Abstract
Human respiratory syncytial virus (RSV), a leading cause of respiratory tract infections in infants, inhibits type I interferon (IFN)-dependent signalling, as well as IFN synthesis. RSV non-structural protein NS1 plays a significant role in this inhibition; however, the mechanism(s) responsible is not fully known. The transcription factor interferon regulatory factor (IRF)-3 is essential for viral-induced IFN-β synthesis. In this study, we found that NS1 protein inhibits IRF-3-dependent gene transcription in constitutively active IRF-3 overexpressing cells, demonstrating that NS1 directly targets IRF-3. Our data also demonstrate that NS1 associates with IRF-3 and its transcriptional coactivator CBP, leading to disrupted association of IRF-3 to CBP and subsequent reduced binding of IRF-3 to the IFN-β promoter without blocking viral-induced IRF-3 phosphorylation, nuclear translocation and dimerization, thereby identifying a novel molecular mechanism by which RSV inhibits IFN-β synthesis.
Human respiratory syncytial virus (RSV), a member of the family Paramyxoviridae, is a leading cause of respiratory infection in infants and children worldwide (Hall, 2001). Unlike other viruses of the family Paramyxoviridae, RSV is characterized by the presence of two non-structural proteins, NS1 and NS2, which have been shown to control viral replication (Collins & Murphy, 2005) and to antagonize type I interferon (IFN) synthesis, with NS1 playing a greater role in inhibiting IFN production than NS2 (Spann et al., 2004). The mechanism(s) by which NS1 antagonizes IFN-β synthesis is not fully understood. Interferon regulatory factor (IRF)-3, a transcription factor belonging to the IRF family, plays an essential role in viral-induced IFN-β synthesis (Barnes et al., 2002; Taniguchi et al., 2001). Recent studies have shown that RSV NS1 protein affects IRF-3 nuclear translocation (Spann et al., 2005), possibly by decreasing cellular levels of TRAF3 and IKKϵ, two kinases upstream of IRF-3 activation (Swedan et al., 2009).
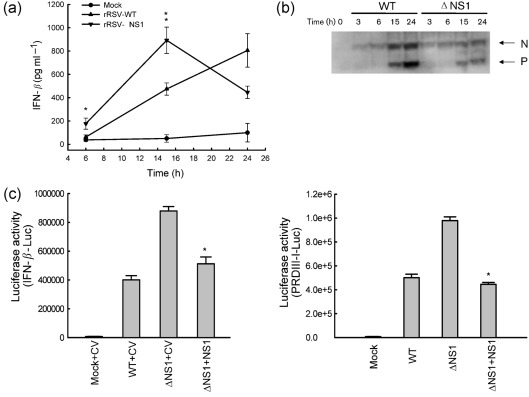
The aim of our study was to identify additional mechanism(s) involved in NS1-dependent inhibition of type I IFN synthesis. To confirm the inhibitory role of NS1 in RSV-induced IFN-β synthesis, A549 cells were infected with recombinant RSV wild-type (rRSV-WT) or recombinant RSV lacking NS1 (rRSV-ΔNS1) at an m.o.i. of 3. Mock infection was used as a control. Both RSV A2 strain-derived recombinant viruses (a gift from MedImmune, Gaithersburg, MD) have been previously characterized (Jin et al., 2000). Viruses were grown in Vero cells and purified on a sucrose gradient. Viral titres were determined by plaque assay (Guerrero-Plata et al., 2006). IFN-β secretion in cell supernatants was determined by ELISA (PBL). There was a significant increase in IFN-β production in cells infected with rRSV-ΔNS1 compared with cells infected with rRSV-WT, both at 6 and 15 h post-infection (p.i.) (Fig. 1a), with no more difference at 24 h p.i., probably due to decreased viral replication of ΔNS1, as shown by a reduction in viral antigen expression at later time points of infection (Fig. 1b).
Fig. 1.
Effect of NS1 protein deletion on type I IFN secretion and IRF-3-dependent gene transcription. (a) A549 cells were infected with rRSV-WT or rRSV-ΔNS1, at an m.o.i. of 3, and harvested at 6, 15 and 24 h p.i. to measure IFN-β secretion in cell supernatants by ELISA. Data shown are representative of three independent experiments. Statistical significance was analysed by ANOVA. P-value of less than 0.05 was considered significant. *P<0.05 and **P<0.01, relative to rRSV-WT-infected A549 cells. (b) Viral protein expression analysis of infected cells. Total cell lysates were prepared after the infections. Equal amounts of protein were subjected to 4–20 % SDS-PAGE, followed by Western blot using an antibody against RSV (AbD Serotec). The results are representative of two independent experiments. (c) A549 cells were cotransfected with a luciferase reporter plasmid IFN-β-Luc (left panel) or PRDIII-I-Luc (right panel), and the expression plasmid containing RSV NS1 protein or the control vector (CV), and infected with rRSV-WT or -ΔNS1, at an m.o.i. of 3. Cells were harvested at 15 h p.i. to measure luciferase activity. Uninfected plates served as controls. For each plate, luciferase was normalized to the β-galactosidase reporter activity. Data are representative of two independent experiments and are expressed as mean±se of normalized luciferase activity. ANOVA was used for statistical analysis. P less than 0.05 was considered significant. *P<0.05 relative to ΔNS1+CV.
To investigate the role of NS1 in regulating IFN-β gene expression, V5-tagged NS1 gene was cloned into the pCAGGS vector (primer information is available upon request). We then compared IRF-3-dependent gene transcription in A549 cells infected with either rRSV-WT or rRSV-ΔNS1 in the presence or absence of NS1 protein expression. Briefly, A549 cells were cotransfected with a reporter plasmid containing the luciferase gene linked to either the IFN-β promoter (IFN-β-Luc) or multimers of the IRF-3-binding site (PRDIII-I-Luc), and the NS1 expression plasmid or its empty vector, as described previously (Bao et al., 2008a, b; Ehrhardt et al., 2004). The next day, cells were mock-infected or infected with rRSV-WT or rRSV-ΔNS1 and lysed to independently measure luciferase and β-galactosidase (internal control) activity, as described previously (Casola et al., 2001). There was a significantly higher induction of IFN-β and IRF-3-dependent gene transcription in cells infected with ΔNS1, compared with those infected with WT (Fig. 1c). Expression of NS1 inhibited the enhanced gene transcription in response to ΔNS1 infection, confirming the ability of NS1 to inhibit virus-induced cellular gene expression (Fig. 1c).
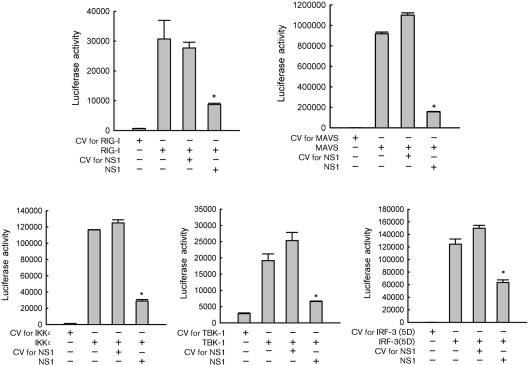
In the context of RNA virus infection, the retinoic acid-inducible gene I (RIG-I)-dependent pathway plays an essential role in initiating cellular signals leading to the activation of transcription factors (Kawai & Akira, 2008). Activated RIG-I interacts with the mitochondrial antiviral signalling (MAVS) protein, resulting in the phosphorylation of IRF-3 by IKKϵ and TBK-1 (Fitzgerald et al., 2003). In response to RSV infection, IRF-3 activation is regulated by the RIG-I-dependent pathway (Liu et al., 2007). To identify potential signalling targets of NS1, 293 cells were cotransfected with RIG-I, or its downstream signalling molecules MAVS/IKKϵ/TBK-1 expression plasmids, a plasmid encoding NS1 or its control vector, and the PRDIII-I-Luc. Individual expression of all molecules significantly induced IRF-3-dependent gene transcription, which was inhibited by NS1 expression (Fig. 2). In addition, IRF-3-dependent gene transcription induced by constitutively active IRF-3 (IRF-3 5D, a gift from Dr Rongtuan Lin, McGill University, Canada), in which five phosphorylation sites are converted to phosphomimetic aspartic acid residues (Lin et al., 1998), was also significantly reduced by NS1 expression (Fig. 2), suggesting that NS1 directly targets IRF-3. On the other hand, NS1 did not have an inhibitory effect on MAVS-induced activator protein (AP)-1-dependent gene transcription, indicating that the inhibitory effect of NS1 on IRF-3 is specific (Supplementary Fig. S1, available in JGV Online).
Fig. 2.
NS1 protein directly affects IRF-3-dependent gene expression. 293 cells in 24-well plate were transfected with PRDIII-I-Luc, plasmids encoding either RIG-I, MAVS, IKKϵ, TBK-1 or the constitutively active form of IRF-3 (IRF-3 5D), or their control vectors (0.1–0.2 µg per well), and a plasmid expressing RSV NS1 or its CV (0.2 µg per well), as indicated at the bottom of each column. Cells were harvested 30 h post-transfection to measure luciferase activity. For each plate, luciferase was normalized to the β-galactosidase reporter activity. Data are representative of two independent experiments and are expressed as mean±se of normalized luciferase activity. Statistical significance was analysed by ANOVA. *P<0.05 relative to signal inducer in each group with CV for NS1.
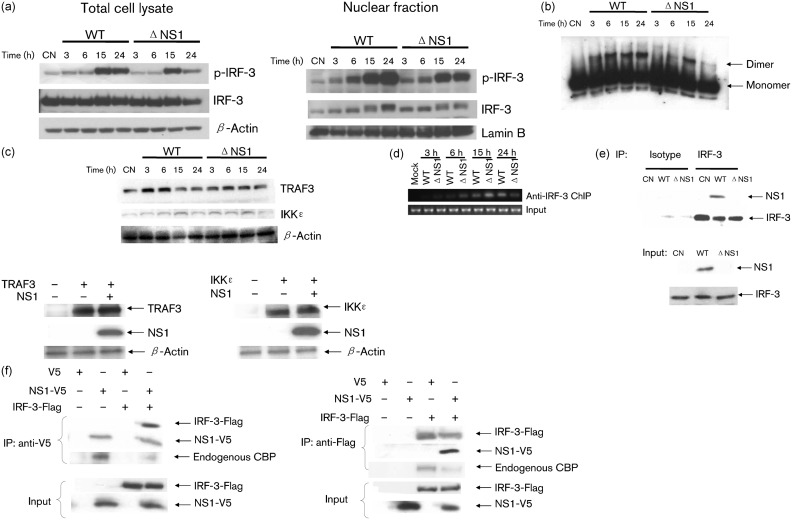
Viral-induced IRF-3 nuclear translocation, dimerization and binding to DNA is regulated through phosphorylation on specific serine residues at its C terminus (Hiscott, 2007; Servant et al., 2001). To investigate whether NS1 inhibited IRF-3 activity through blocking its phosphorylation, we compared levels of IRF-3 serine phosphorylation in total cell lysates of WT- and ΔNS1-infected cells. There was no significant difference in total phospho-IRF-3 levels between WT- and ΔNS1-infected cells (at 24 h p.i. phospho-IRF-3 levels in ΔNS1-infected cells are actually a little lower compared with WT) (Fig. 3a and Supplementary Fig. S2a, left panel, available in JGV Online), suggesting that NS1 does not prevent IRF-3 phosphorylation. Total IRF-3 abundance was also comparable in WT- and ΔNS1-infected cells (Fig. 3a, left panel), suggesting that the NS1 protein does not facilitate IRF-3 degradation, a mechanism used by the rotavirus NS1 protein to inhibit IRF-3 activation (Barro & Patton, 2005). Interestingly, we did not find that NS1 inhibits virus-induced IRF-3 nuclear translocation (Fig. 3a and Supplementary Fig. S2a, right panel), which is in contrast to what has been previously reported (Ling et al., 2009; Spann et al., 2005). Different experimental methods used by the two laboratories may account for the observed discrepancy. Enhanced IFN-β expression by NS1 deletion may result from increased IRF-3dimerization, a key step controlling the binding of IRF-3 to DNA (Dragan et al., 2007). However, we did not observe increased IRF-3 dimerization by NS1 deletion (Fig. 3b). Furthermore, infection of A549 cells with either WT or ΔNS1 viruses or expression of NS1 protein did not cause significant changes in the expression of either TRAF3 or IKKϵ, two kinases involved in RIG-I-dependent activation of IRF-3 (Fig. 3c and Supplementary Fig. S2b), in contrast to a recent report showing that NS1 from Long strain RSV degrades TRAF3 and IKKϵ in an overexpression system (Swedan et al., 2009).
Fig. 3.
NS1 protein prevents IRF-3 binding to IFN-β promoter. (a) NS1 does not affect RSV-induced IRF-3 phosphorylation and nuclear translocation. A549 cells were mock-infected (as a control, CN) or infected with rRSV-WT or rRSV-ΔNS1, at an m.o.i. of 3, for various lengths of time and harvested to prepare either total cell lysates or nuclear extracts as described previously (Bao et al., 2008b). Equal amounts of protein from uninfected and infected cells were analysed by Western blot using antibodies against Ser396 phospho-IRF-3 (pIRF-3) and regular IRF-3. Membranes were stripped and reprobed for β-actin or lamin b, as control for equal loading of the samples of total cell lysate or nuclear fraction, respectively. (b) IRF-3 dimerization is not enhanced by NS1 deletion. Total proteins from cell lysates of A549 cells, mock-infected or infected with recombinant viruses, were separated by native gel electrophoresis and IRF-3 dimerization was analysed by Western blot. (c) NS1 does not facilitate the degradation of TRAF3 and IKKϵ. Upper panel: the abundance of TRAF3 and IKKϵ were compared in WT- versus ΔNS1-infected cells. Membranes were stripped and reprobed for β-actin, as a control for equal loading of the samples. Lower panel: A549 cells were transfected with a plasmid encoding TRAF3 or IKKϵ, or their control vectors, and a plasmid expressing RSV NS1 or its CV, as indicated at the bottom of each column. Cells were harvested 30 h post-transfection to measure the abundance of TRAF3/IKKϵ in the presence or absence of NS1 expression. Membranes were stripped and reprobed for β-actin for equal loading control. (d) NS1 prevents the binding of IRF-3 to the IFN-β promoter. A549 cells were infected with WT or ΔNS1, at an m.o.i. of 3, for 3, 6, 15 and 24 h. ChIP assay was performed using antibody against IRF-3. Equal amounts of DNA samples before ChIP were used for PCR, as control for equal loading of the samples. (e) NS1 associates with IRF-3 in the context of RSV infection. A549 cells were mock-infected or infected with rRSV-WT or rRSV-ΔNS1, at an m.o.i. of 3, and harvested at 6 h p.i. to prepare total cell lysates. Samples were subjected to immunoprecipitation using anti-IRF-3 antibody or control isotype. The immunoprecipitated complexes were then subjected to 4–20 % SDS-PAGE followed by Western blot using anti-NS1 antibody. The membrane was then stripped and reprobed with anti-IRF-3 antibody to determine levels of immunoprecipitated IRF-3, using clean-blot IP detection reagent (HRP) from Thermo Scientific (upper panel). NS1 and IRF-3 from total cell lysates were also measured by Western blot to ensure equal IP input (lower panel). (f) NS1 disrupts the association of IRF-3 with CBP. 293 cells were transfected with plasmids encoding Flag-tagged IRF-3 and V5-tagged NS1 or their control vectors. Total cell lysates were immunoprecipitated with anti-V5 antibody followed by Western blot using anti-Flag antibody to detect Flag-tagged IRF-3. Reverse immunoprecipitation was also done, where Flag-tagged-IRF-3 was immunoprecipitated using anti-Flag antibody, and NS1 protein was then detected using anti-V5 antibody. Membranes were stripped and reprobed to check for properly immunoprecipitated NS1 or IRF-3. The association of endogenous CBP with immunoprecipitated NS1 or IRF-3 was determined by reprobing the membrane with anti-CBP antibodies (Santa Cruz). Representative results are shown from two to four separate experiments.
Therefore, we speculated that NS1 must use an alternative mechanism(s) to inhibit RSV-induced IFN-β synthesis. As NS1 expression occurs in both cytoplasm and nuclear compartments following RSV infection (Munday et al., 2010; Spann et al., 2005; and data not shown), we hypothesized that NS1 inhibits IRF-3-dependent gene transcription by preventing IRF-3 binding to its cognate promoter site. Therefore, we compared IRF-3 binding to the endogenous IFN-β promoter in WT- and ΔNS1-infected cells using a chromatin immunoprecipitation (ChIP) assay. A549 cells, mock-infected or infected with WT or ΔNS1, were cross-linked with 1 % formaldehyde (10 min at room temperature), followed by ChIP assays using a ChIP-IT Express kit (Active Motif). Briefly, chromatin was sheared to an average size of 300 bp with a Vibra Cell (Sonics&Materials Inc.), incubated with 2 µg anti-IRF-3 antibody (Santa Cruz Biotechnology) and 25 µl protein G magnetic beads overnight at 4 °C. After reversal of cross-linking, protein digestion with proteinase K and DNA purification, PCR was performed using RedTag Readymix PCR Reaction Mix (Sigma-Aldrich; reaction conditions are available upon request). The sense and antisense primers for the putative interferon-stimulated reponse element (ISRE) sites present within the IFN-β promoter were referenced from Wagoner et al. (2007), with forward: 5′-CCTCACAGTTTGTAAATCTTTTTCCC-3′, and reverse: 5′-ACGAACAGTGTCGCCTACTACCTG-3′. The results showed that RSV infection induced significant binding of IRF-3 to the IFN-β promoter ISRE site, compared with mock infection. This IRF-3–DNA interaction was further increased by NS1 deletion at 3, 6 and 15 h p.i. (Fig. 3d and Supplementary Fig. S2c), suggesting that NS1 expression impairs IRF-3 DNA-binding activity. At 24 h p.i., less IRF-3–DNA interaction was observed in ΔNS1-infected cells than in WT-infected cells, which may result from restricted replication of ΔNS1 at later time point of infection.
In addition to NS1, the RSV NS2 protein also exhibits IFN antagonist activity (Spann et al., 2004; Teng & Collins, 1999). The mechanism by which NS2 accomplishes its inhibitory effect is via targeting RIG-I. NS1 does not bind to RIG-I (Ling et al., 2009 and data not shown), suggesting that the two NS proteins use different mechanisms to inhibit IFN production. Indeed, we found that NS1 binds to IRF-3 to exert its inhibitory function on IFN-β synthesis (Fig. 3e). A549 cells were mock-infected or infected with WT or ΔNS1. Total cell lysate was immunoprecipitated by anti-IRF-3 antibody, followed by Western blot using anti-NS1 anti-serum (a gift from MedImmune). An isotype antibody was used to control for non-specific binding. NS1 was coimmunoprecipitated by anti-IRF-3 antibody, but not by isotype antibody in WT-infected samples, suggesting an interaction of IRF-3 with NS1 in the context of RSV infection.
There are several possible mechanisms responsible for the attenuated interaction between IRF-3 and IFN-β promoter. NS1 may function as Kaposi’s sarcoma herpevirus’s protein K-bZIP (Lefort et al., 2007), which prevents the attachment of activated IRF-3 to the IFN-β promoter by competitively binding to the same region. CBP/p300, a co-transcriptional activator of IRF-3, is essential for DNA-binding activity of IRF-3 in the context of viral infection (Suhara et al., 2002). Therefore, attenuated binding of IRF-3 to its DNA motif may result from disrupted interaction between IRF-3 with CBP by NS1, similar to what has been reported for human herpes virus (HHV) kinase (Hwang et al., 2009) and HHV-8-encoded vIRF-1 (Lin et al., 2001). Indeed, we found that NS1 binds to both IRF-3 and CBP (Fig. 3f). 293 cells were cotransfected with V5-tagged NS1 and Flag-tagged IRF-3 expression plasmids. Vectors expressing V5 or Flag only were used as negative controls. After 30 h of transfection, cells were lysed followed by immunoprecipitation using anti-V5 antibody (Invitrogen). The immunoprecipitated complex was separated on 4–20 % SDS-PAGE and transferred onto a PVDF membrane. Western blot using anti-Flag antibody (Sigma) revealed that Flag-tagged IRF-3 was pulled down by NS1 (Fig. 3f, upper panel). Clean-blot immunoprecipitation (IP) detection reagent (Thermo Fisher Scientific), which does not bind to denatured IgG, was used to detect IRF-3, as they have a similar molecular mass. NS1 binding to IRF-3 was specific, as NS1 did not bind to overexpressed RIG-I (data not shown), consistent with previous findings (Ling et al., 2009). Overexpressed NS1 was also able to pull down the endogenous CBP, and the NS1–CBP interaction was attenuated when IRF-3 was overexpressed. Reverse immunoprecipitation confirmed that there was an interaction between NS1 and IRF-3, as well as IRF-3 and CBP, which was again attenuated by NS1 overexpression (Fig. 3f, lower panel). Together these results suggest that NS1 affects the interaction between IRF-3 and its coactivator CBP, leading to reduced IRF-3 binding to the IFN-β promoter, identifying a novel mechanism underlying the NS1 inhibitory activity on RSV-induced IFN-β synthesis.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases grants P01 062885 to A. C. and R. P. G., N01-AI-30039 to R. P. G., K22 (KAI074829A) to X. B. and by the American Heart Association (SDG 0835151N) and Parker Francis Foundation (X. B.). The authors thank Animesh Chandra for his assistance in manuscript editing.
Footnotes
Supplementary figures are available with the online version of this paper.
References
- Bao X., Kolli D., Liu T., Shan Y., Garofalo R. P., Casola A. (2008a). Human metapneumovirus small hydrophobic protein inhibits NF-κB transcriptional activity. J Virol 82, 8224–8229 10.1128/JVI.02584-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Liu T., Shan Y., Li K., Garofalo R. P., Casola A. (2008b ). Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog 4, e1000077 10.1371/journal.ppat.1000077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes B., Lubyova B., Pitha P. M. (2002). On the role of IRF in host defense. J Interferon Cytokine Res 22, 59–71 10.1089/107999002753452665 [DOI] [PubMed] [Google Scholar]
- Barro M., Patton J. T. (2005). Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci U S A 102, 4114–4119 10.1073/pnas.0408376102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola A., Burger N., Liu T., Jamaluddin M., Brasier A. R., Garofalo R. P. (2001). Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with respiratory syncytial virus. Role in viral-induced interferon regulatory factor activation. J Biol Chem 276, 19715–19722 10.1074/jbc.M101526200 [DOI] [PubMed] [Google Scholar]
- Collins P. L., Murphy B. R. (2005). New generation live vaccines against human respiratory syncytial virus designed by reverse genetics. Proc Am Thorac Soc 2, 166–173 10.1513/pats.200501-011AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragan A. I., Hargreaves V. V., Makeyeva E. N., Privalov P. L. (2007). Mechanisms of activation of interferon regulator factor 3: the role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucleic Acids Res 35, 3525–3534 10.1093/nar/gkm142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt C., Kardinal C., Wurzer W. J., Wolff T., von Eichel-Streiber C., Pleschka S., Planz O., Ludwig S. (2004). Rac1 and PAK1 are upstream of IKK-ϵ and TBK-1 in the viral activation of interferon regulatory factor-3. FEBS Lett 567, 230–238 10.1016/j.febslet.2004.04.069 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003). IKKϵ and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4, 491–496 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Guerrero-Plata A., Casola A., Suarez G., Yu X., Spetch L., Peeples M. E., Garofalo R. P. (2006). Differential response of dendritic cells to human metapneumovirus and respiratory syncytial virus. Am J Respir Cell Mol Biol 34, 320–329 10.1165/rcmb.2005-0287OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. B. (2001). Respiratory syncytial virus and parainfluenza virus. N Engl J Med 344, 1917–1928 10.1056/NEJM200106213442507 [DOI] [PubMed] [Google Scholar]
- Hiscott J. (2007). Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282, 15325–15329 10.1074/jbc.R700002200 [DOI] [PubMed] [Google Scholar]
- Hwang S., Kim K. S., Flano E., Wu T. T., Tong L. M., Park A. N., Song M. J., Sanchez D. J., O’Connell R. M., et al. (2009). Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5, 166–178 10.1016/j.chom.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Zhou H., Cheng X., Tang R., Munoz M., Nguyen N. (2000). Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273, 210–218 10.1006/viro.2000.0393 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2008). Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci 1143, 1–20 10.1196/annals.1443.020 [DOI] [PubMed] [Google Scholar]
- Lefort S., Soucy-Faulkner A., Grandvaux N., Flamand L. (2007). Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol 81, 10950–10960 10.1128/JVI.00183-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C., Pitha P. M., Hiscott J. (1998). Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Genin P., Mamane Y., Sgarbanti M., Battistini A., Harrington W. J., Jr, Barber G. N., Hiscott J. (2001). HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20, 800–811 10.1038/sj.onc.1204163 [DOI] [PubMed] [Google Scholar]
- Ling Z., Tran K. C., Teng M. N. (2009). Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol 83, 3734–3742 10.1128/JVI.02434-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jamaluddin M., Li K., Garofalo R. P., Casola A., Brasier A. R. (2007). Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol 81, 1401–1411 10.1128/JVI.01740-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday D. C., Emmott E., Surtees R., Lardeau C. H., Wu W., Duprex W. P., Dove B. K., Barr J. N., Hiscox J. A. (2010). Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol Cell Proteomics 9, 2438–2459 10.1074/mcp.M110.001859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant M. J., ten Oever B., LePage C., Conti L., Gessani S., Julkunen I., Lin R., Hiscott J. (2001). Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J Biol Chem 276, 355–363 10.1074/jbc.M007790200 [DOI] [PubMed] [Google Scholar]
- Spann K. M., Tran K. C., Chi B., Rabin R. L., Collins P. L. (2004). Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J Virol 78, 4363–4369 10.1128/JVI.78.8.4363-4369.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann K. M., Tran K. C., Collins P. L. (2005). Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J Virol 79, 5353–5362 10.1128/JVI.79.9.5353-5362.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhara W., Yoneyama M., Kitabayashi I., Fujita T. (2002). Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J Biol Chem 277, 22304–22313 10.1074/jbc.M200192200 [DOI] [PubMed] [Google Scholar]
- Swedan S., Musiyenko A., Barik S. (2009). Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J Virol 83, 9682–9693 10.1128/JVI.00715-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001). IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19, 623–655 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- Teng M. N., Collins P. L. (1999). Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J Virol 73, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagoner J., Austin M., Green J., Imaizumi T., Casola A., Brasier A., Khabar K. S., Wakita T., Gale M., Jr, Polyak S. J. (2007). Regulation of CXCL-8 (interleukin-8) induction by double-stranded RNA signaling pathways during hepatitis C virus infection. J Virol 81, 309–318 10.1128/JVI.01411-06 [DOI] [PMC free article] [PubMed] [Google Scholar]