of the two Ontario haplogroups and appeared to be more closely related to isolates originating in Scandinavia and China.
Figure 1.
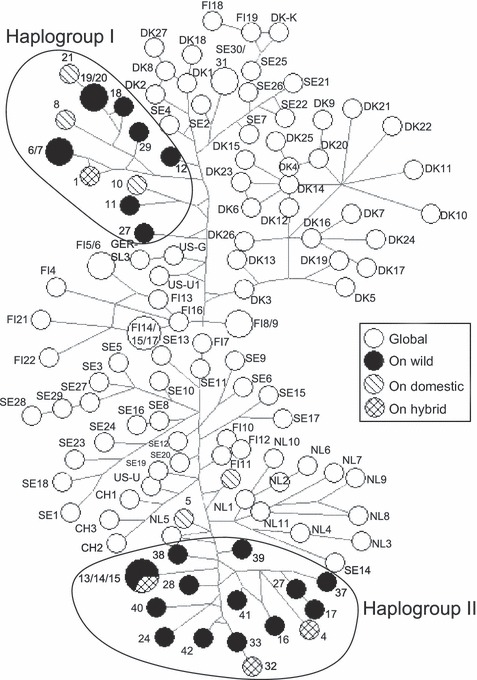
Median-joining network of 127 Aleutian mink disease virus (AMDV) isolates based on alignment of the 322-nucleotide fragment of the AMDV NS1 gene. Network nodes represent AMDV haplotypes, and node size indicates the number of AMDV isolates per haplotype, whereas branch lengths are proportional to the number of mutations between nodes. Most nodes contain only a single isolate. Isolates with only numeric labels were sampled in free-ranging mink from Ontario (ON), Canada, with different genetic ancestries (domestic, wild, and domestic–wild hybrid), and those with alphanumeric labels originated from the global trade of domestic mink. The country codes are CH (China), DK (Denmark), FIN (Finland), NL (Netherlands), SE (Sweden), and US (United States). See Table S1 for isolate descriptions.
Figure 2.
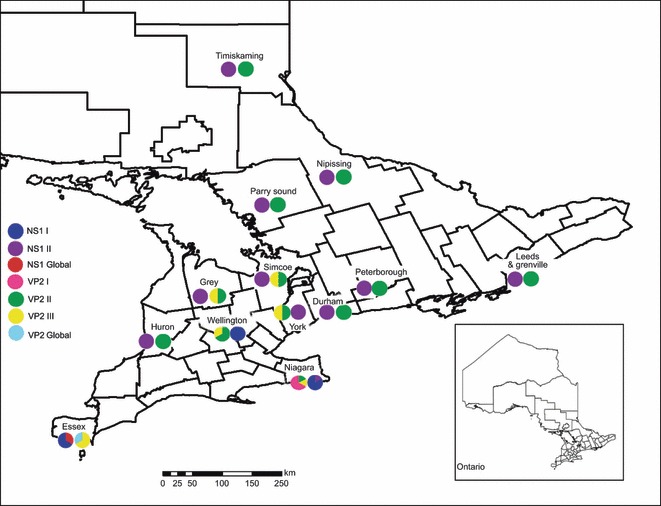
Geographic distribution of Aleutian mink disease virus NS1 and V21 haplogroups sampled in free-ranging mink in Ontario, Canada during 2005–2009. Frequency of haplogroups in each location is indicated by proportion of symbol.
Figure 3.
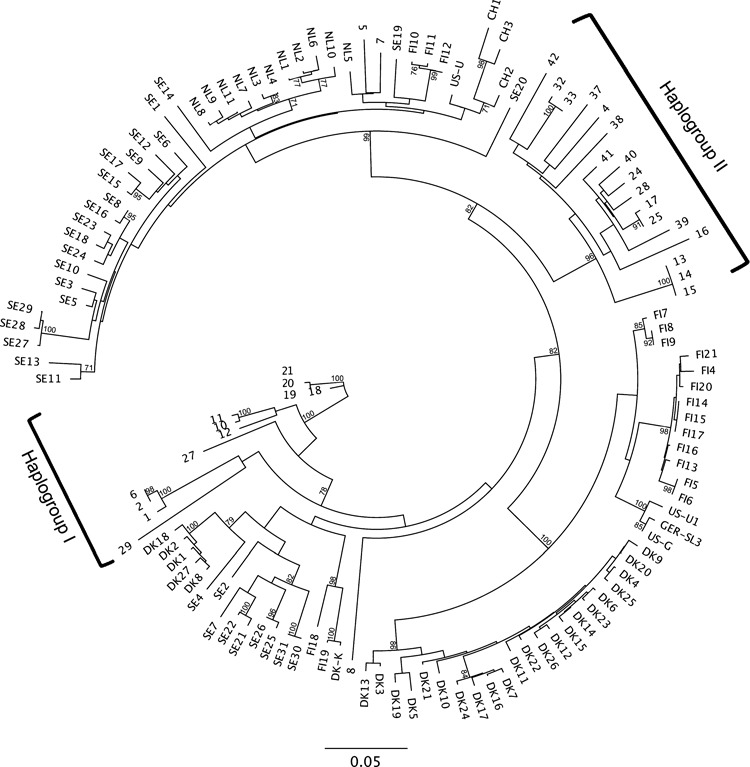
Maximum likelihood phylogeny of 127 Aleutian mink disease virus (AMDV) isolates based on alignment of the 322-nucleotide fragment of the AMDV NS1 gene. Bootstrap percentages (70% and above), from 1000 replications, are shown. Isolates with only numeric labels were sampled in free-ranging mink from Ontario (ON), Canada, with different genetic ancestries (domestic, wild, and domestic–wild hybrid), and those with alphanumeric labels originated from the global trade of domestic mink. The country codes are CH (China), DK (Denmark), FIN (Finland), NL (Netherlands), SE (Sweden), and US (United States). See Table S1 for isolate descriptions.
VP2
The AMDV VP2 median-joining network (Fig. 4) indicated that Ontario AMDV isolates formed three main haplogroups. Ontario haplogroup I contained isolates solely from mink within a single county on the Ontario–New York border (Niagara), haplogroup II consisted of ADV isolates from across the sampling region, and haplogroup III consisted primarily of isolates from southwestern Ontario (Fig. 4). Chinese and Irish AMDV isolates appeared to be closely related. As well, Ontario VP2 isolate 7 appeared to be more closely related to isolates from China, Ireland, and the United States than to other Ontario isolates. Isolate 7 shared 99% sequence identity with United States isolate US-U1 and shared 95% identity with its closest Ontario relative, isolate 5. The AMDV VP2 maximum likelihood phylogeny (Fig. 5) also formed three main Ontario haplogroups, with the same regional grouping as seen in the median-joining network. Isolates 5, 7, and 8, all from Ontario free-ranging domestic mink, did not appear to cluster within the three Ontario VP2 haplogroups.
Figure 4.
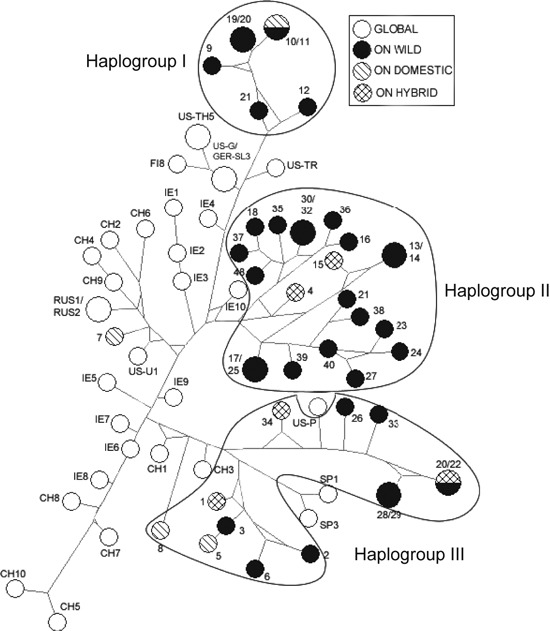
Median-joining network of 75 mink Aleutian mink disease virus (AMDV) isolates based on alignment of the 531-nucleotide fragment of the AMDV VP2 gene. Network nodes represent AMDV haplotypes and node size indicates the number of ADV isolates per haplotype, whereas branch lengths are proportional to the number of mutations between nodes. Most nodes contain only a single isolate. Isolates with only numeric labels were sampled in free-ranging mink from Ontario (ON), Canada, and those with alphanumeric labels originated from the global trade of domestic mink. The country codes are CH (China), FIN (Finland), GER (Germany), IE (Ireland), RUS (Russia), SP (Spain), and US (United States). See Table S1 for isolate descriptions.
Figure 5.
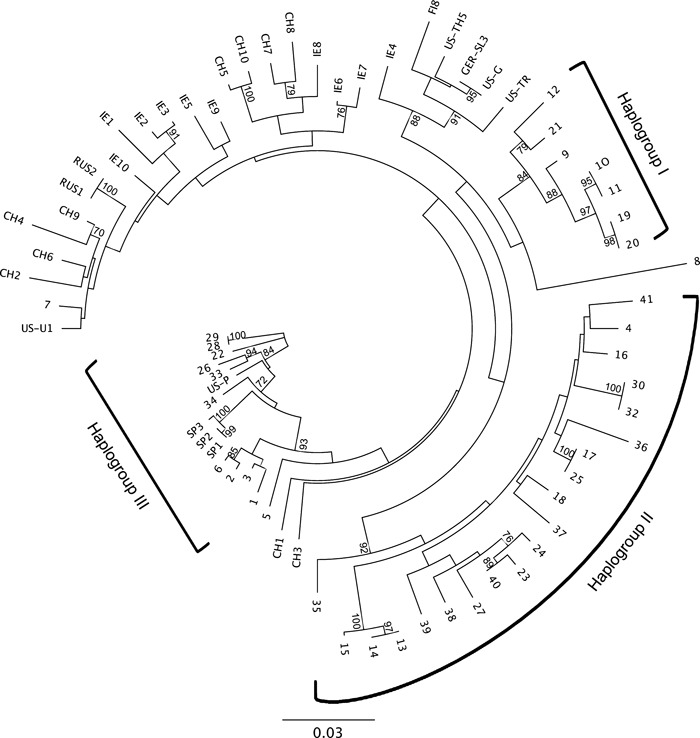
Maximum likelihood phylogeny of 75 Aleutian mink disease virus (AMDV) isolates based on alignment of the 531-nucleotide fragment of the AMDV VP2 gene. Bootstrap percentages (70% and above), from 1000 replications, are shown. Isolates with only numeric labels were sampled in free-ranging mink from Ontario (ON), Canada, and those with alphanumeric labels originated from the global trade of domestic mink. The country codes are CH (China), FIN (Finland), GER (Germany), IE (Ireland), RUS (Russia), SP (Spain), and US (United States). See Table S1 for isolate descriptions.
Discussion
We found evidence consistent with our hypotheses that AMDV transmission occurs between domestic and wild mink, and between mink farms and wild mink. In contrast, however, we also found evidence of previously undescribed haplogroups of AMDV occurring in free-ranging mink from Ontario. Our phylogenetic analysis demonstrated that Ontario AMDV isolates formed two genetic groups at the NS1 region and three genetic groups at the VP2 region. The presence of these Ontario-specific ADV haplogroups suggests clusters of both natural endogenous strains and strains that have evolved largely within the global trade of domestic mink. Ontario haplogroups, however, contained isolates from wild mink, as well as free-ranging domestic, and domestic–wild hybrid mink. This finding is consistent with our hypothesis that AMDV transmission occurs between free-ranging domestic and wild mink. Furthermore, we observed that some escaped domestic mink were infected with AMDV isolates that were closely related to strains from Asian, European, and US domestic mink (e.g. isolates 5 and 7), demonstrating the potential for mink farms to be a source of infection for wild mink. Infection from farms could occur either through contact with escaped mink or through contact with infected materials on farms.
The AMDV isolate from one Ontario mink (isolate 8 from Essex County), an escaped domestic mink, was somewhat divergent from other Ontario isolates, suggesting that an additional, previously unidentified Ontario AMDV haplogroup may exist. As such, we suggest that further molecular analysis of isolates from domestic mink, free-ranging mink, and other wild mustelids in Ontario and in the rest of North America is warranted.
Parvoviruses are commonly proposed to be highly conserved (Parrish 1995); however, our results show unusually high divergence (as much as 20%) between AMDV isolates in the NS1 region. Previous AMDV studies have reported differences of up to 14% in the NS1 region (Knuuttila et al. 2009). High sequence divergence can be suggestive of high mutation rates, as well as genetic recombination and re-assortment (Shackelton and Holmes 2004), which are typical of other ssDNA viruses (Shackelton and Holmes 2006). The degree of variability appears to differ throughout the viral genome, as the VP2 region was more conserved among different AMDV isolates (up to 11% difference).
Overall, there was strong agreement between networks constructed from different AMDV proteins; however, topology was dissimilar between the two networks with respect to a small number of nodes. For example, Essex and Niagara samples were grouped together in the NS1 tree but were placed in different haplogroups in the VP2 tree. Inconsistencies in genetic grouping between protein regions may be due to differential selective pressures on these two regions. As well, fewer isolates have been characterized using the VP2 region; therefore, gaps likely exist in the VP2 tree, as isolates from Denmark, Sweden, and the Netherlands were not available for this region. As such, further characterization of known isolates, using both the NS1 and VP2 regions, is necessary for a more comprehensive comparison of tree topologies from different AMDV proteins.
As in previous analyses, no clustering of AMDV isolates according to pathogenicity was detected, indicating that highly pathogenic strains may appear spontaneously from different genetic origins (Knuuttila et al. 2009; Olofsson et al. 1999), although the sequencing of larger segments of the virus may elucidate sites more critical for virulence. Because AMD pathogenicity does not appear to be related to AMDV genetic grouping, the pathogenicity of isolates circulating in free-ranging Ontario mink remains unknown.
Within Ontario, small-scale phylogeographical clustering of AMDV isolates was observed, with the presence of several regional specific haplogroups, such as VP2 I in Niagara County and NS1 I in Southwestern Ontario. Within Europe, AMD viral isolates also appear to display some regional clustering, however, with incidences of cross-over between countries, as well. The presence of global and several different Ontario-specific AMDV groups suggests that clusters of isolates may reflect the long history of importation and exportation of domestic mink. Mink farming began in the late 1800s in North America, and breeding mink were initially acquired from locally caught wild mink until trade between different farms, regions, and provinces became established (Joergensen 1985). During the 1930s and 1940s, the industry expanded to the Nordic regions of Europe, including Finland, Denmark, and Sweden (Knuuttila et al. 2009). The mink farmed in these regions are thought to have initially been imported from North America, which also likely inadvertently resulted in the transmission of the AMDV to Europe (Mañas et al. 2001). Once mink farming was established in Europe, mink were regularly imported between European and Asian countries, as well as from North America (Knuuttila et al. 2009). Thus, the long-term and extensive trade of mink for the fur farming industry has likely provided repeated opportunities for the transmission of different AMDV genetic groups from Ontario to the rest of the world with instances of importation and disease transmission back to Ontario.
Aleutian mink disease causes severe economic losses in the fur farming industry and has the potential to contribute to wild mink population declines. Molecular epidemiological studies are necessary to assess patterns of disease transmission, to increase our understanding of disease pathogenesis, to improve methods of disease detection, and to develop effective treatments and vaccines. Our study is the first to describe AMDV infection rates and the disease’s molecular epidemiology in free-ranging mink in North America. This is especially important because of the supposed North American origin of AMDV. At present, it remains unclear whether AMDV is native to wild American mink and was transmitted to captive mink populations, or if the virus originated in mink farms and wild mink were infected through accidental escapes and deliberate releases of infected captive animals. To obtain a clearer picture of the geographic origin of the virus, the common ancestor of both global and Ontario AMDV isolates warrants further study. In particular, isolates from North American mink farms and other free-ranging North American populations need to be characterized, ideally using several genes, to construct a more fully resolved AMDV tree and to determine the evolutionary origins of the virus. As well, classification of the pathogenicity of AMDV isolates circulating in free-ranging Ontario mink is required to assess the potential population impacts of the disease.
Aleutian mink disease has the potential to severely affect wild mink populations by reducing both productivity of adult females and survivorship of juveniles and adults; therefore, the identification of infection rates and viral isolates present in the wild is important for mink conservation. Overall, our results suggest the virus has a complex history involving both wild mink and the domestic mink trade. We observed evidence of transmission between free-ranging domestic and wild mink, and between mink farms and wild mink, but also of several separate haplogroups of AMDV from free-ranging mink in Ontario. It seems clear from our results, however, that AMDV transmission is influenced by the long-term global trade in domestic mink and that the disease spills back and forth between domestic and wild mink. As such, biosecurity and AMDV testing on mink farms is warranted to ensure the future sustainability of wild mink populations. This would benefit both wild mink and mink farmers, as there remains uncertainty about the relative directionality of virus transmission between farms and the surrounding wildlife.
Acknowledgments
Support for this research was provided by the Natural Sciences and Engineering Research Council of Canada (J.B. and A.I.S.-H.), the Wildlife Research and Development Section of the Ontario Ministry of Natural Resources (OMNR), and by an NSERC Scholarship to L.A.N. We thank B.A. Pond, C.J. Kyle, and two anonymous reviewers for comments on the work. Finally, we thank the Rabies Research and Development Unit of the OMNR, and the fur trappers who provided samples.
Data archiving statement
All sequence data used in this study are available from GenBank. Accession numbers are available on Supporting Information.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Aleutian mink disease virus NS1 isolates (n = 127) from American mink (Neovisonvison) included in a phylogenetic analysis.
Table S2. Aleutian mink disease virus VP2 isolates (n = 75) from American mink (Neovisonvison) included in a phylogenetic analysis.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Literature cited
- Aasted B, Alexandersen S, Christensen J. Vaccination with Aleutian mink disease parvovirus (AMDV) capsid proteins enhances disease, while vaccination with the major nonstructural AMDV protein causes partial protection from disease. Vaccine. 1998;16:1158–1165. doi: 10.1016/s0264-410x(98)80114-x. [DOI] [PubMed] [Google Scholar]
- Alexandersen S, Uttenthal-Jensen A, Hansen M, Aasted B. Experimental transmission of Aleutian disease virus (ADV) to different animal species. Acta Pathologica, Microbiologica et Immunologica Scandinavica. 1985;93:195–200. doi: 10.1111/j.1699-0463.1985.tb02876.x. [DOI] [PubMed] [Google Scholar]
- Alexandersen S, Larsen S, Aasted B, Uttenthal A, Bloom ME, Hansen M. Acute interstitial pneumonia in mink kits inoculated with defined isolates of Aleutian mink disease parvovirus. Veterinary Pathology. 1994;31:216–228. doi: 10.1177/030098589403100209. [DOI] [PubMed] [Google Scholar]
- Best SM, Shelton JF, Pompey JM, Wolfinbarger JB, Bloom ME. Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. Journal of Virology. 2003;77:5305–5312. doi: 10.1128/JVI.77.9.5305-5312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Race RE, Wolfinbarger JB. Characterization of Aleutian disease virus as a parvovirus. Journal of Virology. 1980;35:836–843. doi: 10.1128/jvi.35.3.836-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Race RE, Aasted B, Wolfinbarger JB. Analysis of Aleutian disease virus infection in vitro and in vivo: demonstration of Aleutian disease virus DNA in tissues of infected mink. Journal of Virology. 1985;55:696–703. doi: 10.1128/jvi.55.3.696-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Kanno H, Mori S. Aleutian mink disease: puzzles and paradigms. Infectious Agents of Disease. 1994;3:279–301. [PubMed] [Google Scholar]
- Bonesi L, Palazon S. The American mink in Europe: status, impacts, and control. Biological Conservation. 2007;134:470–483. [Google Scholar]
- Bowman J, Kidd AG, Gorman Schulte-Hostedde RM. Assessing the potential for impacts by feral mink on wild mink in Canada. Biological Conservation. 2007;139:12–18. [Google Scholar]
- Castelruiz Y, Blixenkrone-Moller M, Aasted B. DNA vaccination with the Aleutian mink disease virus NS1 gene confers partial protection against disease. Vaccine. 2005;23:1225–1231. doi: 10.1016/j.vaccine.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Greenfield J. Eradication of Aleutian disease of mink by eliminating positive counterimmunoelectrophoresis test reactors. Journal of Clinical Microbiology. 1978;7:18–22. doi: 10.1128/jcm.7.1.18-22.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Pedersen M, Aasted B, Alexandersen S. Purification and characterization of the major nonstructural protein (NS-1) of Aleutian mink disease parvovirus. Journal of Virology. 1995;69:1802–1809. doi: 10.1128/jvi.69.3.1802-1809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras P, Macdonald DW. The impact of American mink Mustela vison on water birds in the upper Thames. Journal of Applied Ecology. 1999;36:701–708. [Google Scholar]
- Fournier-Chambrillon C, Aasted B, Perrot A, Pontier D, Sauvage F, Artois M, Cassiede J, et al. Antibodies to Aleutian mink disease parvovirus in free-ranging European mink (Mustela lutreola) and other small carnivores from Southwestern France. Journal of Wildlife Diseases. 2004;40:394–402. doi: 10.7589/0090-3558-40.3.394. [DOI] [PubMed] [Google Scholar]
- Gerell R. Dispersal and acclimatization of the mink (Mustela vison Schreb.) in Sweden. Viltrevy. 1967;4:1–38. [Google Scholar]
- Gottschalck E, Alexandersen S, Storgaard T, Bloom ME, Aasted B. Sequence comparisons of the non-structural genes of four different types of Aleutian mink disease parvovirus indicates an unusual degree of variability. Archives of Virology. 1994;138:213–231. doi: 10.1007/BF01379127. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haas L, Lochelt M, Kaaden OR. Detection of Aleutian disease virus DNA in tissues of naturally infected mink. Journal of General Virology. 1988;69:705–710. doi: 10.1099/0022-1317-69-3-705. [DOI] [PubMed] [Google Scholar]
- Hadlow WJ, Race RE, Kennedy RC. Comparative pathogenicity of four strains of Aleutian disease virus for pastel and sapphire mink. Infection and Immunity. 1983;41:1016–1023. doi: 10.1128/iai.41.3.1016-1023.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Halliwell E, Macdonald D. American mink (Neovison vison) in the Upper Thames catchment: relationship with selected prey species and den availability. Biological Conservation. 1996;76:51–56. [Google Scholar]
- Henson JB, Gorham JR, McGuire TC, Crawford TB. Pathology and pathogenesis of Aleutian disease. Frontiers of Biology. 1976;44:175–205. [PubMed] [Google Scholar]
- Hunter B. Mink health and disease: challenges and accomplishments, a Canadian perspective. Scientifur. 2008;32:200–204. [Google Scholar]
- Joergensen G. Mink Production. Tjele, Denmark: Scientifur; 1985. [Google Scholar]
- Kidd AG, Bowman J, Lesbarrères D, Schulte-Hostedde AI. Hybridization between escaped domestic and wild American mink (Neovison vison) Molecular Ecology. 2009;18:1175–1186. doi: 10.1111/j.1365-294X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- Knuuttila A, Uzcátegui N, Kankkonen J, Vapalahti O, Kinnunen P. Molecular epidemiology of Aleutian mink disease virus in Finland. Veterinary Microbiology. 2009;133:229–238. doi: 10.1016/j.vetmic.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Kruska DCT, Sidorovich VE. Comparative allometric skull morphometrics in mink (Mustela vison Schreber, 1777) of Canadian and Belarus origin; taxonomic status. Mammalian Biology – Zeitschrift für Säugetierkunde. 2003;68:257–276. [Google Scholar]
- Mañas S, Ceaña JC, Ruiz-Olmo J, Palazón S, Domingo M, Wolfinbarger JB, Bloom ME. Aleutian mink disease parvovirus in wild riparian carnivores in Spain. Journal of Wildlife Diseases. 2001;37:138–144. doi: 10.7589/0090-3558-37.1.138. [DOI] [PubMed] [Google Scholar]
- McKenna R, Olson NH, Chipman PR, Baker TS, Booth TF, Christensen J, Aasted B, et al. The three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. Journal of Virology. 1999;73:6882–6891. doi: 10.1128/jvi.73.8.6882-6891.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina G. A comparison of the diet and distribution of southern river otter (Lontra provocax) and mink (Mustela vison) in southern Chile. Journal of Zoology (London) 1997;242:291–297. [Google Scholar]
- Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GR, Mayo MA, et al. Virus taxonomy. In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GR, Mayo MA, et al., editors. Sixth Report of the International Committee on Taxonomy of Viruses. New York, Springer Verlag, Wien, Austria: Plenum Press; 1995. pp. 169–178. [Google Scholar]
- Newman SJ, Reed A. A national survey for Aleutian disease prevalence in ranch mink hers in Canada. Scientifur. 2006;30:33–40. [Google Scholar]
- Nituch LA, Bowman J, Beauclerc KB, Schulte-Hostedde AI. Mink farms predict Aleutian disease exposure in wild American mink. PLoS ONE. 2011;6:e21693. doi: 10.1371/journal.pone.0021693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oie KL, Durrant G, Wolfinbarger JB. The relationship between capsid protein (VP2) sequence and pathogenicity of Aleutian mink disease parvovirus (ADV): a possible role for raccoons in the transmission of ADV infections. Journal of Virology. 1996;70:852–861. doi: 10.1128/jvi.70.2.852-861.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson A, Mittelholzer C, Berndtsson LT, Lind L, Mejerland T, Beldk S. Unusual, high genetic diversity of Aleutian mink disease virus. Journal of Clinical Microbiology. 1999;37:4145–4149. doi: 10.1128/jcm.37.12.4145-4149.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontario Animal Research and Services Committee (OARSC) Ontario Fur Research and Services Sub-Committee Strategic Report. Guelph, ON: Ontario Veterinary College (OVC), University of Guelph; 2004. [Google Scholar]
- Parrish CR. Molecular epidemiology of parvoviruses. Seminars in Virology. 1995;6:415–418. [Google Scholar]
- Parrish CR, Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. The Annual Review of Microbiology. 2005;59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Burger LR, Pintel D. The transcription profile Aleutian mink disease virus in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. Journal of Virology. 2006;80:654–662. doi: 10.1128/JVI.80.2.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuierer S, Bloom ME, Kaaden OR, Truyen U. Sequence analysis of the lymphotropic Aleutian disease parvovirus ADV-SL3. Archives of Virology. 1997;142:157–166. doi: 10.1007/s007050050066. [DOI] [PubMed] [Google Scholar]
- Shackelton LA, Holmes EC. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends in Microbiology. 2004;12:458–465. doi: 10.1016/j.tim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Shackelton LA, Holmes EC. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. Journal of Virology. 2006;80:3666–3669. doi: 10.1128/JVI.80.7.3666-3669.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton LA, Hoelzer K, Parrish CR, Holmes EC. Comparative analysis reveals frequent recombination in the parvoviruses. Journal of General Virology. 2007;88:3294–3301. doi: 10.1099/vir.0.83255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DT, Gorham JR, Harwood RF, Padgett GA. The persistence of Aleutian disease virus in the mosquito Aedes fitchii. Arch Gesamte Virusforsch. 1973;40:375–381. doi: 10.1007/BF01242558. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wildhagen A. Present distribution of North American mink in Norway. Journal of Mammalogy. 1956;37:116–117. [Google Scholar]
- Yamaguchi N, Macdonald DW. Detection of Aleutian disease antibodies in feral American mink in southern England. Veterinary Record. 2001;149:485–488. doi: 10.1136/vr.149.16.485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data used in this study are available from GenBank. Accession numbers are available on Supporting Information.


