Abstract
The attachment organelles of bacterial species belonging to the Mycoplasma pneumoniae phylogenetic cluster are required for host cytadherence, gliding motility and virulence. Despite being closely related, these bacteria possess distinct cellular morphologies and gliding characteristics. The molecular mechanisms for most attachment organelle phenotypes, including shape and ability to power motility, are obscure. The attachment organelle-associated P30 protein of M. pneumoniae is implicated in both adherence and motility, with mutations negatively impacting cell morphology, adherence, gliding and virulence. To test whether the P30 alleles of different mycoplasma species confer species-specific attachment organelle properties, we created an M. pneumoniae strain in which the Mycoplasma genitalium P30 orthologue, P32, was substituted for the native P30. Selected clones were visualized by scanning electron microscopy to assess morphology and by indirect immunofluorescence microscopy to localize P32. Cytadherence ability and gliding motility were assessed by haemadsorption assay and phase-contrast microcinematography, respectively. Cell and attachment organelle morphologies were indistinguishable from wild-type M. pneumoniae as well as M. pneumoniae II-3 expressing a C-terminally 6×His-tagged P30 construct. P32 was localized to the tip of the attachment organelle of transformant cells. Although a specific role for P30 in species-specific phenotypes was not identified, this first test of orthologous gene replacement in different mycoplasma species demonstrates that the differences in the M. pneumoniae and M. genitalium proteins contribute little if anything to the different attachment organelle phenotypes between these species.
Introduction
Mycoplasmas are cell-wall-less bacteria that belong to the class Mollicutes. By virtue of reductive evolution, these organisms have the smallest genomes of any self-replicating cells capable of axenic growth. In nature, these organisms parasitize host cells for nutrients due to limited biosynthetic capabilities, and in the laboratory, they must be provided with a rich growth medium (Razin et al., 1998). Absence of a cell wall imparts pleomorphy to many of these organisms; however, cells of many species of the genus Mycoplasma appear flask-shaped. Polarity is conferred by a differentiated tip structure (Hatchel & Balish, 2008), the attachment organelle, which mediates primary attachment of these organisms to surfaces such as host epithelia. Attachment organelles are required for host colonization and virulence in the human respiratory and genito-urinary tract pathogens Mycoplasma pneumoniae and Mycoplasma genitalium, respectively (Razin & Jacobs, 1992; Ueno et al., 2008; Waites & Talkington, 2004). Unlike many of their walled counterparts, mycoplasmas lack locomotory appendages such as flagella and pili. Instead, the attachment organelle provides the means necessary for an adherence-dependent form of locomotion called gliding motility. The speeds at which species within the M. pneumoniae phylogenetic cluster glide are different (Hatchel & Balish, 2008), implying that some component of the motor apparatus regulates speed. Interestingly, the failure of an M. pneumoniae mutant that moves about as fast as M. genitalium to successfully colonize a normal human bronchial epithelial cell culture (Jordan et al., 2007) suggests that these species-specific speeds are calibrated to other species-specific properties.
Triton X-100-insoluble components of the attachment organelles of each of the species of the M. pneumoniae cluster are visible by electron microscopy (Göbel et al., 1981; Meng & Pfister, 1980; Hatchel & Balish, 2008). These structures prominently include an electron-dense core (Biberfeld & Biberfeld, 1970), which is a bipartite rod (Henderson & Jensen, 2006; Seybert et al., 2006) with a terminal button located at the end that is distal to the cell body, and a base, which physically interacts with the cell chromosome, at the proximal end (Hatchel & Balish, 2008). Scanning electron micrographs of Triton X-100-extracted M. pneumoniae and several of its close relatives demonstrate that core substructures are distinct across species, leading to differences in core length, width and curvature, and conferring distinct morphological properties to the attachment organelle of each species (Hatchel & Balish, 2008). In particular, M. pneumoniae has a straight attachment organelle that is 290 nm in length, whereas that of M. genitalium is only 170 nm long and curves to approximately 20°, with a more prominent terminal knob.
The attachment organelle of M. pneumoniae and its close relatives is composed of many novel proteins (Balish & Krause, 2005; Balish, 2006), including structural proteins such as HMW1 (Stevens & Krause, 1991), HMW2 (Krause et al., 1982), HMW3 (Stevens & Krause, 1992) and P65 (Jordan et al., 2001; Proft et al., 1995), and proteins involved in adherence to host cells, such as P1 (Baseman et al., 1982; Feldner et al., 1982; Hu et al.,1982) and P30 (Morrison-Plummer et al., 1986). The specific relationships between any of these proteins and either attachment organelle morphology or the process of gliding motility are largely obscure. However, analysis of M. pneumoniae cells containing a transposon that disrupts the gene encoding attachment organelle protein P41 indicates clearly that the motor activity for gliding is contained within the attachment organelle (Hasselbring & Krause, 2007).
Henderson & Jensen (2006) have proposed that the electron-dense core drives motility, undergoing conformational changes that move the cells in an inchworm-like fashion. Other evidence suggests that adhesins localized to the attachment organelle may be responsible for gliding motility. Gliding motility and glass binding of M. pneumoniae cells treated with a monoclonal anti-P1 antibody are negatively impacted in an antibody concentration-dependent manner, whereas the antibody minimally affects non-gliding cells (Seto et al., 2005), suggesting a role for the P1 adhesin in gliding. In addition, a spontaneously occurring haemadsorption (HA)-negative mutant of M. pneumoniae, II-3, can neither attach to substrates nor glide because of a frameshift mutation in the gene encoding the 30 kDa attachment organelle protein P30 (Hasselbring et al., 2005; Romero-Arroyo et al., 1999). Also, decreased stability of the attachment organelle protein P65 is evident in these cells (Jordan et al., 2001). An altered stretch of 17 amino acids in P30 as well as truncated versions of P30 allow for very low motility (Hasselbring et al., 2005; Chang et al., 2011).
To test the relationship between gliding speed and sequence features of P30, we undertook an effort to complement the M. pneumoniae P30 null mutant II-3 with P32 from M. genitalium (Reddy et al., 1995), which is approximately 43 % identical to P30 (Fig. 1). We introduced a 6X-His epitope-tagged P32-encoding construct, as well as a non-tagged construct, into mutant II-3. We subsequently examined several transformants in terms of attachment organelle morphology, cytadherence ability and gliding motility parameters, as well as stabilization of P65. Our results rule out a specific role in gliding motility for P30/P32 sequence differences between M. pneumoniae and M. genitalium, but clearly demonstrate the potential for the use of orthologous gene complementation among mycoplasma species to test gene/protein function.
Fig. 1.
Comparison of P30 orthologues from M. pneumoniae strain M129 and M. genitalium strain G37. Shaded amino acids are conserved between the two proteins. Amino acid sequences were aligned using clustal x software.
Methods
Strains and growth conditions.
M. pneumoniae wild-type strain M129, P30 null mutant M. pneumoniae II-3, M. genitalium wild-type strain G37 and M. pneumoniae II-3 transformants were grown in plastic tissue-culture flasks in SP-4 broth (Tully et al., 1979) at 37 °C until mid-exponential phase (phenol red indicator was orange; 3–5 days) in an ambient air atmosphere. For assays requiring plating of strains, cells were grown on SP-4 containing 1 % (w/v) Noble agar (Becton Dickinson). To prepare motility stocks, the protocol detailed by Hatchel et al. (2006) was used. For selection and propagation of transformants only, 18 µg gentamicin ml−1 was included in all media.
Genomic DNA isolation, PCR and cloning.
Mid-exponential phase SP-4 broth-cultures, with or without gentamicin, were harvested by centrifugation for 20 min at 17 400 g. Cell pellets were washed three times with PBS (150 mM NaCl, 3.2 mM NaH2PO4, 13.6 mM Na2HPO4, pH 7.2) and genomic DNA was extracted using the Blood and Body Fluid Spin Protocol of the QIAamp DNA Blood Mini kit (Qiagen). Purified genomic DNA was quantified spectrophotometrically, diluted with sterile ultrapure water and stored at −20 °C until use.
The P30 (MPN453)- or P32 (MG318)-encoding genes of M. pneumoniae M129 and M. genitalium G37, respectively, as well as the genes immediately upstream of these genes (MPN454, encoding P21MP, and MG319, encoding P21MG), were amplified using the primers listed in Table 1, such that they also included the promoter region as identified by Waldo et al. (1999). To make polyhistidine-tagged P30 and P32 proteins, six histidine codons were engineered in-frame into primers that were used to amplify the 3′ end of the gene (Table 1), resulting in production of P30His and P32His. Following PCR, amplicons were cloned using the TA cloning vector pCR2.1 (TA Cloning kit; Invitrogen). Next, clones were screened by restriction analysis and sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) and capillary electrophoresis on an Applied Biosystems 3130xl genetic analyser at the Miami University Center for Bioinformatics and Functional Genomics. Inserts were excised with BamHI and EcoRI and subcloned into BamHI- and EcoRI-digested Tn4001-containing suicide vector pMT85 (Zimmerman & Herrmann, 2005) for transformation of mycoplasmas. These plasmids were designated pOO12, pOO17 and pOO24 (Table 1).
Table 1. Bacterial strains, plasmids and primers used in this study.
| Strain, plasmid or primer | Description | Source or reference |
| Bacterial strains | ||
| Escherichia coli DH5α | Used for plasmid propagation | Laboratory stock |
| E. coli XL-1 Blue | Used for mutagenesis | Agilent Technologies |
| M. genitalium G37 | Wild-type strain, HA+ | Laboratory stock |
| M. pneumoniae M129 | Wild-type strain, HA+ | Laboratory stock |
| M. pneumoniae II-3 | Host for cloning, P30−, HA− | Laboratory stock |
| M. pneumoniae 12-A | M. pneumoniae II-3+M. genitalium G37 P32 operon, HA+ | This study |
| M. pneumoniae 17-B | M. pneumoniae II-3+M. genitalium G37 P32His operon, HA+ | This study |
| M. pneumoniae 24-A | M. pneumoniae II-3+M. pneumoniae M129 P30His operon, HA+ | This study |
| M. pneumoniae 30-C | M. pneumoniae II-3+M. genitalium G37 P32 promoter region+P21 gene, HA− | This study |
| M. pneumoniae 37-D | M. pneumoniae II-3+M. pneumoniae M129 P30 operon, HA+ | This study |
| Plasmids | ||
| pCR2.1 | TA cloning vector | Invitrogen |
| pMT85 | Suicide vector; contains Tn4001 | Laboratory stock/Zimmerman & Herrmann (2005) |
| pOO12 | pMT85+M. genitalium G37 P32 operon | This study |
| pOO17 | pMT85+M. genitalium G37 P32His operon | This study |
| pOO24 | pMT85+M. pneumoniae M129 P30His operon | This study |
| pOO30 | pMT85+M. genitalium G37 P32 promoter region+P21 gene | This study |
| pOO37 | pMT85+M. pneumoniae M129 P30 operon | This study |
| Amplification and mutagenic primers* | ||
| MGP21Bam | TAGAGGATCCTAAAGCTCTATAGTTAAT | This study |
| RFRECORI | AAAGAATTCTAATTAGGGTTTTAAACC | This study |
| RFRECORIHis | TTTGAATTCTCTTTAGTGATGATGGTGGTGATGACCACCATAGGGTTTTAAACC | |
| MPN454Bam | AAGGGATCCAAGCTGTAAGTGGGGAATTAAAGC | This study |
| MPN453EcoHis | AGCGAATTCCTTTTAGTGATGATGGTGGTGATGACCACCATAGCGTTTTGGTGG | This study |
| ClaIMUT† | GGAACTAATTAAGTATCGATAAAGATGGAGTT | This study |
| A_41_T† | GGTGGTGATGACCACCTTAGCGTTTTGGTGGAA | This study |
| A_41_T-antisense† | TTCCACCAAAACGCTAAGGTGGCATCACCACC | This study |
Restriction endonuclease sites are underlined and polyhistidine epitope tag-encoding bases are in bold.
Primers used for site-directed mutagenesis.
To generate pOO30 (Table 1), site-directed mutagenesis was used to introduce a ClaI restriction site in the P21–P32His intergenic region of the construct contained in the plasmid pOO15 (progenitor of pOO17; not shown) using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) and the primer ClaMUT (Table 1). Next, ClaI and MfeI were used to remove the P32His-encoding gene, these restriction sites were blunted with the Klenow fragment and the plasmid was recircularized. This modification allowed for retention of the original EcoRI site, which was exploited to facilitate subcloning of this construct into pMT85 for subsequent transformation of M. pneumoniae II-3. To generate pOO37 (Table 1), site-directed mutagenesis was used to reintroduce the original translational stop codon back into P30His of the pCR2.1 progenitor of pOO24. The resulting construct was next subcloned into pMT85.
Electroporation was used for transformation of M. pneumoniae II-3 cells and was performed as described by Hedreyda et al. (1993). Transformants were selected on gentamicin-containing SP-4 agar and multiple gentamicin-resistant colonies were picked to control for transposon insertion site positional effects in transformants (Hedreyda et al., 1993). To ensure genetic homogeneity of transformants, colonies were grown in selective broth, filtered through a 0.22 µm syringe filter and then plated onto selective agar. This filter-cloning approach was carried out three times. Transformants were next grown in 10 ml selective broth, and aliquots were frozen at −80 °C for further use.
HA assay.
To assess the cytadherence ability of the mycoplasmas examined in this study, the HA assay was performed on plate cultures following 7 days of incubation as described by Sobeslavsky et al. (1968), except that sheep blood in Alsever’s solution was used, and an additional PBS wash was also carried out. Colonies were visualized with an inverted microscope to determine whether colonies haemadsorbed sheep red blood cells.
Immunoblot analysis.
Cell pellets were prepared from 50 ml SP-4 broth cultures with or without gentamicin by centrifugation for 20 min at 17 400 g at 4 °C, followed by three washes with PBS. Pellets were resuspended in 150 mM NaCl plus 20 mM Tris/HCl, pH 7.5, and protein concentration was determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific). For P30 and polyhistidine immunoblots, cellular material equivalent to 5 µg of protein was used, and for P65 immunoblotting, 2.5 µg of protein was used. In all cases, lysates were electrophoresed through 12 % SDS-polyacrylamide gels (Laemmli, 1970) and transferred to nitrocellulose membranes overnight at room temperature for immunoblotting (Towbin et al., 1979). Blots were blocked for 2 h at room temperature in 5 % (w/v) skimmed milk and blotted with a 1 : 250 dilution of P30 antiserum (Romero-Arroyo et al., 1999), a 1 : 1000 dilution of anti-P65 (Proft et al., 1995) or a 1 : 1000 dilution of anti-6×-His (Immunology Laboratory Consultants). Membranes were subsequently probed with alkaline phosphatase-conjugated goat anti-rabbit IgG (Fc) (Promega). Between each antibody incubation step, membranes were washed five times for 5 min each with Tris-buffered saline containing Tween-20 [50 mM Tris base, pH 7.5, 150 mM NaCl, 0.05 % (v/v) Tween 20]. Protein bands were detected using a solution of nitro blue tetrazolium and 5-bromo-4-chloro-3′-indoylphosphate. To test for cross-reactivity of the anti-6×-His antibody with non-transformed M. pneumoniae, 5 µg total cell lysate of M. pneumoniae wild-type strain M129 was probed with the anti-6×His antibody along with 5 µg of M. pneumoniae 24-A lysate.
Immunofluorescence microscopy.
To localize P30His and P32His within transformant cells, a modified version of the protocol detailed by Jordan et al. (2001) was used. Briefly, 200 µl aliquots of cells were allowed to adhere to glass coverslips for 2 h at 37 °C. Following incubation and fixation, cells were probed with 2 µg anti-6×His antibody ml−1 (Immunology Consultants Laboratories) followed by a 1 : 100 dilution of a Cy2-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). Coverslips were rinsed once with sterile water before being mounted on glass microscope slides using Vectashield (Vector Laboratories). Cells were examined using epifluorescence illumination with a Leica DM IRB inverted microscope (Leica Microsystems). Images were taken and manipulated with spot v4.1.1 software (Diagnostic Instrument) and Adobe Photoshop software to reduce background fluorescence while maintaining P30His and P32His focus integrity.
Scanning electron microscopy (SEM).
Strains were grown on glass coverslips in a 24-well tissue culture plate until mid-exponential phase (phenol red indicator was orange). To avoid disrupting non-adherent microcolonies and cells of M. pneumoniae II-3 and M. pneumoniae 30-C that settled onto the surface of coverslips and adherent cells of other strains, broth was gently aspirated from each well. Cells were next fixed to the coverslips with a solution of 1.5 % glutaraldehyde, 1 % formaldehyde and 0.1 M sodium cacodylate, pH 7.2, for 30 min at room temperature. Coverslips were then rinsed with 0.1 M sodium cacodylate, pH 7.2, for 30 min and were subsequently dehydrated in increasing concentrations of ethanol (25–100 %, v/v). Coverslips were critical-point dried and gold sputter-coated as previously described (Hatchel et al., 2006). Cells were examined using a Zeiss Supra 35 FEG-VP scanning electron microscope (Carl Zeiss) at the Miami University Center for Advanced Microscopy and Imaging.
Time-lapse microcinematography.
Motility stocks (50 µl) were inoculated into 750 µl SP-4 broth containing 3 % (w/v) gelatin and, for transformants, gentamicin was included. Samples were next syringe-triturated seven times through a 25-gauge needle and passed into chamber slides (Thermo Fisher Scientific). Inoculated chamber slides were incubated for 3 h at 37 °C. Phase-contrast microcinematography was performed as described previously (Hatchel et al., 2006; Relich et al., 2009) using 1 s intervals for 27 consecutive frames. Motility parameters were assessed using images manipulated with both spot v4.1.1 and Adobe Photoshop software. Mean gliding speeds of strains were compared using a one-way analysis of variance with Tukey multiple comparisons. A significance level of 0.05 (family-wise error rate for multiple comparisons) was used for all analyses.
Results
Construct engineering and transformation of M. pneumoniae II-3
The Tn4001-containing plasmid pMT85 (Zimmerman & Herrmann, 2005) was used to introduce constructs for production of M. pneumoniae P30 and its M. genitalium orthologue, P32, into the P30 mutant M. pneumoniae II-3 (Krause et al., 1982) using established protocols (Hedreyda et al., 1993). Mutant II-3 has a frameshift mutation near the middle of the gene, but antisera against polypeptides corresponding to the region upstream of the frameshift fail to detect a truncated protein (Romero-Arroyo et al., 1999; Chang et al., 2011; data not shown), suggesting that II-3 is a null mutant. The relevant transformants are listed in Table 1.
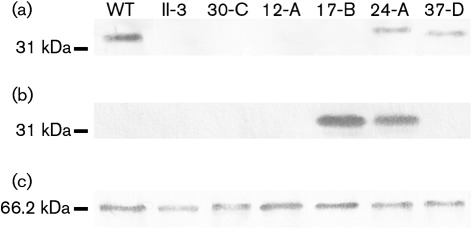
Attempts to drive production of P32 from the promoter in Tn4001 were unsuccessful (data not shown). Therefore, to produce P30 or P32 in M. pneumoniae II-3, we engineered constructs that included the native promoter region (Waldo et al., 1999) and the gene located between the promoter and the P30 (MPN453)- or P32 (MG318)-encoding gene, designated as the P21MP (MPN454)- or P21MG (MG319)-encoding gene (Fig. 2). To enable detection of P32 by a commercially available antibody, we created a P32-encoding construct that carried a C-terminal 6×His tag (Fig. 2b). To eliminate the possibility of tag-associated artefacts, parallel constructs without 6×His tags were also introduced into M. pneumoniae II-3. To confirm that the 6×His antibody did not cross-react with M. pneumoniae, whole-cell lysate from the parental strain was blotted in parallel with the positive control M. pneumoniae 24-A. No bands were detected (Fig. 3), confirming that all signal from this antibody could be attributed to P30His or P32His.
Fig. 2.
Constructs generated for this study. PCR was used to amplify the genes of interest from genomic DNA of either wild-type M. pneumoniae strain M129 or wild-type M. genitalium strain G37. Amplicons were cloned in the TA cloning vector pCR2.1, sequenced and subcloned into the Tn4001-containing vector pMT85 for delivery of the constructs into M. pneumoniae II-3 or M. pneumoniae M129. Constructs in pOO12 (a), pOO17 (b), pOO30 (c) and pOO24 (d). The construct in pOO37 is a derivative of that which is in pOO24; it contains the original MPN453 translational stop codon (the position of which is indicated by the vertical arrow) and does not include the 6×His epitope tag.
Fig. 3.

Immunoblot confirmation that the 6×His antibody does not cross-react with any proteins in non-transformed M. pneumoniae. Wild-type M. pneumoniae strain M129 (WT) total cell lysate was probed with the 6×-His antibody alongside the transformant M. pneumoniae 24-A. Molecular mass markers are indicated to the left.
P30 was detected by immunoblot analysis in wild-type M. pneumoniae and the II-3 transformants containing the P30His and P30 constructs, strains 24-A and 37-D, respectively. P30 was not detected in either the P32His- or the P32-producing strains (Fig. 4a), indicating a lack of reversion to the wild-type P30 allele. The 6×His epitope tag was detected in all strains containing tagged constructs, but not in strains lacking such constructs (Fig. 4b).
Fig. 4.
Immunoblot analysis for the visualization of P30, P30His, P32His and P65. Whole-cell lysates of wild-type M. pneumoniae strain M129 (WT), M. pneumoniae II-3 and the transformant strains M. pneumoniae 30-C, 12-A, 17-B, 24-A and 37-D were probed with anti-P30 (a), anti-6×His (b) or anti-P65 (c) antibodies. Molecular mass markers are indicated to the left.
P21MG does not complement phenotypes associated with P30 loss
To control for possible phenotypes conferred by P21MG or by additional P21MP, we created a plasmid, pOO30, which contains a derivative of the pOO17 insert and lacks the P32His-encoding gene (Fig. 2c). M. pneumoniae pOO30 transformant 30-C, which was representative of several transformants, was non-cytadherent (Table 1) and its cellular morphology was indistinguishable from II-3 (Fig. 5). Like mutant II-3, transformant 30-C did not attach to the flask during growth. Also, full stabilization of P65 was not evident (Fig. 4c). Overall, P21MG did not complement any aberrant phenotype of mutant II-3, eliminating the possibility that our results were meaningfully impacted by the presence of this protein or its M. pneumoniae orthologue in our experimental constructs.
Fig. 5.
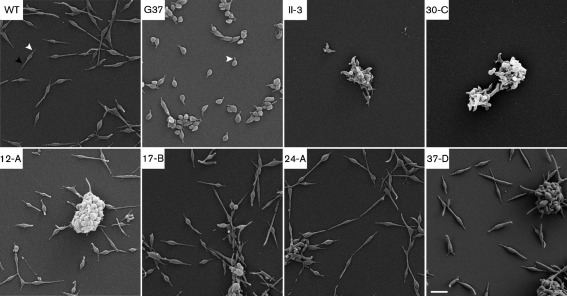
Morphology of strains used in this assay. SEM reveals that transformants containing either P30 or P32, with or without the C-terminal 6×His tag, are indistinguishable from wild-type M. pneumoniae M129 (WT). Strains are indicated in the upper left of each panel, with G37 indicating M. genitalium. White arrowheads indicate attachment organelles and the black arrowhead indicates a trailing filament. Bar, 1 µm.
M. genitalium P32 complements P30 loss in mutant II-3
As a result of lacking P30, M. pneumoniae II-3 is non-cytadherent and possesses morphology quite distinct from that of wild-type M. pneumoniae. Rather than having a straight and narrow attachment organelle at the anterior end of the cell and a long trailing filament at the posterior end, II-3 cells exhibit extensive pleomorphy, and most cells appear ovoid with one or more tip structures protruding from the cell body (Fig. 5; Romero-Arroyo et al., 1999). In addition, when grown in broth, II-3 cells form aggregates that are suspended in the medium. In contrast, wild-type M. pneumoniae cells are cytadherent and are dispersed individually on the surface of culture vessels (Fig. 5) and associated with attached microcolonies. The correlation between the absence of P30 production and the loss of both cytadherence and wild-type morphology has been well documented (Hasselbring et al., 2005; Romero-Arroyo et al., 1999). To determine the extent to which P32 could complement these phenotypes, HA assays and SEM were performed to gauge cytadherence and morphology, respectively. Also, we examined the site of P32His and P30His localization by epifluorescence microscopy.
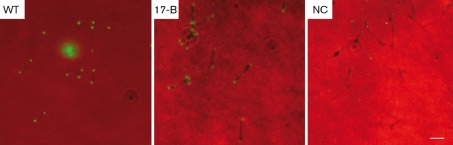
All P30- or P32-producing transformants grew attached to the surface of the flask, and they were all cytadherent, as indicated by the adherence of sheep erythrocytes to colonies (Table 1 and data not shown). The presence of the 6×His tag did not interfere with HA, and all colonies evaluated were indistinguishable from one another. SEM of P30- or P32-producing transformants revealed adherent cells with straight and slender attachment organelles, relatively thin bodies, and long trailing filaments, which are all characteristic features of wild-type M. pneumoniae cells and distinct from wild-type M. genitalium cells (Fig. 5). When examined by indirect immunofluorescence microscopy using a 6×His primary antibody for detection, P32His and P30His were found at discrete foci at the tips of the attachment organelles of transformed cells (Fig. 6). Together, these data suggest that P32 is trafficked to the same intracellular site as P30 within M. pneumoniae, and that P32 fully complements wild-type M. pneumoniae morphology and does not confer the curvature of the attachment organelle characteristic of M. genitalium. These data do not support a specific role for P30 in differential morphogenesis of the attachment organelle across species.
Fig. 6.
P30His and P32His localize to the attachment organelle tip, the site of localization of native P30. Whole cells grown on glass coverslips (red channel) were probed with a monoclonal anti-6×His antibody and an FITC-conjugated secondary antibody (green channel). Strains are indicated in the upper left of each panel, except that NC indicates wild-type (WT) cells processed with only secondary antibody as a negative control. Bar, 2 µm.
The attachment organelle protein P65 is stabilized in P32- and P32His-expressing transformants
The protein P65 is localized within the attachment organelle of M. pneumoniae and is unstable in the absence of P30 (Jordan et al., 2001). To determine whether P32 could restore P65 stabilization, transformant lysates were immunoblotted with a polyclonal P65 antiserum (Proft et al., 1995). Transformants expressing P32 produced more P65 than was present in mutant II-3 cells (Fig. 4c), suggesting stabilization of P65.
P32-expressing transformants glide at speeds similar to that of wild-type M. pneumoniae
The mean gliding speeds of both the P30- and the P32-expressing transformants were similar to the mean gliding speed of the wild-type M. pneumoniae strain M129, although a small statistical difference, independent of the origin of the P30/P32 allele, was evident (Table 2). We suspect that this difference could be attributed to somewhat lower levels of P30 or P32 produced in transformants. Giving credence to this possibility are recent observations by us suggesting that low levels of P30 production by M. pneumoniae II-3 transformants correlate with lower gliding speed. However, the 6×His tag does not interfere with the function of P30 or P32 in gliding, as all transformants glided at similar speeds (Table 2).
Table 2. Gliding motility parameters of wild-type and transformant strains.
| Strain | Gliding motility speed (nm s−1; mean±sd, n = 100) | Range (nm s−1) | Percentage of moving cells (total cells analysed) | Percentage of frames containing moving cells (total frames = 2600) |
| M. genitalium G37 | 111±26 | 53–192 | 87 (575) | 94 |
| M. pneumoniae M129 | 269±62 | 129–503 | 45 (463) | 82 |
| M. pneumoniae II-3 | − | − | − | – |
| M. pneumoniae 12-A | 237±52 | 111–352 | 43 (255) | 71 |
| M. pneumoniae 17-B | 236±57 | 114–406 | 43 (553) | 75 |
| M. pneumoniae 24-A | 252±66 | 118–396 | 39 (788) | 78 |
| M. pneumoniae 30-C | − | − | − | – |
| M. pneumoniae 37-D | 237±68 | 116–404 | 39 (554) | 67 |
Discussion
The mechanisms underlying differences in gliding speed and other attachment organelle properties in members of the M. pneumoniae phylogenetic cluster are not understood. For this reason, we investigated the contribution of the P30 family of transmembrane attachment organelle proteins by using orthologous gene replacement, a technique that has not previously been reported using mycoplasmas. This approach allowed for evaluation of phenotypes conferred by the M. genitalium P32 protein to the P30 null mutant M. pneumoniae II-3, allowing us to address aspects of the specific role of P30 in attachment organelle function. We focused on whether the substantial sequence differences between the two orthologues (Fig. 1) contributed in any observable way to the distinct attachment organelle phenotypes of the two organisms.
Transformants with P32 constructs were cytadherent and morphologically indistinguishable from wild-type M. pneumoniae cells. Moreover, gliding speeds and stabilization of attachment organelle protein P65 in transformants producing P32 and P32His were very similar to wild-type M. pneumoniae and M. pneumoniae II-3 producing P30 and P30His, although in some transformants there was a subtle decrease in speed that could be attributable to the somewhat reduced amount of P30 or P32 present in these cells. Like P30, P32 localized to the attachment organelle, suggesting that they are equivalent substrates for the unknown trafficking mechanism responsible for M. pneumoniae cell polarity. These data suggest that the sequence differences between P30 and P32 are not responsible for the obvious morphological differences seen in the attachment organelles of M. pneumoniae and M. genitalium (Fig. 5; Hatchel & Balish, 2008). Overall, these analyses indicate that P32 functions in M. pneumoniae about as well as P30.
Conceivably, the high similarity between P30 and P32 in and near the transmembrane domain provides the basis for the ability of P32 to substitute for P30, supporting an important role for this region in protein function, as suggested by Chang et al. (2011). Additionally, P30 and P32 have in common a proline-rich repeat region at the C terminus, although the repeats themselves are different and less regular in P32. The fact that alterations to this region in P30 lead to significant compromise of function (Jordan et al., 2007; Chang et al., 2011) makes it surprising that the divergent corresponding region in P32 is fully substitutable. These results suggest that the overall structure of the domain is similar in P30 and P32, and loss of repeat elements is deleterious to this structure.
The specific role of P30 in the process of cellular gliding is not yet known, except that our evidence suggests that its activity is not rate-limiting in terms of gliding speed. We envision a scenario in which the M. pneumoniae gliding machinery is at least partly analogous to that of Mycoplasma mobile, in which an adhesin, Gli349 (Adan-Kubo et al., 2006; Nakane et al., 2011), is proposed to undergo conformational changes with respect to the substrate, directly generating movement; these changes are driven ultimately by an ATPase and proximately by proteins such as Gli521 that bridge the adhesin and the ATPase (Miyata, 2010). One possibility for P30 function is that it is analogous to Gli349, which is supported by its known adherence function (Morrison-Plummer et al., 1986). Another possibility is that P30 interacts with and promotes the activity of proteins that directly drive motility themselves. In M. mobile, the protein Gli521 is proposed to transfer the propulsive force to Gli349 (Miyata, 2010). By analogy, P30 might perform this role in M. pneumoniae, with some other attachment organelle protein such as the major adhesin P1 in the role of Gli349. In neither case is P30 rate-limiting, leaving other components of the M. pneumoniae gliding apparatus responsible for species-specific differences in speed, possibly including other proteins involved in adherence, such as P1, as well as any unidentified proteins that provide energy to drive conformational changes. As an additional possibility, the number of P30 molecules, rather than the individual activity of each one, might control gliding speed, as suggested by the slight reduction in speed in transformants producing slightly less P30His or P32His (unpublished data). P30 sequence differences also appear not to have a specific role in attachment organelle morphology, although MG217, the orthologue of the P30-associated protein P65, is linked to attachment organelle curvature in M. genitalium (Burgos et al., 2008).
The work presented here demonstrates for the first time the usefulness of orthologous gene transformation as a means for assessing protein function in mycoplasmas. Experiments to further characterize the contribution of P30 proteins from other related Mycoplasma species as well as the roles of other attachment organelle proteins are currently under way using methods adapted from this research.
Acknowledgements
This work was supported by the National Institutes of Health (Public Health Service grant R15 AI073994) and by the Miami University Doctoral-Undergraduate Opportunities in Scholarship program. We thank Dr Duncan Krause (University of Georgia) for the generous gift of anti-P30 antiserum, Mr Michael Hughes (Miami University) for help with statistical analysis of gliding motility and Dr Iddo Friedberg (Miami University) for advice about protein comparison software. We also thank the Miami University Center for Bioinformatics and Functional Genomics and the Miami University Center for Advanced Microscopy and Imaging for use of their equipment and facilities.
Abbreviations:
- HA
haemadsorption
- SEM
scanning electron microscopy
References
- Adan-Kubo J., Uenoyama A., Arata T., Miyata M. (2006). Morphology of isolated Gli349, a leg protein responsible for Mycoplasma mobile gliding via glass binding, revealed by rotary shadowing electron microscopy. J Bacteriol 188, 2821–2828. 10.1128/JB.188.8.2821-2828.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish M. F. (2006). Subcellular structures of mycoplasmas. Front Biosci 11, 2017–2027. 10.2741/1943 [DOI] [PubMed] [Google Scholar]
- Balish M. F., Krause D. C. (2005). Mycoplasma attachment organelle and cell division. In Mycoplasmas: Molecular Biology, Pathogenicity, and Strategies for Control, pp. 189–237. Edited by Blanchard A., Browning G. Norwich: Horizon Bioscience. [Google Scholar]
- Baseman J. B., Cole R. M., Krause D. C., Leith D. K. (1982). Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol 151, 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biberfeld G., Biberfeld P. (1970). Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol 102, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos R., Pich O. Q., Querol E., Piñol J. (2008). Deletion of the Mycoplasma genitalium MG_217 gene modifies cell gliding behaviour by altering terminal organelle curvature. Mol Microbiol 69, 1029–1040. 10.1111/j.1365-2958.2008.06343.x [DOI] [PubMed] [Google Scholar]
- Chang H.-Y., Jordan J. L., Krause D. C. (2011). Domain analysis of protein P30 in Mycoplasma pneumoniae cytadherence and gliding motility. J Bacteriol 193, 1726–1733. 10.1128/JB.01228-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldner J., Göbel U., Bredt W. (1982). Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature 298, 765–767. 10.1038/298765a0 [DOI] [PubMed] [Google Scholar]
- Göbel U., Speth V., Bredt W. (1981). Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J Cell Biol 91, 537–543. 10.1083/jcb.91.2.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring B. M., Krause D. C. (2007). Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol Microbiol 63, 44–53. 10.1111/j.1365-2958.2006.05507.x [DOI] [PubMed] [Google Scholar]
- Hasselbring B. M., Jordan J. L., Krause D. C. (2005). Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J Bacteriol 187, 6281–6289. 10.1128/JB.187.18.6281-6289.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchel J. M., Balish M. F. (2008). Attachment organelle ultrastructure correlates with phylogeny, not gliding motility properties, in Mycoplasma pneumoniae relatives. Microbiology 154, 286–295. 10.1099/mic.0.2007/012765-0 [DOI] [PubMed] [Google Scholar]
- Hatchel J. M., Balish R. S., Duley M. L., Balish M. F. (2006). Ultrastructure and gliding motility of Mycoplasma amphoriforme, a possible human respiratory pathogen. Microbiology 152, 2181–2189. 10.1099/mic.0.28905-0 [DOI] [PubMed] [Google Scholar]
- Hedreyda C. T., Lee K. K., Krause D. C. (1993). Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30, 170–175. 10.1006/plas.1993.1047 [DOI] [PubMed] [Google Scholar]
- Henderson G. P., Jensen G. J. (2006). Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol Microbiol 60, 376–385. 10.1111/j.1365-2958.2006.05113.x [DOI] [PubMed] [Google Scholar]
- Hu P.-C., Cole R. M., Huang Y. S., Graham J. A., Gardner D. E., Collier A. M., Clyde W. A., Jr (1982). Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216, 313–315. 10.1126/science.6801766 [DOI] [PubMed] [Google Scholar]
- Jordan J. L., Berry K. M., Balish M. F., Krause D. C. (2001). Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J Bacteriol 183, 7387–7391. 10.1128/JB.183.24.7387-7891.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan J. L., Chang H.-Y., Balish M. F., Holt L. S., Bose S. R., Hasselbring B. M., Waldo R. H., III, Krunkosky T. M., Krause D. C. (2007). Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect Immun 75, 518–522. 10.1128/IAI.01344-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause D. C., Leith D. K., Wilson R. M., Baseman J. B. (1982). Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun 35, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Meng K. E., Pfister R. M. (1980). Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J Bacteriol 144, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M. (2010). Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol 64, 519–537. 10.1146/annurev.micro.112408.134116 [DOI] [PubMed] [Google Scholar]
- Morrison-Plummer J., Leith D. K., Baseman J. B. (1986). Biological effects of anti-lipid and anti-protein monoclonal antibodies on Mycoplasma pneumoniae. Infect Immun 53, 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D., Adan-Kubo J., Kenri T., Miyata M. (2011). Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J Bacteriol 193, 715–722. 10.1128/JB.00796-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T., Hilbert H., Layh-Schmitt G., Herrmann R. (1995). The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J Bacteriol 177, 3370–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S., Jacobs E. (1992). Mycoplasma adhesion. J Gen Microbiol 138, 407–422. [DOI] [PubMed] [Google Scholar]
- Razin S. D., Yogev D., Naot Y. (1998). Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 62, 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. P., Rasmussen W. G., Baseman J. B. (1995). Molecular cloning and characterization of an adherence-related operon of Mycoplasma genitalium. J Bacteriol 177, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relich R. F., Friedberg A. J., Balish M. F. (2009). Novel cellular organization in a gliding mycoplasma, Mycoplasma insons. J Bacteriol 191, 5312–5314. 10.1128/JB.00474-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Arroyo C. E., Jordan J., Peacock S. J., Willby M. J., Farmer M. A., Krause D. C. (1999). Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J Bacteriol 181, 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S., Kenri T., Tomiyama T., Miyata M. (2005). Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J Bacteriol 187, 1875–1877. 10.1128/JB.187.5.1875-1877.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Herrmann R., Frangakis A. S. (2006). Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J Struct Biol 156, 342–354. 10.1016/j.jsb.2006.04.010 [DOI] [PubMed] [Google Scholar]
- Sobeslavsky O., Prescott B., Chanock R. M. (1968). Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J Bacteriol 96, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. K., Krause D. C. (1991). Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J Bacteriol 173, 1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. K., Krause D. C. (1992). Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J Bacteriol 174, 4265–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76, 4350–4354. 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully J. G., Rose D. L., Whitcomb R. F., Wenzel R. P. (1979). Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J Infect Dis 139, 478–482. 10.1093/infdis/139.4.478 [DOI] [PubMed] [Google Scholar]
- Ueno P. M., Timenetsky J., Centonze V. E., Wewer J. J., Cagle M., Stein M. A., Krishnan M., Baseman J. B. (2008). Interaction of Mycoplasma genitalium with host cells: evidence for nuclear localization. Microbiology 154, 3033–3041. 10.1099/mic.0.2008/020735-0 [DOI] [PubMed] [Google Scholar]
- Waites K. B., Talkington D. F. (2004). Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 17, 697–728. 10.1128/CMR.17.4.697-728.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo R. H., III, Popham P. L., Romero-Arroyo C. E., Mothershed E. A., Lee K. K., Krause D. C. (1999). Transcriptional analysis of the hmw gene cluster of Mycoplasma pneumoniae. J Bacteriol 181, 4978–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman C. U., Herrmann R. (2005). Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol Lett 253, 315–321. 10.1016/j.femsle.2005.09.054 [DOI] [PubMed] [Google Scholar]