Abstract
Cytomegalovirus (CMV) major immediate–early protein 1 (IE1) has multiple functions and is important for efficient viral infection. As does its counterpart in human CMV, murine CMV (MCMV) IE1 also functions as a disruptor of mouse-cell nuclear domain 10 (ND10), where many different gene-regulation proteins congregate. It still remains unclear how MCMV IE1 disperses ND10 and whether this dispersion could have any effect on viral replication. MCMV IE1 is 595 aa long and has multiple functional domains that have not yet been fully analysed. In this study, we dissected the IE1 molecule by truncation and/or deletion and found that the H2B homology domain (amino acid sequence NDIFERI) is required for the dispersion of ND10 by IE1. Furthermore, we made additional deletions and point mutations and found that the minimal truncation in the H2B homology domain required for IE1 to lose the ability to disperse ND10 is just 3 aa (IFE). Surprisingly, the mutated IE1 still interacted with PML and co-localized with ND10 but failed to disperse ND10. This suggests that binding to ND10 key protein is essential to, but not sufficient for, the dispersal of ND10, and that some other unknown mechanism must be involved in this biological procedure. Finally, we generated MCMV with IFE-deleted IE1 (MCMVdlIFE) and its revertant (MCMVIFERQ). Although MCMVdlIFE lost the ability to disperse ND10, plaque assays and viral gene production assays showed that the deletion of IFE did not increase viral replication in cell culture. We conclude that the dispersion of ND10 appears not to be important for MCMV replication in a mouse-cell culture.
Introduction
Biological and pathogenic similarities between murine cytomegalovirus (MCMV) and other CMVs make MCMV infection in mice the most commonly used small-animal model for CMV studies (Reddehase et al., 2008). Interactions of MCMV with infected host cells start immediately after viral particles come into contact with cells, with much attention paid to and study of the immediate–early (IE) stage (Maul & Negorev, 2008; Münch et al., 1988, 1991, 1992). At this stage, input tegument proteins and IE gene products are required to shut off host gene expression and replication and cause cell-cycle arrest (Wiebusch & Hagemeier, 1999; Wiebusch et al., 2008). The large IE transcription unit is activated by tegument proteins, possibly through the sequestration of Daxx and histone deacetylase (HDAC) (Hwang & Kalejta, 2007; Saffert & Kalejta, 2006, 2007; Tang et al., 2003; Taylor & Bresnahan, 2005), and the IE transcript is alternatively spliced (Stenberg, 1996). The two major spliced products, IE1/2 in human cytomegalovirus (HCMV) and IE1/3 in MCMV (Keil et al., 1987; Messerle et al., 1992; Stenberg, 1996), appear to be necessary for the activation of early promoters in the tightly regulated transcription cascade (Ghazal et al., 2005; Sinclair & Sissons, 1996). Most of the early proteins play an important role in the replicative process and are synthesized before the start of viral DNA replication. IE proteins have been shown to autoregulate their promoter, with IE1 enhancing transcription from the major IE promoter (MIEP) and IE2/IE3 downregulating it (Bühler et al., 1990; Cherrington & Mocarski, 1989; McElroy et al., 2000; Messerle et al., 1992; Scully et al., 1995; Stenberg, 1996; Stenberg & Stinski, 1985; Stenberg et al., 1990). In many of the DNA viruses studied, the transcription of the major IE (MIE) gene occurs at nuclear domain 10 (ND10), and viral DNA replication shows a preference for initiating at this nuclear substructure (Tang et al., 2000, 2003).
First observed by electron microscopy almost 50 years ago, ND10 [also called promyelocytic leukaemia (PML) oncogenic domain – POD – or PML bodies] was originally identified as being nuclear dense granular bodies (Kremer bodies) (Van Damme et al., 2010). Many ND10 components have been demonstrated to have a repressive effect on viral gene expression and viral replication (reviewed by Saffert & Kalejta, 2008). However, the exact nature of the function of ND10 as a nuclear structure is still unknown (Tavalai & Stamminger, 2008). PML is a scaffold protein with a tripartite motif that consists of an RBCC motif with a conserved RING domain, one or two B-box zinc-finger domains, and an α-helical coiled-coil domain (Borden, 2008; Borden & Culjkovic, 2009). PML is essential for the formation of ND10 and is the matrix protein of the structure; PML-knockout cells lack ND10, while PML-rescued cells restore the ND10 structures (Ishov et al., 1999; Zhong et al., 2000). SUMOlaytion (Sumo, small ubiquitin-like molecule) plays an important role in the interaction of PML with other ND10 components (Ishov et al., 1999). Sumoylated PML is able to recruit other proteins (such as SP100, Daxx, CBP, etc.) into ND10, and the desumoylation of PML by SENPs disperses ND10 (Ishov et al., 1999; Shen et al., 2006). Previously, we and others found that ND10s were the preferred places for some DNA viruses to dock their DNA, replicate their genome and transcribe some genes (Everett et al., 2004, 2007; Tang et al., 2000, 2003), which supports the hypothesis that ND10 might have protective effects on virus-replicated DNA and transcribed pre-mRNA. On the other hand, several lines of study have shown that ND10 components, including SP100, PML and Daxx, are all viral gene repressors and negatively affect viral replication (Cosme et al., 2011; Everett & Chelbi-Alix, 2007; Everett et al., 2006; Jiang et al., 2011; Ling et al., 2005, 2008; Tavalai et al., 2008), which suggests that ND10 is a defensive focus against viral replication. Therefore, the effects of ND10 as nuclear structures on viral infection are still unclear.
Several viruses encode gene products that disperse ND10. Herpes simplex virus (HSV-1)-encoded ICP0 disperses ND10 by degrading PML (Everett et al., 1998). For CMVs (including MCMV and HCMV), IE1 has been identified as dispersing ND10 by an as-yet unknown mechanism, but it is not able to degrade PML (Ahn & Hayward, 1997; Kang et al., 2006; Kelly et al., 1995; Korioth et al., 1996; Lee et al., 2004). The induction of PML desumoylation by HCMV IE1 reported by Lee et al. (2004) needs to be investigated for MCMV IE1.
IE1 of CMV has been shown to be non-essential for viral replication for both MCMV and HCMV (Ghazal et al., 2005; Greaves & Mocarski, 1998) at a high m.o.i., although the MCMV IE3 protein or the HCMV-equivalent, IE2, is essential for the activation of early proteins and for viral replication (Bühler et al., 1990; Cherrington & Mocarski, 1989; McElroy et al., 2000; Scully et al., 1995; Stenberg, 1996; Stenberg & Stinski; 1985; Stenberg et al., 1990). We previously showed that the interaction of MCMV IE1 with HDAC reduces HDAC deacetylation activity, suggesting a mechanism to silence viral transcription (Tang & Maul, 2003). It has been reported that HCMV IE1 has the same effect on HDAC (Nevels et al., 2004). Therefore, IE1 may counter the cellular repression of the MCMV MIEP by mechanisms distinct from the previously proposed direct regulation of transcription (Cherrington & Mocarski, 1989). MCMV IE1 has the ability to enhance viral replication in vivo (Ghazal et al., 2005), and its regulatory effects on cellular genes involved in nucleotide metabolism are not critical for viral replication (Wilhelmi et al., 2008). MCMV IE1 contains a 7 aa region (aa 135–141, NDIFERI) that is similar to that of histone protein H2B (aa 68–74) (Münch et al., 1991; Wang et al., 1996) but is not related to the DNA and chromatin binding properties of IE1 (Münch et al., 1991). In the present study, we show that the NDIFERI region is required for IE1 to disperse ND10. We further dissect IE1 and identify other domains that are important for ND10 dispersal. The minimal motif required for the dispersal of ND10 by IE1 is aa 137–139 (IFE), and the dispersive effect of IE1 on ND10 is not important for either viral replication or gene expression.
Results
Gene mutational analysis of the binding and dispersion of ND10 by IE1
The ability to disperse ND10 is one of the properties of IE1 (Ahn & Hayward, 1997; Ishov et al., 1997; Kelly et al., 1995; Korioth et al., 1996; Tang & Maul, 2003). To determine the IE1 motif required for this property, several IE1 deletion mutants were produced (Fig. 1, left; Supplementary Methods). Since the mAbs used to identify IE1 recognize an epitope between amino acids 430 and 530, some mutants with deletions in this area were detected using a GFP tag at the amino terminus of all constructs. A Western blot was performed to confirm the expression of mutant IE1 products after transfection into NIH/3T3 cells. After stripping, the same membrane was probed first with anti-IE1 antibody (top) and then with anti-GFP antibody (bottom). The products of the IE1 mutants were always seen as multiple bands, regardless of which antibody was used as a probe (Fig. 1, right). It is not clear whether these products arose from differential splicing or were post-translational effects.
Fig. 1.
(Left) Schematic outline of the IE1 deletion mutants produced. (Middle) Different activities tested. Numbers in the ‘dispersion’ column indicate whether the respective mutants dispersed ND10 after transfection; numbers in the ‘binding’ column indicate whether the IE1 dots colocalized with ND10. Fifty IE1-expressing cells were selected for each mutant. (Right) Expression of each plasmid in NIH/3T3 cells after transfection is shown by Western blot by using anti-IE1 antibody followed by stripping and reprobing with anti-GFP antibody.
We then analysed the ability of the mutants to bind to and disperse ND10 (Fig. 1, centre, which summarizes the data from 50 analysed cells). We transfected each mutant IE1 plasmid into NIH/3T3 cells overnight. After fixation and permeabilization, cells were stained for ND10 by using anti-PML antibody. Fig. 2(a) shows that wild-type (wt) IE1 colocalizes with ND10 when less IE1 is produced (Fig. 2a, rightmost cell; seen in 10/50 cells) and disperses ND10 (left cell) when IE1 is present at higher concentrations (seen in 40/50 cells). The fact that ND10 was dispersed in all cells that were transfected with wt IE1 for 72 h (not shown) confirmed our notion that the 20 % of cells where IE1 co-localized with ND10 had lower levels of IE1 expression. Deleting exon 2 (dl1–36, Fig. 2b) and nearly 200 aa at the carboxy terminus (dl400–595, Fig. 2h) had no effect on the normal co-localization and dispersion of ND10.
Fig. 2.
Immunohistochemical analysis of ND10-dispersion capabilities of IE1 deletion mutants in transfected NIH/3T3 cells. Cells were stained for the GFP-tagged IE1 deletion mutants (green) and PML (red) as an indicator of ND10 integrity. (a) Cells transfected with the wt IE1 plasmid show that IE1 binds to PML at the beginning of IE1 production (rightmost cell) but disperses ND10 when more IE1 is produced (leftmost cell). (b) Deleted exon 2 (aa 1–36) indicating the ability of wt IE1 to disperse ND10. (c–g and i) Cells transfected with the indicated mutants show loss of binding (no colocalization of red and green signals during early IE1 production) and dispersion capability, as indicated by the red puncta remaining in the cells that are bright green during late IE1 production. (h) Cells transfected with mutants carrying a deletion of approximately 200 aa from the carboxy terminus show the same capability of binding to PML and dispersing ND10 as does wt IE1. (j–l) Cells transfected with IE1 with aa 135–141 deleted show binding in the upper-left cell (as indicated by co-localized red and green signals in the puncta) but no dispersion of ND10, as indicated by the red puncta still visible in the leftmost cell despite high overexpression of this mutant. Bars, 10 µm.
Any deletions made between aa 37–400, except the deletion of aa 135–141, resulted in the loss of both binding and dispersion capabilities (Fig. 2c–g and i). Most deletions appeared to affect both binding and dispersion. Interestingly, the small 7 aa histone 2B homology-region deletion mutant (pgfpie1_dl135–141, Fig. 2j, k and l) retained ND10 binding ability [as shown in the nucleus on the right top of Fig. 2(j, k and l), the lower part of the nucleus is magnified in the boxes to show the co-localization of the IE1 dots and ND10], but did not, even when highly overexpressed, disperse this structure. These findings suggest that two domains, aa 37–99 and aa 350–400, determine the ND10 binding ability of IE1, and the domain affected by the 7 aa deletion (aa 135–141) determined the dispersion of ND10. Also, the binding of IE1 to PML (or intermediaries) seems necessary for the dispersion of ND10, but it is not sufficient for this dispersion.
Pinpointing the IE1 motif within the H2B homology region that is essential for ND10 dispersion
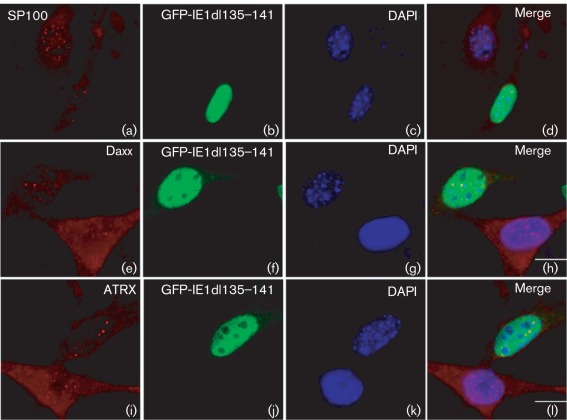
Previous studies of IE1 function by our and other groups have shown its activities of binding to non-specific DNA, in vitro interaction with histones, interaction with ND10 proteins and dispersion of ND10, and transactivation of cellular genes (such as the thymidylate synthase and ribonucleotide reductase genes) involved in dNTP biosynthesis (Gribaudo et al., 2000; Lembo et al., 2000; Münch et al., 1988, 1991, 1992; Tang & Maul, 2003; Wilhelmi et al., 2008). None of these functions have been mapped to the H2B homology region of IE1. Here we show the importance of the H2B homology region of IE1 for dispersing ND10 (Fig. 2). We further confirmed that IE1 with deletions in the H2B homology region also lost the ability to disperse other ND10 components, such as SP100 (Fig. 3a–d), Daxx (Fig. 3e—h), and ATRX (Fig. 3i—l).
Fig. 3.
Immunohistochemical analysis of ND10 component dispersion capabilities of IE1 with deletion of H2B homology domain (aa 135–141) in transfected NIH/3T3 cells. (a–d) Effect of IE1dl135–141 on SP100; (e–h) effect of IE1dl135–141 on Daxx; (i–l) effect of dl135–141 on ATRX. Bars, 10 µm.
Since the H2B homology region contains 7 aa (NDIFERI), we used deletion mutations to further map out the minimal region required for dispersing ND10 (Fig. 4, Table 1 and Supplementary Table S1, available in JGV Online). We made a series of deletion mutants by using overlapping PCR (Table 1 and Supplementary Table S1), and immunofluorescence assays (IFA) were performed to analyse the effects of these smaller deletion mutations on ND10 dispersal. As can be seen in Fig. 4, pgfpie1_dl137–139 lost the ability to disperse ND10, whereas the dual deletion mutations pgfpie1_dl137–138 and pgfpie1_dl138–139 did not. Therefore, the minimal region that is required for dispersing ND10 was mapped to aa 137–139 (IFE).
Fig. 4.
Determination of the minimal motif in the H2B homology region required for IE1 to disperse ND10 in transfected NIH/3T3 cells. (a) The H2B homology domain is shown with nucleotide sequence numbers 135–141 (top); nucleotides and their encoded amino acids (NDIFERI) with various deletion mutations in the H2B homology region of IE1 are shown below. (b) Immunohistochemical analysis of ND10 dispersion capabilities of IE1 deletion mutants in transfected NIH/3T3 cells. PML puncta (left) show the existence of ND10 that can be dispersed by pgfpie1_dl137–138 (upper panel) and pgfpie1_dl_138–139 (lower panel) but not by pgfpie1_dl137–139 (middle panel), as indicated by PML puncta remaining when the GFP–IE1 signal is strong (middle panels). DAPI staining (right) shows the nucleic acid outline of each of the cells highlighted in this figure. Bar, 10 µm.
Table 1. Mutagenesis within the H2B homology region of IE1.
The wt nucleotide sequence of the H2B homology region and sequences flanking it are shown in the header row of the table. The amino acids of the H2B homology domain and their positions within IE1 are also shown in the header row. The nucleotide sequences altered in the mutants indicated are shown in boldface type.
| Mutant | ctctgtcagctagcc | aat gat atc ttc gag cgc atc | gaaagacaacgc | ND10 dispersion |
| 135 136 137 138 139 140 141 | ||||
| N D I F E R I | ||||
| pgfpie1_dl137–139 | ctctgtcagctagcc | aat gat cgc atc | gaaagacaacgc | − |
| pgfpie1_dl135–139 | ctctgtcagctagcc | cgc atc | gaaagacaacgc | − |
| pgfpie1_dl139–141 | ctctgtcagctagcc | aat gat atc ttc | gaaagacaacgc | + |
| pgfpie1_dl137 | ctctgtcagctagcc | aat gat ttc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_dl138 | ctctgtcagctagcc | aat gat atc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_dl139 | ctctgtcagctagcc | aat gat atc ttc gtg cgc atc | gaaagacaacgc | + |
| pgfpie1_I137F | ctctgtcagctagcc | aat gat gtc ttc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_F138V | ctctgtcagctagcc | aat gat atc gtc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_E139V | ctctgtcagctagcc | aat gat atc ttc gtg cgc atc | gaaagacaacgc | + |
| pgfpie1_F138C | ctctgtcagctagcc | aat gat atc tgc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_F138I | ctctgtcagctagcc | aat gat atc atc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_F138L | ctctgtcagctagcc | aat gat atc ctc gag cgc atc | gaaagacaacgc | + |
| pgfpie1_F138Y | ctctgtcagctagcc | aat gat atc tac gag cgc atc | gaaagacaacgc | + |
| pgfpie1_F138S | ctctgtcagctagcc | aat gat atc tcc gag cgc atc | gaaagacaacgc | + |
Next, we were curious as to whether a single amino acid deletion or a point mutation could cause IE1 to lose its ability to disperse ND10. We first utilized an overlapping PCR method by designing internal primers, in which the desired single deletion or point mutation was included, to construct different plasmids, as shown as in Table 1. After transfecting the plasmids into NIH/3T3 cells overnight, the cells were fixed and an IFA was performed. We found that all of the single amino acid mutants of IE1 could still disperse ND10 (results summarized on the right of Table 1).
Interaction with PML, but not dispersion of ND10, is important for the activation of the MCMV MIEP by IE1
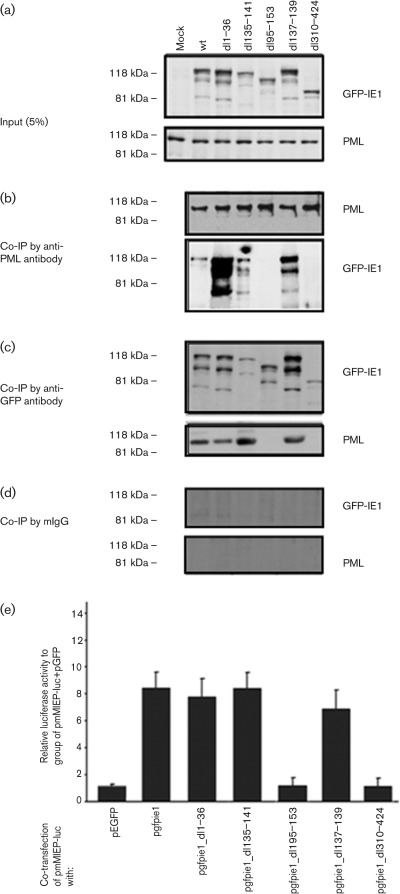
Previously, we found that IE1 can interact with PML and other ND10 components (Tang & Maul, 2003). To determine the domains that are important for this interaction with PML, we transfected our mutated IE1-expressing plasmids (including wt, dl1–36, dl135–141, dl95–153, dl137–139 and dl310–424) into NIH/3T3 cells. Twenty-four hours post-transfection, nuclear extracts were made, and anti-PML and anti-GFP antibodies were employed for co-immunoprecipitation (co-IP) assays. The expression of IE1 (or its mutants) and PML was detected by Western blot, as shown in Fig. 5(a). As can be seen in Fig. 5(b and c), PML interacts with the mutants of dl1–36, dl135–141 and dl137–139, but not dl95–153 or dl310–424; PML is able to pull down wt IE1 and the three longer mutants (Fig. 5b), and conversely, only the three longer mutants are able to pull down PML (Fig. 5c). Antibodies against GFP and PML were made from mouse, so mouse IgG (mIgG) was used as control (Fig. 5d), showing that neither PML nor IE1 was pulled down by mIgG beads. Therefore, elements in regions 195–153 and 310–424 are important for interacting with PML, either by direct binding or via some intermediary protein(s).
Fig. 5.
(a–d) Interaction of PML with different IE1 deletion mutants. NIH/3T3 cells were transfected with different IE1 deletion mutant-expressing plasmids, and nuclear extracts were prepared. (a) Five per cent of the input cell extract probed with anti-GFP antibodies reveals the proteins expressed from different IE1 deletion mutant-expressing plasmids. PML from the input samples is shown below. (b) Bands in the upper panel show the immunoprecipitation of PML by anti-PML antibody. Bands in the lower panel show the immunoprecipitation of IE1 by anti-PML antibody. (c) Bands in the upper panel show the immunoprecipitation of IE1 by anti-GFP antibody. Bands in the lower panel show the immunoprecipitation of PML immunoprecipitated by anti-GFP antibody. (d) Nuclear extract samples were reacted with mouse IgG and beads, and the beads were washed and probed with anti-PML or anti-GFP antibodies. (e) Effects of different IE1 deletion mutants on MIEP. Whole NIH/3T3 cell lysates were collected 24 hours after co-transfection of the luciferase expression plasmid with different IE1 mutants. Lysate concentrations were equalized for total protein amount so as to normalize samples for luciferase assays. Renilla luciferase plasmid was included in each transfection as an internal control. At 24 h post-transfection, dual-luciferase assays were performed with the cell lysates of the transfected cells. Relative luciferase activities were calculated by dividing the normalized firefly-luciferase activity of each reporter by that of the pGL3 plasmid in pcDNA3-transfected cells. The relative activity of luciferase was obtained by comparing it with that of a group of pmMIEP_luc+pGFP.
Next, we tested whether these mutants of IE1 retain their ability to activate MIEP. We co-transfected the NIH/3T3 cells with the firefly (Photinus)-luciferase reporter plasmid (in which the luciferase expression is directed by the MCMV MIEP and pgfpie1 (wt) or its mutants, including dl1–36, dl135–141, dl95–153, dl137–139 and dl310–424. pGFP was used as a control against possible GFP-induced activation (Fig. 5e, lane 1). As shown in Fig. 5(e), wt IE1 and all other mutants that are able to interact with PML can activate MIEP. However, the IE1 mutants that lost the ability to interact with PML failed to activate the promoter. The results suggest that the interaction of IE1 with PML, but not the dispersion of ND10, is important for the activating function of IE1.
Generation of MCMV mutants with IFE-deleted IE1
To determine whether the disruption of ND10 by IE1 could be important for viral replication, we generated mutant MCMV [by using MCMV in which IE3 was tagged with GFP at the carboxy terminus (Martinez et al., 2010)] with IFE-deleted IE1 and its revertant by using the galK counter-selection BAC system (Martinez et al., 2010). MCMVdlIFE, MCMVIFERQ, MCMVdlIE1 and MCMVIE1RQ were prepared as described in Methods.
MCMVdlIFE fails to disperse ND10, and IFE-deleted IE1 is attracted to IE3 domains
As can be seen in Fig. 4, IFE-deleted IE1 did not disperse the ND10 of NIH/3T3 cells. To test whether a whole virus with the same deletion would also be unable to disperse ND10, we infected NIH/3T3 with MCMVdlIFE at an m.o.i. of 0.5 for 24 h, and ND10s were visualized by IFA using anti-PML antibody. As shown in Fig. 6(a), ND10s were not dispersed, and PML tended to be attracted to IE3 domains (Fig. 6b), which have previously been demonstrated to be pre-replication compartments (Martinez et al., 2010). The revertant virus (MCMVIFERQ) was consistent with the wt MCMV infection in mouse cells (Fig. 6i–p), as ND10s were dispersed (Fig. 6i–l). Thus, we found that for MCMV to disperse ND10, the H2B homology region (minimized to IFE) of IE1 is necessary.
Fig. 6.
IFA to determine effects of MCMVdlIFE on ND10 in NIH/3T3 cells, and the distribution of IE1 and IE3. NIH/3T3 cells seeded on coverslips were infected with MCMVdlIFE (a)–(d) or revertant MCMVdlIFERQ (i)–(l) and (m)–(p) for 24 h. The cells were fixed and permeabilized, then stained for ND10 with anti-PML antibody (a) and (i) and IE1 with anti-IE1 antibody (e) and (m) followed by reaction with Texas red-conjugated secondary antibody. IE3 was tagged with GFP, so it is shown in green (b), (f), (j) and (n). DAPI staining was done to show the total number of cells in the field of the microscope (c), (g), (k) and (o). Green, blue (DAPI) and red signals were merged as shown in (d), (h), (l) and (p). Bar, 10 µm.
IFE-deleted IE1 produced from MCMVdlIFE-infected cells was detected by anti-IE1 antibody (Fig. 6e). Interestingly, IE1 with deleted IFE colocalized with IE3 in MCMVdlIFE-infected cells, as shown in Fig. 6(d–g). This is in stark contrast to wt MCMV infection in NIH/3T3 cells or the revertant (Fig. 6m–p). Whether the ability to disperse ND10 is related to the colocalization of IE3 with IE1 needs to be explored further.
To determine the physical relationship of IE3 with IE1dlIFE and PML over the course of infection, we performed IFA to show the distributions of IE3, IE1dlIFE, and PML throughout the time course of infection. NIH/3T3 cells were infected with MCMVdlIFE at an m.o.i. of 0.5 and fixed at the indicated times post-infection (p.i.) (Supplementary Figures S1 and S2, available in JGV Online). As can be seen, IE3 diffused in the nucleus at an early time of infection [Supplementary Figs S1(A2) and S2(A2)], while IE1dlIFE also diffused in the nucleus [Supplementary Fig. S1(A1)]. At the early time of infection [4 h p.i.; Supplementary Fig. S1(A1–A4)], E1dlIFE diffused in all the nuclei of the IE1dlIFE positively expressed cells. No punctate dots were seen, which is different from what was seen in pIEdlIFE-transfected cells. Later, at 8 h p.i., IE3 formed small dots in the nucleus [bottom of Supplementary Fig. S1(B2), and top of Supplementary Fig. S2(B2)]. IE3 formed domains in the nucleus when the infection time was extended, as shown in Supplementary Fig. S1 C1–C4 (16 h p.i.) and D1–D4 (24 h p.i.). IE1 distributed as both diffusion and dots that co-localize with IE3 during the entire time of infection (Supplementary Fig. S1, B1–B4, C1–C4, and D1–D4). When IE3 formed small dots at 4 h p.i., it distributed side by side with ND10 (Supplementary Fig. S2(B1–B4)]. At a later time of infection, when IE3 formed domains in the nucleus, ND10 appeared to be partially attracted to IE3 domains. However, ND10 remained intact throughout the time course of infection.
Dispersing ND10 by IE1 is not important for MCMV gene expression or viral replication
The biological function of ND10 in relation to viral replication has been controversial. The observations that major components such as PML, Daxx and Sp100 have inhibitory effects on viral gene expression and replication support the notion that ND10s are defensive foci (Everett & Chelbi-Alix, 2007; Everett et al., 2006; Ling et al., 2005, 2008; Tavalai et al., 2008), but the fact that DNA viruses replicate DNA and transcribe important viral RNA at ND10 argues that ND10 might favour viral replication (Everett et al., 2004, 2007; Tang et al., 2000, 2003). If the dispersion of ND10 has positive effects on viral gene expression and viral replication, then MCMVdlIFE infection in NIH/3T3 cells should have significant defects in both viral gene expression and viral replication. We performed Western blot and p.f.u. assays to test these assumptions.
First, we infected MCMVdlIFE and its revertant (MCMVdlIFERQ) into NIH/3T3 cells at an m.o.i. of 0.5; the total-cell-lysate samples were collected at different time points p.i. (mock infected, 6, 12, 24, 48 and 72 h p.i., as shown in Fig. 7a), and then different viral products were detected by Western blot. The following proteins were detected: viral IE proteins, including IE1 and IE3; early proteins, including E1 (also called m112/113) and M44; and M25, containing three products (one is an early protein and the other two are late proteins). Tubulin was used as a sample loading control. Thus, no significant defect in viral gene production was observed between the infections of MCMVdlIFE and its revertant.
Fig. 7.
Viral gene expression and replication. (a) NIH/3T3 cells were mock infected (lane 1 in each column) or infected with MCMVdlIFE or rescued revertant MCMdlIFERQ for 6 h (lane 2), 12 h (lane 3), 24 h (lane 4), 48 h (lane 5) and 72 h (lane 6) at an m.o.i. of 0.5. The whole cell-lysis samples were separated by PAGE and transferred to nitrocellulose membranes for Western blot assays. Blots were first probed with anti-GFP to detect ~108 kDa IE3–EGFP (top) then stripped and reprobed with antibodies against the proteins indicated on the right. (b and c) Viral growth curve in NIH/3T3 cells. NIH/3T3 cells seeded in 24-well plates were infected with MCMVdlIFE mutant, MCMVdlIFERQ (rescued) and MCMV wt, as indicated, at an m.o.i. of either 0.01 (b) or 0.1 (c). Samples of cells with medium were collected at the indicated times and utilized for viral plaque assays. The detailed protocol is described in Methods.
We then performed p.f.u. assays to detect viral replication. NIH/3T3 cells seeded in 24-well plates were infected with either MCMVdlIFE or wt MCMV at an m.o.i. of either 0.01 (Fig. 7b) or 0.1 (Fig. 7c). Revertants were also infected as controls. The infected cells were collected (together with medium) at the number of days p.i. indicated in Fig. 7(b) and (c) (x-axis). No significant differences were observed between MCMVdlIFE and MCMVdlIFERQ or wt MCMV regarding viral growth in cell culture. Therefore, we conclude that the dispersion of ND10 might not be important for the replication of MCMV in mouse cells. However, it is not clear whether the growth defect resulting from the failure of ND10 dispersal in MCMVdlIFE infection might be compensated for by that of the IE1dlIFE co-localized with IE3. In addition, it still remains to be determined whether the dispersion of ND10 might affect viral replication in vivo.
Discussion
The IE1 protein of HCMV and MCMV is often referred to as a promiscuous transactivator because of its effects on transcription directly at the promoter level. In general, three properties have been defined for IE1: IE1 can disperse ND10 (Ahn et al., 1998; Tang & Maul, 2003), it can bind to and inactivate HDAC (Nevels et al., 2004; Tang & Maul, 2003) and it can activate the MIEP. All of these might induce the augmentation of early-gene transcription. Our previous results showed that IE1 dispersed ND10 and interacted with ND10 components to counter gene suppression, thus resulting in gene activation (Tang & Maul, 2003). Here, we used mutational analysis of MCMV IE1 to determine whether the properties of IE1 could be parsed into separate domains, and to discover which domain is essential in the immediate–early gene transcriptional augmentation. First, we dissected the functional domains of IE1 to determine which domain is responsible for dispersing ND10. We found that a large area of IE1 (aa 37–400) is important for the dispersal of ND10 by IE1, because all of the deletions in the region caused failures in the dispersing of ND10 (Figs 1, 2). The finding that the deletion of 7 aa (H2B homology region: NDIFERI) within the PML-binding region of IE1 allows binding to PML but not dispersion of ND10 indicates the need for an additional dispersion function – possibly a post-translational modification affected by IE1 on ND10-associated proteins, or perhaps interaction with an intermediate protein or proteins with the NDIFERI region that affect a dispersion cascade. Furthermore, we observed that the interaction with PML, but not the dispersion of ND10, is required if IE1 is to activate the MIE promoter (as shown in Fig. 5). We identified discontinuous domains for the binding of IE1 to ND10. This suggests that dispersion is an active process and is not solely dependent on the binding properties of IE1 with ND10 proteins.
The IE1 proteins of HCMV and MCMV modify ND10s (Ahn & Hayward, 1997; Ishov & Maul, 1996; Korioth et al., 1996; Tang & Maul, 2003; Wilkinson et al., 1998), which have themselves been implicated in the viral life cycle; the IE transcripts of large DNA viruses emerge from ND10s and start replication there when the virus is mutated to eliminate the ND10-dispersing protein (Ishov & Maul, 1996; Ishov et al., 1997). For HSV-1, the elimination of ND10 appears to rest in the ubiquitination capability of the virus ICP0 protein (Everett et al., 1998; Parkinson et al., 1999). For CMV, the binding of IE1 to the ND10 matrix protein PML appears to deprive the nuclear domain of enough proteins to maintain its recognizable structure (Ahn et al., 1998; Ahn & Hayward, 1997; Korioth et al., 1996; Wilkinson et al., 1998; Xu et al., 2001). However, ND10-associated proteins such as PML accumulate upon MCMV infection (Tang & Maul, 2003), suggesting that it is not the presence or absence of ND10-associated proteins that is important to the progression of the viral replication cycle, but rather their functional elimination independent of location (Tavalai et al., 2006). Since IE1 disperses ND10 and the absence of IE1 induces a severe replicative disadvantage, it is of interest to determine whether the elimination of the ND10 dispersive ability can be dissociated from the transcriptional co-activation of early proteins by IE1.
To determine whether the dispersal of ND10 is important for IE1 to affect viral replication, we need to have a mutated IE1 that contains minimal mutations (thereby ensuring that other functions are not modified), and the IFE deletion appears, thus far, to fit this criterion. All the point mutations of the IFE locus failed to disable the capacity of IE1 to disperse ND10 (Table 1). The IFE deletion retained other functions, including its interaction with PML and its activation of the MIE promoter (Fig. 5). Therefore, it is possible that IE1 with IFE deleted could be used to construct a MCMV that will not disperse ND10. After comparing the MCMVdlIFE with its revertant, we showed that MCMVdlIFE lost the ability to disperse ND10 and, surprisingly, facilitated the co-localization of IE1 with IE3 during the entirety of the infection time course (Supplementary Figures S1 and S2).
Of late, MCMV IE1 has been widely studied. First, it was reported not to be important for MCMV replication in cell culture, though it was found to be important for viral replication and pathogenesis in mice (Ghazal et al., 2005). Later, the role of IE1 in the transactivation of those cellular genes involved in nucleotide metabolism was deemed unimportant for MCMV replication and pathogenesis (Wilhelmi et al., 2008). Whether the dispersion of ND10 is important for the replication of MCMV, thereby causing disease in mice, could be tested in vivo by using MCMVdlIFE.
In summary, we have, for the first time, dissected the MCMV IE1 functional domains regarding dispersing ND10 and interacting with PML. We found that the sequence IFE in the H2B homology region of IE1 is a minimal motif required by IE1 if it is to disperse ND10. The mutated IE1 still interacts with PML. The MCMVdlIFE mutant virus lost the ability to disperse ND10 but kept the same replication phenotype as its revertant and the wt. Therefore, we conclude that the dispersion of ND10 might not be important for MCMV replication in mouse cell culture. However, the question as to whether the co-localization of IE1dlIFE with IE3 could compensate for the growth defect resulting from the failure of ND10 dispersal during MCMVdlIFE infection is still open.
Methods
Tissue culture and transfection.
NIH/3T3 cells (ATCC accession no. CRL-1658) were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10 % FCS (Sigma) and 1 % penicillin–streptomycin (PS; Sigma). For immunohistochemical staining, cells were grown on round coverslips (Corning) in 24-well plates (Falcon; Becton Dickinson Labware). Plasmid DNA was transfected into NIH/3T3 cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Antibodies.
The antibodies used for Western blot and the IFA are listed. mAb against tubulin (T-9026) was purchased from Sigma-Aldrich (1 : 1000 for Western blot); polyclonal antibodies against PML (sc-5621), mouse SP100 (M-75, sc-25569), mouse Daxx (M-112, sc-7152), ATRX (sc-15048) and anti-GFP mAb (sc-9996) were purchased from Santa Cruz Biotechnology and used at the dilutions given in the manufacturer’s manual; mAbs against MCMV IE1 and E1 were provided by Dr Stipan Jonjic (University of Rijeka, Croatia) (dilutions of 1 : 50 for IFA and 1 : 200 for Western blot) (Tang et al., 2005).
Immunohistochemistry.
The localization of ND10 by immunohistochemistry has been described (Martinez et al., 2010). Briefly, cells were seeded on coverslips and 24 h later were washed twice with PBS (pH 7.0), fixed in 1 % paraformaldehyde for 10 min at room temperature, washed again (twice) with PBS and and permeabilized with 0.2 % Triton X-100 on ice for 20 min. Primary antibody was added and incubated for 30 min at room temperature. Cells were then washed twice with PBS. Secondary antibody (either anti-rabbit or anti-mouse IgG) labelled with Texas red or FITC (green) was added and incubated for an additional 30 min at room temperature. After a final wash with PBS, cells were stained with Hoechst 33258.
Western blot analysis.
Proteins were separated by SDS-PAGE (10–20 µg loaded in each lane; 7.5 % acrylamide gel), transferred to nitrocellulose membranes (Amersham), and blocked with 5 % (w/v) dried non-fat milk for 60 min at room temperature. Membranes were incubated overnight at 4 °C with primary antibody, followed by incubation with HRP-coupled secondary antibody and detection with enhanced chemiluminescence (Pierce), according to standard methods (for regular Western blot, we used secondary antibody from Amersham; for the detection of protein in the immunoprecipitation, we used TrueBlot ULTRA secondary antibodies from eBioscience, cat. no. 18-8817 for mouse or 18-8816 for rabbit). To detect additional proteins, membranes were stripped with stripping buffer (100 mM β-mercaptoethanol, 2 % SDS, 62.5 mM Tris/HCl, pH 6.8), washed with PBS/0.1 % Tween 20, and reprobed as described above.
Preparation of nuclear extracts.
Nuclear extracts were obtained essentially as described previously (Tang & Maul, 2003). Briefly, monolayer cells were washed with PBS and scraped into fresh Eppendorf tubes. Cell pellets were resuspended in cold (4 °C) buffer A (10 mM HEPES/KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF) and incubated at 4 °C for 10 min. Then cells were homogenized with 10–20 plunges in a Kontes-B (Wheaton) Dounce homogenizer (pestle B) and poured into new bottles after centrifugation (at 12 000 g for 10 min). Pellets were resuspended in cold buffer C (20 mM HEPES/KOH, pH 7.9, 25 % glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF) by vortexing, and incubated at 4 °C for 30 min. Pellets were homogenized again with 10–20 plunges in a Kontes-B Dounce homogenizer (pestle B), and the clarified extracts were transferred to fresh tubes and stored at −70 °C until use.
Co-immunoprecipitation.
Antibodies were coupled to protein G–Sepharose beads (Amersham Pharmacia Biotech), according to the manufacturer's instructions. After a wash with PBS/0.1 % BSA, the beads were incubated overnight at 4 °C with clarified extracts. They were then washed again in PBS/0.1 % BSA, and resuspended in a mixture of PBS and 2× Laemmli buffer (20 µl of each). After heating at 95 °C for 5 min, the beads were removed by centrifugation and the supernatants were analysed by SDS-PAGE and immunoblotting.
Luciferase assay.
Cells were collected 20 h after transfection, and luciferase activity was determined using the Luciferase Assay System (Promega), according to the manufacturer's instructions. Each assay was performed in triplicate, and luciferase activity was normalized to the total protein amount.
Confocal microscopy.
Cells were examined at ×100 magnification with a Leica TCS SPII confocal laser scanning system equipped with a water-cooled argon–krypton laser. Two wavelength channels (495 and 590 nm) were recorded simultaneously or sequentially. Power and integration were adjusted to minimize bleed-through between the green and far-red channels prior to data acquisition. The digital images obtained were cropped and adjusted for contrast with Photoshop (Adobe).
P.f.u. assay.
Viral titres were determined by plaque assay, largely as described by Tang & Maul (2003), but with a slight modification. Supernatants containing serially diluted virus particles were added to confluent NIH/3T3 cell monolayers in six-well plates. After adsorption for 2 h, medium was removed and cells were washed twice with serum-free DMEM and overlaid with phenol-free DMEM containing 5 % FCS, 0.5 % low-melting-point agarose (GIBCO) and PS. Mean p.f.u. were determined after averaging from different dilutions.
Acknowledgements
We would like to thank Drs S. Jonjic (University of Rijeka, Croatia) and M. Messerle (Hannover Medical School, Germany) for donating reagents. This study was supported by a Pilot Grant from the Research Center for Minority Institutes (RCMI) program (2G12RR003050-24) (Q. T.), an American Cancer Society grant (RSG-090289-01-MPC) (Q. T.), an ACS-IRG grant (IRG-92-032-13, subaward # 60-14599-01-01-S6) (Q. T.) and NIH/NCRR U54RR022762 (Q. T.). We acknowledge the PSM Molecular Biology Core Laboratory for instrument support and Drs Richard Noel and Andrew Boileau for critical reading. We acknowledge Bob Ritchie of the Ponce School of Medicine and Health Sciences/RCMI Publications Office (Grant #5G12RR003050-25) for his help with manuscript preparation.
Footnotes
Two supplementary figures, a supplementary table and supplementary methods are available with the online version of this paper.
References
- Ahn J. H., Hayward G. S. (1997). The major immediate–early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol 71, 4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J. H., Brignole E. J., III, Hayward G. S. (1998). Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol 18, 4899–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L. (2008). Pondering the puzzle of PML (promyelocytic leukemia) nuclear bodies: can we fit the pieces together using an RNA regulon? Biochim Biophys Acta 1783, 2145–2154 10.1016/j.bbamcr.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L., Culjkovic B. (2009). Perspectives in PML: a unifying framework for PML function. Front Biosci 14, 497–509 10.2741/3258 [DOI] [PubMed] [Google Scholar]
- Bühler B., Keil G. M., Weiland F., Koszinowski U. H. (1990). Characterization of the murine cytomegalovirus early transcription unit E1 that is induced by immediate–early proteins. J Virol 64, 1907–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington J. M., Mocarski E. S. (1989). Human cytomegalovirus IE1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol 63, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosme R. C., Martínez F. P., Tang Q. (2011). Functional interaction of nuclear domain 10 and its components with cytomegalovirus after infections: cross-species host cells versus native cells. PLoS One 6, e19187 10.1371/journal.pone.0019187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Chelbi-Alix M. K. (2007). PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89, 819–830 10.1016/j.biochi.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Everett R. D., Freemont P., Saitoh H., Dasso M., Orr A., Kathoria M., Parkinson J. (1998). The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol 72, 6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Sourvinos G., Leiper C., Clements J. B., Orr A. (2004). Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J Virol 78, 1903–1917 10.1128/JVI.78.4.1903-1917.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Rechter S., Papior P., Tavalai N., Stamminger T., Orr A. (2006). PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80, 7995–8005 10.1128/JVI.00734-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Murray J., Orr A., Preston C. M. (2007). Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol 81, 10991–11004 10.1128/JVI.00705-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazal P., Visser A. E., Gustems M., García R., Borst E. M., Sullivan K., Messerle M., Angulo A. (2005). Elimination of IE1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture. J Virol 79, 7182–7194 10.1128/JVI.79.11.7182-7194.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves R. F., Mocarski E. S. (1998). Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus IE1 mutant. J Virol 72, 366–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaudo G., Riera L., Lembo D., De Andrea M., Gariglio M., Rudge T. L., Johnson L. F., Landolfo S. (2000). Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J Virol 74, 4979–4987 10.1128/JVI.74.11.4979-4987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Kalejta R. F. (2007). Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367, 334–338 10.1016/j.virol.2007.05.037 [DOI] [PubMed] [Google Scholar]
- Ishov A. M., Maul G. G. (1996). The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol 134, 815–826 10.1083/jcb.134.4.815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A. M., Stenberg R. M., Maul G. G. (1997). Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol 138, 5–16 10.1083/jcb.138.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., III, Maul G. G. (1999). PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147, 221–234 10.1083/jcb.147.2.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Entezami P., Gamez M., Stamminger T., Imperiale M. J. (2011). Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection. MBio 2, e00281-10 10.1128/mBio.00281-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Kim E. T., Lee H. R., Park J. J., Go Y. Y., Choi C. Y., Ahn J. H. (2006). Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J Gen Virol 87, 2181–2190 10.1099/vir.0.81787-0 [DOI] [PubMed] [Google Scholar]
- Keil G. M., Ebeling-Keil A., Koszinowski U. H. (1987). Immediate–early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol 61, 526–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C., Van Driel R., Wilkinson G. W. (1995). Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol 76, 2887–2893 10.1099/0022-1317-76-11-2887 [DOI] [PubMed] [Google Scholar]
- Korioth F., Maul G. G., Plachter B., Stamminger T., Frey J. (1996). The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res 229, 155–158 10.1006/excr.1996.0353 [DOI] [PubMed] [Google Scholar]
- Lee H. R., Kim D. J., Lee J. M., Choi C. Y., Ahn B. Y., Hayward G. S., Ahn J. H. (2004). Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol 78, 6527–6542 10.1128/JVI.78.12.6527-6542.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo D., Gribaudo G., Hofer A., Riera L., Cornaglia M., Mondo A., Angeretti A., Gariglio M., Thelander L., Landolfo S. (2000). Expression of an altered ribonucleotide reductase activity associated with the replication of murine cytomegalovirus in quiescent fibroblasts. J Virol 74, 11557–11565 10.1128/JVI.74.24.11557-11565.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P. D., Peng R. S., Nakajima A., Yu J. H., Tan J., Moses S. M., Yang W. H., Zhao B., Kieff E., et al. (2005). Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J 24, 3565–3575 10.1038/sj.emboj.7600820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P. D., Tan J., Sewatanon J., Peng R. (2008). Murine gammaherpesvirus 68 open reading frame 75c tegument protein induces the degradation of PML and is essential for production of infectious virus. J Virol 82, 8000–8012 10.1128/JVI.02752-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F. P., Cosme R. S., Tang Q. (2010). Murine cytomegalovirus major immediate–early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J Gen Virol 91, 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G. G., Negorev D. (2008). Differences between mouse and human cytomegalovirus interactions with their respective hosts at immediate early times of the replication cycle. Med Microbiol Immunol (Berl) 197, 241–249 10.1007/s00430-008-0078-1 [DOI] [PubMed] [Google Scholar]
- McElroy A. K., Dwarakanath R. S., Spector D. H. (2000). Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J Virol 74, 4192–4206 10.1128/JVI.74.9.4192-4206.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerle M., Bühler B., Keil G. M., Koszinowski U. H. (1992). Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate–early gene 3. J Virol 66, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch K., Keil G. M., Messerle M., Koszinowski U. H. (1988). Interaction of the 89K murine cytomegalovirus immediate–early protein with core histones. Virology 163, 405–412 10.1016/0042-6822(88)90281-4 [DOI] [PubMed] [Google Scholar]
- Münch K., Bühler B., Messerle M., Koszinowski U. H. (1991). The core histone-binding region of the murine cytomegalovirus 89K immediate early protein. J Gen Virol 72, 1967–1974 10.1099/0022-1317-72-8-1967 [DOI] [PubMed] [Google Scholar]
- Münch K., Messerle M., Plachter B., Koszinowski U. H. (1992). An acidic region of the 89K murine cytomegalovirus immediate–early protein interacts with DNA. J Gen Virol 73, 499–506 10.1099/0022-1317-73-3-499 [DOI] [PubMed] [Google Scholar]
- Nevels M., Paulus C., Shenk T. (2004). Human cytomegalovirus immediate–early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A 101, 17234–17239 10.1073/pnas.0407933101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J., Lees-Miller S. P., Everett R. D. (1999). Herpes simplex virus type 1 immediate–early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol 73, 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase M. J., Simon C. O., Seckert C. K., Lemmermann N., Grzimek N. K. (2008). Murine model of cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol 325, 315–331 10.1007/978-3-540-77349-8_18 [DOI] [PubMed] [Google Scholar]
- Saffert R. T., Kalejta R. F. (2006). Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate–early gene expression. J Virol 80, 3863–3871 10.1128/JVI.80.8.3863-3871.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert R. T., Kalejta R. F. (2007). Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J Virol 81, 9109–9120 10.1128/JVI.00827-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert R. T., Kalejta R. F. (2008). Promyelocytic leukemia-nuclear body proteins: herpesvirus enemies, accomplices, or both? Future Virol 3, 265–277 10.2217/17460794.3.3.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully A. L., Sommer M. H., Schwartz R., Spector D. H. (1995). The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein–protein associations. J Virol 69, 6533–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006). The mechanisms of PML-nuclear body formation. Mol Cell 24, 331–339 10.1016/j.molcel.2006.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J., Sissons P. (1996). Latent and persistent infections of monocytes and macrophages. Intervirology 39, 293–301 [DOI] [PubMed] [Google Scholar]
- Stenberg R. M. (1996). The human cytomegalovirus major immediate–early gene. Intervirology 39, 343–349 [DOI] [PubMed] [Google Scholar]
- Stenberg R. M., Stinski M. F. (1985). Autoregulation of the human cytomegalovirus major immediate–early gene. J Virol 56, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg R. M., Fortney J., Barlow S. W., Magrane B. P., Nelson J. A., Ghazal P. (1990). Promoter-specific trans activation and repression by human cytomegalovirus immediate–early proteins involves common and unique protein domains. J Virol 64, 1556–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Maul G. G. (2003). Mouse cytomegalovirus immediate–early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J Virol 77, 1357–1367 10.1128/JVI.77.2.1357-1367.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Bell P., Tegtmeyer P., Maul G. G. (2000). Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol 74, 9694–9700 10.1128/JVI.74.20.9694-9700.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Li L., Ishov A. M., Revol V., Epstein A. L., Maul G. G. (2003). Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J Virol 77, 5821–5828 10.1128/JVI.77.10.5821-5828.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Li L., Maul G. G. (2005). Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate–early promoter. J Virol 79, 257–263 10.1128/JVI.79.1.257-263.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N., Stamminger T. (2008). New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta 1783, 2207–2221 10.1016/j.bbamcr.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Tavalai N., Papior P., Rechter S., Leis M., Stamminger T. (2006). Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80, 8006–8018 10.1128/JVI.00743-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N., Papior P., Rechter S., Stamminger T. (2008). Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol 82, 126–137 10.1128/JVI.01685-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. T., Bresnahan W. A. (2005). Human cytomegalovirus immediate–early 2 gene expression blocks virus-induced beta interferon production. J Virol 79, 3873–3877 10.1128/JVI.79.6.3873-3877.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E., Laukens K., Dang T. H., Van Ostade X. (2010). A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci 6, 51–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. F., Krasikov T., Frey M. R., Wang J., Matera A. G., Marzluff W. F. (1996). Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1Mb. Genome Res 6, 688–701 10.1101/gr.6.8.688 [DOI] [PubMed] [Google Scholar]
- Wiebusch L., Hagemeier C. (1999). Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol 73, 9274–9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebusch L., Neuwirth A., Grabenhenrich L., Voigt S., Hagemeier C. (2008). Cell cycle-independent expression of immediate–early gene 3 results in G1 and G2 arrest in murine cytomegalovirus-infected cells. J Virol 82, 10188–10198 10.1128/JVI.01212-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi V., Simon C. O., Podlech J., Böhm V., Däubner T., Emde S., Strand D., Renzaho A., Lemmermann N. A., et al. (2008). Transactivation of cellular genes involved in nucleotide metabolism by the regulatory IE1 protein of murine cytomegalovirus is not critical for viral replicative fitness in quiescent cells and host tissues. J Virol 82, 9900–9916 10.1128/JVI.00928-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G. W., Kelly C., Sinclair J. H., Rickards C. (1998). Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol 79, 1233–1245 [DOI] [PubMed] [Google Scholar]
- Xu Y., Ahn J. H., Cheng M., apRhys C. M., Chiou C. J., Zong J., Matunis M. J., Hayward G. S. (2001). Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate–early protein IE1 to inhibit PML-mediated transcriptional repression. J Virol 75, 10683–10695 10.1128/JVI.75.22.10683-10695.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Müller S., Ronchetti S., Freemont P. S., Dejean A., Pandolfi P. P. (2000). Role of SUMO-1-modified PML in nuclear body formation. Blood 95, 2748–2752 [PubMed] [Google Scholar]