Abstract
Chlamydia trachomatis is the most common bacterial infection of the human reproductive tract globally; however, the mechanisms underlying the adaptation of the organism to its natural target cells, human endocervical epithelial cells, are not clearly understood. To secure its intracellular niche, C. trachomatis must modulate the host cellular machinery by secreting virulence factors into the host cytosol to facilitate bacterial growth and survival. Here we used primary human endocervical epithelial cells and HeLa cells infected with C. trachomatis to examine the secretion of bacterial proteins during productive growth and persistent growth induced by ampicillin. Specifically, we observed a decrease in secretable chlamydial protease-like activity factor (CPAF) in the cytosol of host epithelial cells exposed to ampicillin with no evident reduction of CPAF product by C. trachomatis. In contrast, the expression of CopN and Tarp was downregulated, suggesting that C. trachomatis responds to ampicillin exposure by selectively altering the expression of secretable proteins. In addition, we observed a greater accumulation of outer-membrane vesicles from C. trachomatis in persistently infected cells. Taken together, these results suggest that the regulation of both gene expression and the secretion of chlamydial virulence proteins is involved in the adaptation of the bacteria to a persistent infection state in human genital epithelial cells.
Introduction
Genital serovariants (serovars D–K) of the obligate intracellular bacterium Chlamydia trachomatis are the world’s most common sexually transmitted bacterial pathogens, accounting for an estimated 90 million new cases annually (Brunham & Rey-Ladino, 2005). These serovariants have an exclusive tropism for the columnar epithelial cells of the genital mucosae. A typical infection cycle is initiated when infectious elementary bodies (EBs) attach to and enter into these cells. Subsequently, bacteria reside within a membrane-bound vacuole termed an inclusion and undergo a programmed biphasic transition from EBs to metabolically active replicating reticulate bodies (RBs), and then from RBs to EBs (Hatch, 1999; Moulder, 1991). Eventually, EBs and RBs exit the cell by extrusion or cell lysis (Hybiske & Stephens, 2007). Chlamydia species can also enter into an altered growth state that has been termed persistence. Chlamydial persistence in vitro is defined as a viable but non-cultivable growth state and is typified by enlarged, pleiomorphic RBs that cannot undergo binary fission and differentiation into EBs; however, they do continue chromosomal and plasmid replication (Beatty et al., 1994; Hogan et al., 2004; Lambden et al., 2006). Although antibiotics usually resolve infection, these aberrant persistent forms are more refractory to antibiotic treatment in culture (Reveneau et al., 2005; Wyrick & Knight, 2004). Several inducers have been shown to cause persistent forms in vitro (Wyrick, 2010), including exposure to penicillin or ampicillin (Matsumoto & Manire, 1970; Wolf et al., 2000), exposure to interferon-gamma (IFN-γ) (Beatty et al., 1994; Belland et al., 2003), nutrient depletion (Harper et al., 2000; Raulston, 1997) and co-infection of epithelial cells with herpes simplex virus (Deka et al., 2006).
The most common genital site of infection in women is the endocervix. Infection may result in the clinical syndrome of mucopurulent cervicitis but it is asymptomatic in over 80 % of individuals (Brunham & Rey-Ladino, 2005). Natural history studies suggest that most women can eventually clear the infection, but this can take up to several years (Brunham & Rey-Ladino, 2005; Gottlieb et al., 2010; Molano et al., 2005). In some women, C. trachomatis may ascend into the endometrium and fallopian tubes, where the bacteria can establish a chronic infection that results in pelvic inflammatory disease. Why Chlamydia infections take so long to clear is not known, but it is thought to be the consequence of the numerous evasion strategies used by the organism, including the ability to persist as an altered intracellular form (Brunham & Rey-Ladino, 2005).
To secure an intracellular niche, Chlamydia must secrete virulence proteins into the host cell, where they modulate the host cellular machinery to promote infection (Betts et al., 2009). Chlamydia species utilize not only the type III secretion system (T3SS), but also other diverse secretion mechanisms to translocate bacterial proteins into host cells. A major virulence factor, chlamydial protease-like activity factor (CPAF), has been shown to translocate via the Sec-dependent pathway (Chen et al., 2010). CPAF primarily interferes with cellular processes by cleaving host proteins, including the transcription factors RFX and USF-1 that are required for major histocompatibility complex (MHC) antigen expression (Zhong et al., 2001). Some T3SS effectors are also known to target various cellular processes. For example, secretion of the T3SS effectors Tarp (Clifton et al., 2004) and CT694 (Hower et al., 2009) from EBs into the host cytosol facilitates C. trachomatis entry via actin recruitment. Additionally, CopN serves as a virulence factor (Huang et al., 2008) and as an exported T3SS regulator that modulates the secretion of T3SS translocators and effectors (Fields & Hackstadt, 2000). Members of the family of inclusion-membrane proteins (Inc), such as IncA, directly participate in inclusion biogenesis, acquisition of host nutrients and interaction with the host protein factors (Bannantine et al., 1998; Rockey et al., 2002). Elucidation of the machinery and the conditions that trigger secretion of virulence proteins from C. trachomatis into the cytosol of host cells would provide new insights for understanding the pathogenesis of the disease.
Previous studies have indicated that in vitro inducers of persistence, including iron depletion or IFN-γ treatment of infected HEp-2 cells, decrease the secretion of CPAF protein by C. pneumoniae (Heuer et al., 2003; Shaw et al., 2002). However, relationships between secretion activities, gene regulation, and the mechanism by which C. trachomatis survives under conditions that induce persistence, remain to be determined. Here, using primary epithelial cells derived from the human endocervix, the most common natural tissue site of C. trachomatis, and the classic HeLa cell infection model, we determined whether the secretion activity of C. trachomatis changed in the presence of ampicillin and explored some of the possible mechanisms underlying these changes. As a commonly prescribed antibiotic for treatment of bacterial infections, ampicillin has been used as an inducer of persistence in vitro and it simulates an inadequate antimicrobial treatment of Chlamydia infection (Wyrick, 2010). We demonstrated that ampicillin exposure of infected epithelial cells interrupted the development of C. trachomatis and redistributed CPAF. We also confirmed our observations in the IFN-γ-mediated model of C. trachomatis persistence (Belland et al., 2003). These changes in the bacteria–host cell interaction during persistence could provide an advantage to C. trachomatis for survival and adaptation in human endocervical epithelial cells.
Methods
Cell culture and C. trachomatis infection.
HeLa 229 cells were cultured in RPMI 1640 medium supplemented with 10 % (v/v) fetal bovine serum and 10 µg gentamicin ml−1. Human primary endocervical epithelial cell cultures were established from endocervical tissue explants obtained from women undergoing hysterectomies for benign gynaecological conditions under a protocol approved by the LSU Health Sciences Center Institutional Review Board as previously described (Herbst-Kralovetz et al., 2008). Primary endocervical epithelial cells were maintained at 37 °C in 5 % CO2 in a serum-free medium that is highly selective for epithelial cells: EpiLife medium containing EpiLife defined growth supplement (Cascade Biologics). Monolayers of HeLa 229 cells or primary endocervical cells were inoculated with C. trachomatis serovar F/Cal-I-13 with a dose that results in 50 % of cells being infected, and centrifuged at 1600 g for 40 min at 37 °C. Fresh medium was added to the infected cells and incubated at 37 °C for various time periods as indicated in each experimental result.
Ampicillin and IFN-γ exposure of C. trachomatis-infected cells and persistence reactivation assays.
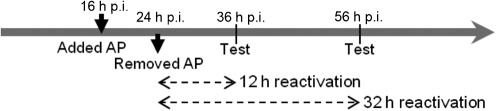
Infected HeLa and primary human endocervical epithelial cells were exposed to ampicillin (10 µg ml−1) at 16 h post-infection (p.i.). A reactivation experiment was performed in both HeLa and primary endocervical cells by removing ampicillin-containing medium from the culture after 8 h of ampicillin exposure (at 24 h p.i.), followed by incubation with ampicillin-free medium for an additional 12 or 32 h (Fig. 1). Subculture was performed to enumerate infectious progeny (i.e. EBs) through the calculation of inclusion-forming units (IFUs).
Fig. 1.
Experimental schematic for the reactivation assay.
The IFN-γ model of chlamydial persistence was performed as described by Belland et al. (2003), but with minor modifications. Briefly, HeLa and primary human endocervical epithelial cells were exposed to 50 or 200 U IFN-γ ml−1, respectively, for 24 h prior to infection. After infection, fresh IFN-γ-containing medium was added and maintained until fixation at 36 h p.i. A concentration of 50 U IFN-γ ml−1 was used for infected HeLa cells, and 200 U IFN-γ ml−1 was used for infected primary endocervical cells since there is a high concentration of tryptophan in the primary culture medium. The optimal IFN-γ concentrations that resulted in a persistent phenotype were determined using morphological analyses and subculture techniques to enumerate infectious progeny. The ability to reactivate persistent bacteria was confirmed using the standard methodology of Belland et al. (2003).
Purification of C. trachomatis particles from infected HeLa cells.
RBs and EBs were purified by centrifugation on Renografin density gradients as previously described (Shen et al., 2000). To isolate fragile persistent forms (PBs), cell disruption by nitrogen cavitation was used, which is a technique based on the rapid decompression of a cell suspension from a pressure vessel (Gottlieb & Adachi, 2000). For 2.5×108 infected HeLa cells, we used 125 p.s.i. (862 kPa) for 5 min intervals until more than 90 % host cell lysis was achieved, as determined by trypan blue staining. A solution of 25 % Renografin in sucrose/phosphate/glutamic acid buffer (SPG: 0.2 M sucrose, 3.8 mM KH2PO4, 6.7 mM Na2HPO4, 5 mM l-glutamic acid, pH 7.4) was overlaid with a 5 ml sample of infected cells and centrifuged in a Beckman SW 28 rotor at 18 000 r.p.m. for 1 h at 4 °C. The resuspended bacterial mixture was then overlaid on and centrifuged through a discontinuous gradient consisting of 28 %, 32 %, 40 %, 44 % and 52 % Renografin in a SW 28 rotor at 24 000 r.p.m. for 1 h at 4 °C. The bands of turbidity at interfaces of 28–34 % and 34–40 % Renografin solution were collected, sedimented by centrifugation, and suspended in 2 ml SPG for analysis by transmission electron microscopy (TEM) and immunoblotting.
Antibodies.
The primary antibodies used to detect chlamydial antigens in this study were (i) rabbit polyclonal antibodies directed against CopN (kind gift of Dr Ken Fields, University of Miami, FL, USA), IncA and Tarp (kind gifts of Dr Ted Hackstadt, Rocky Mountain Laboratories, NIH, NIAID, Hamilton, MT, USA) and OmcB (a kind gift of Dr Patrick Bavoil, University of Maryland, MD, USA); (ii) mouse monoclonal antibodies against the major outer-membrane protein (MOMP) (Wang et al., 2006) and EF-Tu (kind gifts of Dr You-xun Zhang, Boston University, Boston, MA, USA), and the C-terminal region of CPAF (clone 100) (Zhong et al., 2001).
Indirect immunofluorescence assays.
C. trachomatis-infected HeLa and primary endocervical cells on 12 mm-diameter glass coverslips were fixed with 2 % paraformaldehyde for 45 min and permeabilized with 4 % saponin for 30 min, followed by blocking with iTFX (Invitrogen) overnight at 4 °C. Primary antibodies against chlamydial MOMP, IncA, CPAF, CopN or Tarp were incubated with cells overnight at 4 °C, followed by either Alexa 568- or FITC-conjugated secondary antibodies (Molecular Probes) for 45 min at 37 °C. All cells were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Molecular Probes). Images were obtained with a Nikon E600 and Leica DMRXA automated upright epifluorescent microscope. Z-axis plane capture, deconvolution and analyses were performed with SlidebookTM deconvolution software (Intelligent Imaging Innovations).
Immunoprecipitation and immunoblot analysis.
A Subcellular Protein Fractionation kit (Thermo Fisher Scientific) was used to separate and extract different cellular contents stepwise from C. trachomatis-infected cells at 4 °C, according to the manufacturer’s instructions. Briefly, cytoplasmic extraction buffer was added to the cell pellet to cause selective membrane permeabilization, releasing the soluble host cell cytoplasmic fraction. Membrane extraction buffer was used to solubilize plasma, mitochondrial and endoplasmic reticulum–Golgi membranes. Nuclear extraction buffer was used to solubilize nuclear and chlamydial inclusion membranes and inclusion contents. The remaining pellet contained mostly actin-bound protein and insoluble chlamydial proteins. For simplification of nomenclature in this study, these fractions were designated cytosol, membrane, inclusion fraction and pellet, respectively. For immunoprecipitation, soluble cellular fractions dialysed against buffer containing 10 mM Tris/HCl (pH 8.0), 200 mM NaCl, 1 µM DTT and 5 % (v/v) glycerol were mixed with different specific antibodies and incubated at 4 °C overnight. Protein and antibody complexes were captured with protein A Sepharose (Sigma) for an additional 6 h at 4 °C. After extensive washing to remove non-specifically bound material, protein complexes were then separated by 12 % SDS-PAGE, transferred to Immobilon-PVDF membranes and subjected to immunoblot analysis as described previously (Hua et al., 2009). Blots were developed using a horseradish peroxidase-conjugated secondary antibody and the SuperSignalP Chemiluminescent Detection kit (Pierce). Density of the signals on the immunoblot was determined using Quantity One software (Bio-Rad) (Shen et al., 2004).
Total RNA extraction and RT-PCR analysis.
Total RNA was isolated from cells harvested at 24, 36, 40 and 48 h p.i. using TRIzol (Invitrogen), according to the manufacturer’s instructions. Samples were treated with DNase I (USB) to remove contaminating DNA, as described previously (Rao et al., 2009). Primer pairs were designed to amplify products of 332 bp for the tuf gene (CT322), 200 bp for the cpaf gene (CT858), 213 bp for the copN gene (CT089), and 164 bp for the tarp gene (CT456). They are tufrtF and tufrtR (5′-GACAAGTTGGGGTTCCTTAC-3′and 5′-ACGCTCAATACGTCCAGTTA-3′), cpafrtF and cpafrtR (5′-CTCAGGATGAAGTGGTTGAT-3′ and 5′-GAATAGGCGTTGATAACTCG-3′), copNrtF and copNrtR (5′-AACTGGTAGGCCCAGATACT-3′ and 5′-GAGGCGTACTAGAGAAGGAAG-3′) and tarprtR3 and tarprtR3 (5′-AGTTACACAAACAGCAAACG-3′ and 5′-ACACTTCCTGAATCATCTCC-3). Five micrograms of total RNA was reverse transcribed in a reaction containing reverse primers and 5 U AMV reverse transcriptase (USB) at 42 °C for 1 h. PCR was performed on the cDNA products, with the primer pairs as above, and Taq DNA polymerase (New England Biolabs) using the following protocol: 94 °C 0.5 min, 54 °C 0.5 min, and 72 °C 0.5 min for 30 cycles. PCR products were separated on a 2.0 % agarose gel, stained with ethidium bromide, photographed with a Gel Doc 3000 (Bio-Rad), and then quantified using Quantity One software (Bio-Rad) as described previously (Shen et al., 2004).
Electron microscopy.
For ultrastructural analysis, infected HeLa and primary endocervical epithelial cells or Renografin-purified C. trachomatis forms were fixed and processed as described previously (Beatty, 2006).
Results
Establishment of C. trachomatis infection in primary human endocervical epithelial cells
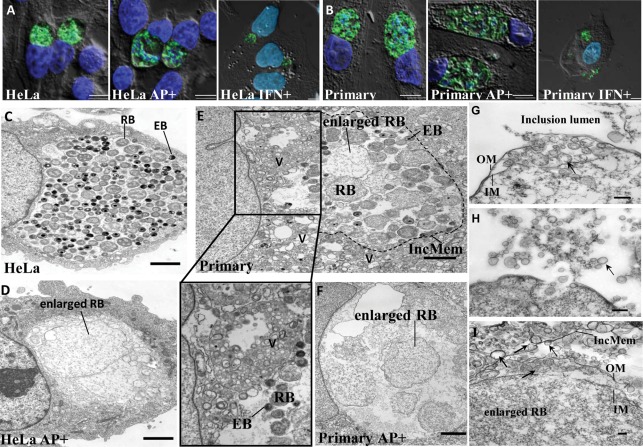
Previous studies have indicated that cultured primary human endometrial and ectocervical epithelial cells are less susceptible to C. trachomatis infection compared to McCoy cells (Moorman et al., 1986). To establish whether normal C. trachomatis forms could develop in primary human endocervical epithelial cells, we used 10 times more EBs than the number needed for a 50 % infection in HeLa cells to achieve equivalent infection rates in both cell types. Depending on the donor (n = 7), 30–70 % of cells were infected as determined by immunofluorescence analysis (IFA) using an anti-MOMP antibody. C. trachomatis inclusions in primary endocervical cell cultures exhibited an irregular staining pattern of MOMP compared to that observed in HeLa cells (Fig. 2A, B). The size and morphology of inclusions in these primary cell cultures varied from donor to donor, but no correlation was found between the size of the inclusion and the size of the cell when the inclusion area was normalized to the area of the cell (data not shown). TEM analysis revealed that inclusions generally contained fewer chlamydial particles in primary endocervical cells (Fig. 2E) than those seen in HeLa cells (Fig. 2C). In infected primary endocervical cells fixed at 36 h p.i., there was a mixture of normal EBs, RBs and intermediate forms, features suggestive of productive chlamydial forms. Additionally, enlarged RB-like forms with lower electron density often co-existed in C. trachomatis-infected primary endocervical cells. We also observed that some of the membranes of the enlarged forms as well as the inclusions were disrupted in the cross-section view. These data indicate that primary human endocervical epithelial cells are susceptible to C. trachomatis infection ex vivo. Interestingly, there were abundant multivesicular structures in the cytosol of infected primary endocervical epithelial cells that were adjacent to the inclusion (Fig. 2E). These multivesicular structures were also observed in C. trachomatis-infected HeLa cells, but in much lower numbers (data not shown). In the cross-section view of cells these multivesicular structures were generally round, with diameters ranging from 100 to 300 nm. No similar structures were observed in uninfected cells, suggesting that the formation of these multivesicular structures was associated with C. trachomatis infection.
Fig. 2.
C. trachomatis infection in genital tract epithelial cells. (A, B) Morphological analysis by epifluorescent microscopy and deconvolution of C. trachomatis-infected cells unexposed or exposed to ampicillin (AP+) or IFN-γ (IFN+). Infected HeLa cells (A) and primary human endocervical epithelial cells (B) were fixed at 36 h p.i. MOMP was visualized as green. Host nuclei and bacteria were visualized as blue by DAPI staining. All images shown are overlays of fluorescence images (green and blue) on differential interference contrast (DIC) images (grayscale). Scale bars, 5 µm. (C, D) TEM of C. trachomatis inclusions reveals the presence of productive forms in the absence of ampicillin (C) and the accumulation of enlarged persistent forms in the presence of ampicillin (D) in C. trachomatis-infected HeLa cells. Scale bars, 2 µm. (E, F) TEM of C. trachomatis inclusions in infected primary human endocervical epithelial cells unexposed (E) or exposed to ampicillin (F). An enlarged image of multivesicular structures from (E) is shown below. V, multivesicular structures; IncMem, inclusion membrane (outlined with dotted line). Scale bars, 2 µm. (G–I) C. trachomatis produces OMVs in cultured primary human endocervical epithelial cells exposed to ampicillin as determined by TEM. OM/IM, outer/inner membrane; IncMem, inclusion membrane. Representative OMVs are indicated by arrows. Scale bars, 200 nm (G, H) or 100 nm (I).
Ampicillin and IFN-γ induce viable but non-cultivable persistent forms in HeLa and primary endocervical epithelial cells
Exposure of C. trachomatis serovar F-infected HeLa and primary endocervical cells to ampicillin or IFN-γ resulted in aberrant, enlarged pleiomorphic RBs as determined by IFA with anti-MOMP staining (Fig. 2A, B). TEM imaging was also utilized to visualize ampicillin-mediated persistent forms (Fig. 2C, D). These results are consistent with observations from other investigators (Matsumoto & Manire, 1970; Wolf et al., 2000; Wyrick & Knight, 2004).
Infection assays were performed to evaluate infectious chlamydial forms in both C. trachomatis-infected HeLa and primary endocervical cells exposed to ampicillin. IFUs recovered from exposed cultures were negligible, consistent with a viable but non-cultivable phenotype (Table 1). Reactivation from persistence was effective in recovering infectious forms after removal of ampicillin. The recovered IFUs from reactivation were similar in number to those found in unexposed cultures (Table 1). The reactivated inclusions contained normal RBs, EBs and intermediate bodies, similar to those observed in the normal infected cultures.
Table 1. Reactivation assay and recovery of IFUs from infected cells.
See Methods and Fig. 1 for details of the reactivation assay. IFU counts are means±sd from three replicates for each sample. AP, ampicillin exposure; RA, reactivation.
| Sample | No. of IFUs recovered | |||
| DTCRC2* | DTCRC3* | DTCRC66* | HeLa cells | |
| 36 h | (3.88±2.52)×102 | (4.96±2.8)×105 | (1.01±0.13)×105 | (3.05±0.21)×106 |
| 36 h AP | 2.82±0.58 | 3.10±0.30 | 4.50±0.20 | 8.95±2.90 |
| 36 h RA | (3.84±2.16)×102 | (4.62±0.63)×105 | (2.70±1.4)×105 | (2.88±0.14)×106 |
| 56 h | (3.91±1.9)×102 | (3.44±3.1)×105 | (2.42±0.24)×105 | (3.21±0.71)×106 |
| 56 h AP | 4.42±1.6 | 3.15±0.10 | 4.30±0.10 | 6.60±1.12 |
| 56 h RA | (4.21±2.5)×102 | (4.62±0.41)×105 | (3.25±2.3)×105 | (2.89±0.71)×106 |
Primary epithelial cells from three donors, DTCRC2, DTCRC3 and DTCRC66.
Together these data indicate that despite slight phenotypic differences, persistent C. trachomatis forms can be induced by ampicillin and IFN-γ in both primary endocervical and HeLa cells. Thus, the generation of abnormal persistent forms of C. trachomatis is not an artefact of chlamydial growth in transformed cell lines.
Ampicillin exposure increases the formation of OMVs in HeLa and primary endocervical epithelial cells
TEM analysis showed that ampicillin exposure induced an accumulation of small spherical bilayered vesicles in the size range of 30 to 200 nm as viewed in cross-section (Fig. 2G–I). The presence of these vesicles corresponded to the reported OMVs or blebs observed in other cell lines infected with Chlamydia spp. (Giles et al., 2006; Matsumoto & Manire, 1970; Stirling & Richmond, 1980). The vesicles were observed between the inner and outer bacterial membranes, and this observation was highly reproducible in both C. trachomatis-infected primary endocervical cells and HeLa cells. TEM images suggest that OMVs may be released from the bacterial cells into the inclusion lumen (Fig. 2H, I). These vesicles appear to also be able to attach to the inclusion membrane (Fig. 2I) and can also be found in the host cell cytosol (Fig. 2I). In contrast, few OMVs were observed in C. trachomatis-infected HeLa and primary endocervical cells unexposed to ampicillin, and were more commonly associated with RBs rather than the inclusion membrane (data not shown).
Generation of persistent forms of C. trachomatis is coupled with altered localization of secretable bacterial proteins in infected cells
As a step to determine the relationship between bacterial secretion and the persistent state, we assessed the cellular localization of secretable proteins, namely CPAF and two T3SS effectors, Tarp (Clifton et al., 2004) and CopN (Fields & Hackstadt, 2000), during persistence. The T3SS effector IncA (Bannantine et al., 1998; Rockey et al., 2002) was used to visualize the inclusion membrane.
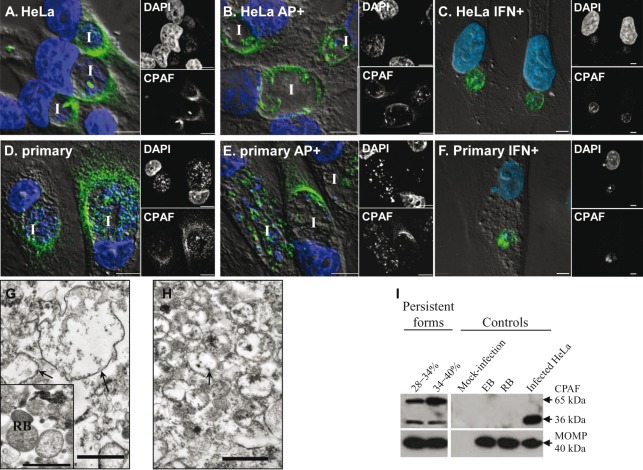
Time-course experiments of CPAF protein expression in normal C. trachomatis serovar F-infected HeLa cells indicated that CPAF-specific signals were present in the inclusion by 16 h p.i. By 24 h p.i., CPAF had clearly translocated into the host cell cytosol, where it remained for up to 48 h p.i. (36 h time point shown in Fig. 3A). This is consistent with previous observations using HeLa cells infected with C. trachomatis serovars A, D and L2 (Heuer et al., 2003; Zhong et al., 2001). Similarly, CPAF localized to the host cell cytosol in primary human endocervical epithelial cells at 36 h p.i. (Fig. 3D). Exposure to ampicillin or IFN-γ significantly reduced the amount of CPAF located in the host cytosol in the majority of infected HeLa (Fig. 3B, C) and primary endocervical cells (Fig. 3E, F). CPAF was distributed in two different patterns in exposed cells. In approximately 89 % of infected HeLa cells and 53 % of infected primary endocervical cells CPAF was retained within inclusions. In the rest of the cells, CPAF was generally located at the inclusion periphery in the host cytosol. The similarity in the altered CPAF localization observed in the two persistence models suggests that this observation is unlikely to be an artefact.
Fig. 3.
Localization of CPAF protein in C. trachomatis-infected cells. Infected HeLa cells (A–C) and primary endocervical cells (D–F) cultured in the absence or presence of persistence inducers (ampicillin, AP+; or IFN-γ, IFN+) as indicated were fixed at 36 h p.i. and immunostained with anti-CPAF antibody 100a. Overlays of DIC, CPAF (green) and DAPI (blue) images are shown in the large panels to the left, with separate small images of the single channels to the right. I, inclusion. Scale bars, 5 µm. (G, H) Representative TEM images of persistent forms isolated from C. trachomatis-infected HeLa cells exposed to ampicillin at the 28–34 % Renografin interface (G) and the 34–40 % Renografin interface (H). The control image of isolated normal RBs is shown in the inset in (G). Representative persistent forms are indicated by arrows. Note the difference in electron density between normal RBs and the persistent forms. Scale bars, 1 µm. (I) Immunoblot analysis of isolated C. trachomatis forms against CPAF. Lysates of isolated persistent forms at the 28–34 % and 34–40 % interfaces (left panels), and control RBs and EBs (right panels), were fractioned by SDS-PAGE, followed by immunoblotting with anti-CPAF antibody 100a and anti-MOMP antibody. Mock-infected and C. trachomatis-infected cells were used as negative and positive controls. Each panel is representative of at least three independent experiments.
To further study the localization of CPAF protein with C. trachomatis exposed to ampicillin, persistent forms that accumulated at the interfaces of 28–34 % and 34–40 % Renografin solutions were collected. TEM analysis revealed that the majority of the chlamydial persistent forms in these preparations were intact and all had a lower electron density compared to normal RBs (Fig. 3G, H). While some larger persistent forms (equal or larger than 1 µm in diameter) were present with smaller forms in the 28–34 % Renografin sample (Fig. 3G), relatively uniform-sized persistent forms (average ~0.5 µm in diameter) were observed in the 34–40 % Renografin interface sample (Fig. 3H). Immunoblot analysis with lysates of these samples revealed the presence of CPAF protein corresponding to CPAFc (36 kDa) and its zymogen (~65 kDa) (Fig. 3I). In control experiments, CPAF was undetectable in both isolated normal RBs and EBs. These results suggest that CPAF was associated with the isolated ampicillin-induced persistent forms. Therefore, the observed decrease in host cytosolic CPAF is not due to either decreased synthesis or increased protein degradation in ampicillin-induced C. trachomatis persistence.
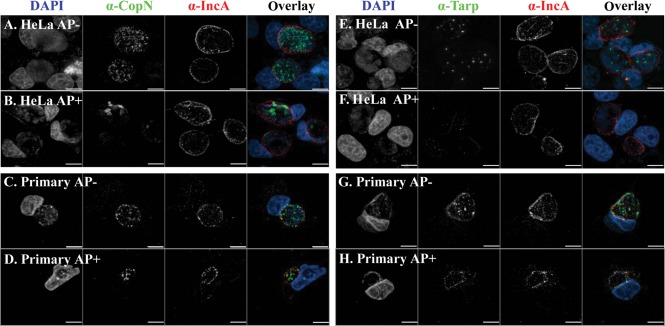
In infected HeLa cells, CopN-specific immunostaining was not detected until 20 h p.i., and high levels of CopN were observed up to 48 h p.i. (not shown). In normal C. trachomatis infection of primary endocervical and HeLa cells at 36 h p.i., the majority of CopN was located inside the inclusion or associated with the inclusion membrane (Fig. 4). This localization is consistent with the observations reported by Fields & Hackstadt (2000) with C. trachomatis serovar L2. However, ampicillin exposure altered the staining pattern of CopN, which remained in the inclusion, but displayed a weaker staining signal in the infected cells.
Fig. 4.
Analysis of CopN and Tarp proteins in C. trachomatis-infected cells. Immunostaining patterns of CopN (left panels) and Tarp (right panels) in infected HeLa (A, B, E, F) and primary human endocervical epithelial cells (C, D, G H) as indicated. Cells were fixed at 36 h p.i. CopN or Tarp proteins were visualized as green. Host nuclei and bacteria were visualized as blue by counterstaining with DAPI. The inclusion membrane is labelled as red with an anti-IncA antibody. Scale bars, 5 µm. α-, anti-.
Tarp-specific immunostaining became detectable at 28 h p.i. in infected HeLa cells and was observed at higher levels up to 48 h p.i. (not shown). Tarp was localized within the inclusion and distributed in a punctate pattern in normal infected HeLa (Fig. 4E) and primary endocervical cells (Fig. 4G). In contrast, exposure to ampicillin decreased levels of Tarp, and many inclusions displayed little or no detectable Tarp-specific staining in infected HeLa (Fig. 4F) and primary endocervical cells (Fig. 4H). Unlike CopN and Tarp, the levels and distribution of IncA in the inclusion membrane were not significantly changed upon ampicillin exposure (Fig. 4). Taken together, these results indicate that ampicillin exposure induced different and specific changes in these secretable chlamydial proteins.
C. trachomatis responds to ampicillin exposure by selectively altering the expression of secretable proteins in infected cells
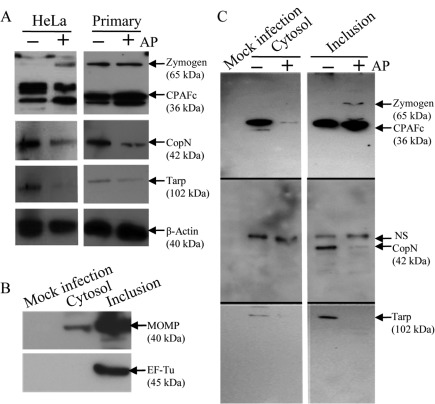
To more precisely identify the mechanisms underlying the observed ampicillin-induced phenotypic changes, whole cellular lysates of ampicillin-exposed and normal C. trachomatis-infected cells were assayed by immunoblot analysis for levels of CPAF, Tarp and CopN. Because of the variability in C. trachomatis infection in primary endocervical cells from donor to donor, cells derived from four different donors were examined. In all infected primary endocervical cell cultures, bands corresponding to CPAFc (36 kDa) and occasionally its zymogen (~65 kDa) were observed. A representative result is shown in Fig. 5(A), in which the overall levels of CPAF under ampicillin exposure or normal growth conditions were equivalent. A similar result was also observed in HeLa cells. In contrast, levels of Tarp and CopN proteins were clearly decreased in both infected primary endocervical cells and HeLa cells exposed to ampicillin (Fig. 5A).
Fig. 5.
Quantification of the level and subcellular distribution of C. trachomatis CPAF, CopN and Tarp. (A) Levels of CPAF, CopN and Tarp were examined in whole-cell lysates of C. trachomatis-infected HeLa and primary human endocervical epithelial cells exposed to ampicillin or under normal growth conditions. Cells were harvested at 36 h p.i. Antibodies specific to chlamydial CPAF, CopN and Tarp were used to visualize relevant proteins. Host β-actin was probed with an anti-β-actin antibody and used to normalize the protein loading amounts. Results are shown of a representative experiment from three HeLa cell experiments, and a representative experiment with primary endocervical epithelial cells derived from one of four donors. (B) Evaluation of cytosol and inclusion fractions from normal infected HeLa cells using immunoprecipitation, followed by immunoblotting with antibodies specific to chlamydial MOMP or EF-Tu. The lysate of mock-infected HeLa cells was used as a negative control. (C) Levels of CPAF, CopN and Tarp proteins in cytosol and inclusion fractions extracted from C. trachomatis-infected HeLa cells either under normal growth conditions (AP−) or exposed to ampicillin (AP+), determined by immunoprecipitation followed by immunoblot analysis. A lysate of mock-infected HeLa cells was used as a negative control. The same amounts of C. trachomatis-infected HeLa cells were used in the presence of an excess amount of antibody for immunoprecipitation. Results shown are a representative experiment of three independent experiments.
We then used C. trachomatis-infected HeLa cells harvested at 36 h p.i. to separate the host and bacterial protein contents into specific cellular fractions for chlamydial protein distribution analysis. It is difficult to obtain sufficient primary human endocervical epithelial cells from a single donor for a reproducible cellular fractionation study, and IFA indicates similarity in the localization of CPAF, CopN and Tarp between C. trachomatis-infected HeLa and primary endocervical cells (Figs 3 and 4). Cellular fractions defined as cytosol, membrane and inclusion (see Methods) were analysed by immunoprecipitation, followed by immunoblotting. As expected, host β-actin was present in the all three fractions and chlamydial EF-Tu was only present in the inclusion fraction. While chlamydial MOMP was detected at a high level in the inclusion fraction, it was present at a low level in the cytosol fraction (Fig. 5B). Cytosolic MOMP has been described previously and likely associates with the inclusion-membrane vesicles (Giles & Wyrick, 2008). These results suggest that there was no obvious chlamydial cytoplasmic protein contamination in the host-cell fractions.
We next measured the levels of the secretable chlamydial proteins CPAF, CopN and Tarp from cytosol and inclusion fractions using immunoprecipation, followed by immunoblotting. CPAF was significantly enriched in the cytosol fraction and present at a low level in the inclusion fraction in cultures with normal productive C. trachomatis growth (Fig. 5C). In contrast, ampicillin exposure reduced the level of CPAF obtained from the cytosol fraction. Instead, a higher level of CPAF was obtained from the inclusion fraction (Fig. 5C). Nevertheless, the overall quantity of CPAF from both the cytosol and inclusion fractions in ampicillin-exposed cells was approximately equivalent to that found in the infected cells unexposed to ampicillin. These results substantiate our IFA and immunoblot data (Figs 3 and 5A), further suggesting that the total level of CPAF is unchanged, and the CPAF translocation into the host cell cytosol is decreased in ampicillin-exposed cells.
CopN protein was detected only in the inclusion fraction (Fig. 5C). A higher level of CopN was found in normal infected cells, while a reduced level of CopN was observed in ampicillin-exposed cells. The CopN protein band was observed at approximately 42 kDa, close to the predicted molecular mass. A larger band reactive to CopN antibody on the immunoblot was resolved; however, the signal was constant regardless of presence or absence of ampicillin and was absent in mock-infected HeLa cells. Thus, we considered this band non-specific and due to a cross-reaction with an unknown chlamydial protein. A low level of Tarp protein was immunoprecipitated from the cytosol fraction, and a much higher level of Tarp was detected in the inclusion fraction of normal C. trachomatis-infected cells (Fig. 5C). Detection of the host-cytosol-localized Tarp at 36 h p.i. was unexpected, as Tarp protein has been reported to be rapidly secreted upon C. trachomatis–host cell contact (Clifton et al., 2004). We failed to detect any Tarp signal in samples from ampicillin-exposed HeLa cells. Accordingly, the overall levels of CopN or Tarp from both cytosol and inclusion fractions of ampicillin-exposed cells were substantially lower compared to those from normal infected cells.
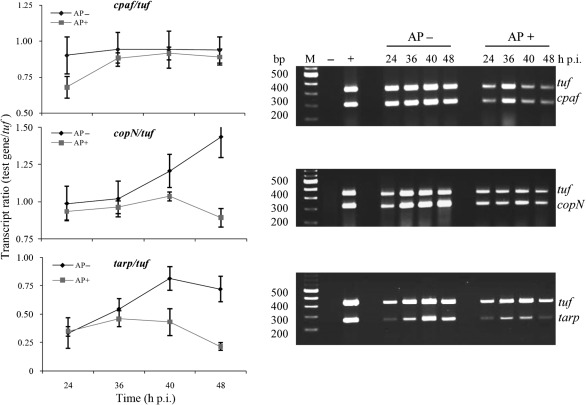
To determine whether the levels of protein analysed by immunoblot analysis were in concordance with their mRNA levels, we investigated the transcription of the copN, tarp and cpaf genes in C. trachomatis-infected HeLa cells. We used a dual-complex RT-PCR, in which two primer pairs were designed to amplify partial coding regions of copN, tarp or cpaf, as well as tuf, which encodes the translation elongation factor EF-Tu. We optimized the conditions that generated compatible amounts of amplicon production with the primers using varied amounts of C. trachomatis genomic DNA as templates. No amplified products were observed in the absence of a cDNA synthesis step with C. trachomatis RNA as a template. This demonstrated that no detectable DNA contamination was present in the samples. We performed RT-PCR experiments measuring the transcription of copN, tarp and cpaf and then normalized these gene transcript levels to tuf transcript levels. The tuf gene was chosen as a control for normalization because its product EF-Tu is essential for protein synthesis during all stages of C. trachomatis growth (Shen et al., 2000). The transcript ratio was used to compare the relative changes in transcription levels over time. Fig. 6 demonstrates that ampicillin exposure reduced the overall transcription levels of copN, tarp, cpaf and tuf in infected cells from 24 to 48 h p.i. compared to those in normal infected cells, consistent with slow-growing bacteria. After normalization to tuf, the relative transcript ratio of cpaf in ampicillin-exposed cells was equivalent to that in normal infected cells and consistent with the level of total CPAF protein as determined by immunoblot analysis (Fig. 5A). In contrast, relative transcript ratios from copN and tarp decreased (Fig. 6) in ampicillin-exposed cells harvested at 24, 36 and 48 h p.i. Taken together these results indicate that ampicillin exposure resulted in the selective downregulation of Tarp and CopN expression, but not CPAF expression, at both the transcriptional and translational level. Thus, the decreased protein levels of CopN and Tarp (Fig. 5C) can be attributed, at least in part, to decreased mRNA levels of copN and tarp.
Fig. 6.
Transcription analysis of chlamydial genes in C. trachomatis-infected HeLa cells by dual-complex RT-PCR. The right panels show ethidium-bromide-stained agarose gels indicating the transcript levels of the tuf, cpaf, copN and tarp genes from C. trachomatis-infected HeLa cells exposed to ampicillin (AP+) or under normal growth conditions (AP−). Genomic DNA extracted from purified C. trachomatis EBs was used as a positive control to demonstrate that in this dual-complex reaction the two primer pairs amplified the targets with similar efficiencies (lane +). RNA isolated from uninfected HeLa cells was used as a negative control (lane −). A fixed quantity of total RNA (host cell RNA+C. trachomatis RNA) isolated at the indicated times was treated with DNase I and subjected to RT-PCR. Graphs depicting the mean±sd of three independent experiments for each primer set are shown on the left. These show the ratio of cpaf, copN and tarp transcripts to the tuf transcripts at different time points p.i. as revealed by the RT-PCR analysis in normal infected cultures or cultures exposed to ampicillin. Note the decrease in transcript levels that occurred in ampicillin-exposed samples due to bacterial growth retardation.
Discussion
In this study, we investigated key events that occur during persistence of C. trachomatis growth in vitro: the developmental cycle, secretion activity, and gene expression. Our work suggests that these events may be involved in the adaptation of C. trachomatis to a persistent state in human endocervical epithelial cells.
The value of using primary endocervical cells for biological studies of C. trachomatis infection is becoming widely appreciated (Moorman et al., 1986; Rasmussen et al., 1997; Roth et al., 2010). We were able to demonstrate, for the first time, that primary human endocervical epithelial cells cultured ex vivo are permissive for C. trachomatis infection. Appearance of persistent-like-forms in primary endocervical cell cultures might be the result of the bacteria’s ability to adapt to a stricter intracellular niche, as primary endocervical cells, unlike many transformed cell lines, maintain appropriate expression and regulation of tissue-specific molecules (Fichorova et al., 1997; Rasmussen et al., 1997; Wyrick, 2006). Additionally, this pleiomorphic population of primary endocervical cells at multiple stages of differentiation most likely interacts with C. trachomatis differently, resulting in the heterogeneity of infection observed in this study. Therefore, the primary human endocervical epithelial cell model is likely to provide a more accurate representation of in vivo infection, and is recommended for validation of work performed in transformed cell lines. One limitation of using primary human endocervical epithelial cells for studies of C. trachomatis infection is that it is difficult to grow them in large numbers. In this study, this problem was rectified by the combined use of and comparison with the HeLa infection model. We established that the generation of abnormal persistent forms and the redistribution of secretable CPAF when C. trachomatis is exposed to ampicillin or IFN-γ was not an artefact of transformed cell lines and validated our novel experimental findings observed in HeLa cells.
Our data demonstrate that the vesiculation phenotype is common during productive C. trachomatis infection in primary endocervical cells (Fig. 2E). Fewer multivesicular structures are observed in C. trachomatis-infected HeLa cells. Although the origin of the multivesicular structures is unclear, it seems likely that they are involved in processes driven by both the bacteria and the host cell. First, they are present only in C. trachomatis-infected cells. Second, their localization is closely associated with both the inclusion and the host cytosol. Third, it has been shown that chlamydiae use numerous intracellular trafficking pathways to acquire nutrients from the host cell, and multiple vesicles have been shown to be important organelles in separating host cell protein and lipid droplets (Beatty, 2006; Cocchiaro et al., 2008). Further analyses are needed to determine their origin, dynamics and roles during both productive and persistent chlamydial growth.
It has been previously shown that C. trachomatis produces OMVs during the transition from RBs to EBs throughout the developmental cycle (Giles et al., 2006; Stirling & Richmond, 1980). The presence of relatively few and small OMVs in normal productively infected cells and the abundance of OMVs in ampicillin-exposed cells suggest that the formation of OMVs is ampicillin responsive. It is possible that OMV formation is one strategy utilized by C. trachomatis in response to stress. In other bacteria, overproduction of OMVs occurs in the presence of stress factors, such as antibiotics (Kadurugamuwa & Beveridge, 1995), phage infection (Loeb, 1974) and envelope stress due to the accumulation of unfolded proteins in the periplasm (McBroom et al., 2006). Increasing evidence indicates that OMVs provide a specific secretory mechanism in Gram-negative pathogens by which diverse virulence factors packaged in OMVs are distantly delivered, allowing pathogens to interact with the host (Amano et al., 2010; Ellis & Kuehn, 2010; Ünal et al., 2010). Giles & Wyrick (2008) reported that vesicles everting from the inclusion membrane can deliver diverse chlamydial antigens into human endometrial HEC-1B epithelial cells. The subsequent impact of chlamydial OMV–host cell interactions on the immune response or disease outcome remains to be elucidated. Since OMV components from other bacteria have proven to be valid vaccine candidates (Amano et al., 2010; Ellis & Kuehn, 2010; Ünal et al., 2010), analysis of immunogenic candidate proteins in chlamydial OMVs may aid in vaccine development for C. trachomatis.
The observed decrease of host-cytosol-localized CPAF induced by ampicillin or IFN-γ could result from decreased protein expression, increased degradation or altered transport. Unaltered levels of CPAF transcript and protein in ampicillin-exposed cells suggest the latter is the case. Our data are consistent with previous reports that the transcription (Belland et al., 2003) and protein levels of CPAF (Shaw et al., 2002) are also unchanged in an IFN-γ model of persistence. The finding that ampicillin exposure of C. trachomatis-infected primary endocervical epithelial cells alters CPAF localization provides additional support for our observations of CPAF localization in infected HeLa cells. Additionally, we developed the first IFN-γ-mediated persistence model in primary endocervical epithelial cells, which also confirms our findings in HeLa cells as well as findings reported by others (Heuer et al., 2003; Shaw et al., 2002).
In contrast to CPAF, we found that ampicillin induced a decrease in expression of CopN and Tarp. This differs from the findings by Belland et al. (2003) in the IFN-γ model of C. trachomatis persistence in HeLa cells, in which transcription of the tarp gene was downregulated, while copN transcription remained unchanged. These results suggest that transcriptional changes of certain chlamydial genes encoding secretable proteins vary in a stressor-dependent manner, as noted previously (Belland et al., 2003; Klos et al., 2009; Ouellette et al., 2006), perhaps due to differences in the mechanisms by which persistence is induced by different stressors.
Essential to the pathogenesis of C. trachomatis is the establishment and maintenance of an intracellular niche and the evasion of host immune responses during infection (Betts et al., 2009). A well-established function of CPAF is the modulation of MHC class I, MHC class II and CD1d expression (Kawana et al., 2007; Zhong et al., 1999, 2000). Because protein function is mostly coupled to protein localization and its levels, altered CPAF localization and decreased levels of T3SS components induced by ampicillin might (directly or indirectly) change the host cell environment. In addition, components of OMVs may also modulate the host signal transduction and inflammatory response (Ellis & Kuehn, 2010; Giles & Wyrick, 2008; Ünal et al., 2010). Alterations of chlamydial protein secretion may allow C. trachomatis to achieve intracellular survival upon ampicillin exposure, therefore protecting the bacteria from innate and acquired immunity. Our in vitro primary human endocervical epithelial cell model provides a valuable tool to investigate host cell factors that are relevant to the establishment of persistence, as this model most likely retains the appropriate responsiveness to endogenous factors, such as female steroid hormones that can be altered in transformed cervical cell lines (Guseva et al., 2005). While this study has focused on selective chlamydial secretable proteins, a broader proteomic analysis is warranted to elucidate how the overall secretome affects the infection process.
Acknowledgements
This work was supported, in part, by the Louisiana Vaccine Center and the South Louisiana Institute for Infectious Disease Research sponsored by the Louisiana Board of Regents (A. J. Q. and L. S.) and by NIH grants AI055869 (L. S.) and U19AI061972 (A. J. Q.). We are grateful to Drs David Martin, Robert Schoborg and Priscilla Wyrick for critical review of the manuscript and to Kristen Krup for her assistance in editing.
Abbreviations:
- CPAF
chlamydial protease-like activity factor
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- DIC
differential interference contrast
- EB
elementary body
- IFA
immunofluorescence analysis
- IFN-γ
interferon-gamma
- IFU
inclusion-forming unit
- MHC
major histocompatibility complex
- MOMP
major outer-membrane protein
- OMV
outer-membrane vesicle
- p.i.
post-infection
- RB
reticulate body
- TEM
transmission electron microscopy
- TTSS
type III secretion system
References
- Amano A., Takeuchi H., Furuta N. (2010). Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect 12, 791–798. 10.1016/j.micinf.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Bannantine J. P., Stamm W. E., Suchland R. J., Rockey D. D. (1998). Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect Immun 66, 6017–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty W. L. (2006). Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J Cell Sci 119, 350–359. 10.1242/jcs.02733 [DOI] [PubMed] [Google Scholar]
- Beatty W. L., Morrison R. P., Byrne G. I. (1994). Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev 58, 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R. J., Nelson D. E., Virok D., Crane D. D., Hogan D., Sturdevant D., Beatty W. L., Caldwell H. D. (2003). Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci U S A 100, 15971–15976. 10.1073/pnas.2535394100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts H. J., Wolf K., Fields K. A. (2009). Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr Opin Microbiol 12, 81–87. 10.1016/j.mib.2008.11.009 [DOI] [PubMed] [Google Scholar]
- Brunham R. C., Rey-Ladino J. (2005). Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 5, 149–161. 10.1038/nri1551 [DOI] [PubMed] [Google Scholar]
- Chen D., Lei L., Lu C., Flores R., DeLisa M. P., Roberts T. C., Romesberg F. E., Zhong G. (2010). Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology 156, 3031–3040. 10.1099/mic.0.040527-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D. R., Fields K. A., Grieshaber S. S., Dooley C. A., Fischer E. R., Mead D. J., Carabeo R. A., Hackstadt T. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A 101, 10166–10171. 10.1073/pnas.0402829101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro J. L., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H. (2008). Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A 105, 9379–9384. 10.1073/pnas.0712241105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka S., Vanover J., Dessus-Babus S., Whittimore J., Howett M. K., Wyrick P. B., Schoborg R. V. (2006). Chlamydia trachomatis enters a viable but non-cultivable (persistent) state within herpes simplex virus type 2 (HSV-2) co-infected host cells. Cell Microbiol 8, 149–162. 10.1111/j.1462-5822.2005.00608.x [DOI] [PubMed] [Google Scholar]
- Ellis T. N., Kuehn M. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74, 81–94. 10.1128/MMBR.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichorova R. N., Rheinwald J. G., Anderson D. J. (1997). Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol Reprod 57, 847–855. 10.1095/biolreprod57.4.847 [DOI] [PubMed] [Google Scholar]
- Fields K. A., Hackstadt T. (2000). Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol 38, 1048–1060. 10.1046/j.1365-2958.2000.02212.x [DOI] [PubMed] [Google Scholar]
- Giles D. K., Wyrick P. B. (2008). Trafficking of chlamydial antigens to the endoplasmic reticulum of infected epithelial cells. Microbes Infect 10, 1494–1503. 10.1016/j.micinf.2008.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles D. K., Whittimore J. D., LaRue R. W., Raulston J. E., Wyrick P. B. (2006). Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microbes Infect 8, 1579–1591. 10.1016/j.micinf.2006.01.018 [DOI] [PubMed] [Google Scholar]
- Gottlieb R. A., Adachi S. (2000). Nitrogen cavitation for cell disruption to obtain mitochondria from cultured cells. Methods Enzymol 322, 213–221. 10.1016/S0076-6879(00)22022-3 [DOI] [PubMed] [Google Scholar]
- Gottlieb S. L., Brunham R. C., Byrne G. I., Martin D. H., Xu F., Berman S. M. (2010). Introduction: The natural history and immunobiology of Chlamydia trachomatis genital infection and implications for Chlamydia control. J Infect Dis 201 (Suppl. 2), S85–S87. 10.1086/652392 [DOI] [PubMed] [Google Scholar]
- Guseva N. V., Dessus-Babus S. C., Whittimore J. D., Moore C. G., Wyrick P. B. (2005). Characterization of estrogen-responsive epithelial cell lines and their infectivity by genital Chlamydia trachomatis. Microbes Infect 7, 1469–1481. 10.1016/j.micinf.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Harper A., Pogson C. I., Jones M. L., Pearce J. H. (2000). Chlamydial development is adversely affected by minor changes in amino acid supply, blood plasma amino acid levels, and glucose deprivation. Infect Immun 68, 1457–1464. 10.1128/IAI.68.3.1457-1464.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch T. P. (1999). Developmental biology. In Chlamydia. Intracellular Biology, Pathogenesis, and Immunity. Edited by Stephens R. S., pp. 29–67. Washington, DC: American Society for Microbiology. [Google Scholar]
- Herbst-Kralovetz M. M., Quayle A. J., Ficarra M., Greene S., Rose W. A., II, Chesson R., Spagnuolo R. A., Pyles R. B. (2008). Quantification and comparison of Toll-like receptor expression and responsiveness in primary and immortalized human female lower genital tract epithelia. Am J Reprod Immunol 59, 212–224. 10.1111/j.1600-0897.2007.00566.x [DOI] [PubMed] [Google Scholar]
- Heuer D., Brinkmann V., Meyer T. F., Szczepek A. J. (2003). Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell Microbiol 5, 315–322. 10.1046/j.1462-5822.2003.00278.x [DOI] [PubMed] [Google Scholar]
- Hogan R. J., Mathews S. A., Mukhopadhyay S., Summersgill J. T., Timms P. (2004). Chlamydial persistence: beyond the biphasic paradigm. Infect Immun 72, 1843–1855. 10.1128/IAI.72.4.1843-1855.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hower S., Wolf K., Fields K. A. (2009). Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol 72, 1423–1437. 10.1111/j.1365-2958.2009.06732.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Rao X., Feng X., Luo X., Liang Y., Shen L. (2009). Mutagenesis of region 4 of sigma 28 from Chlamydia trachomatis defines determinants for protein-protein and protein-DNA interactions. J Bacteriol 191, 651–660. 10.1128/JB.01083-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Lesser C. F., Lory S. (2008). The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456, 112–115. 10.1038/nature07355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K., Stephens R. S. (2007). Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A 104, 11430–11435. 10.1073/pnas.0703218104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa J. L., Beveridge T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 177, 3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawana K., Quayle A. J., Ficarra M., Ibana J. A., Shen L., Kawana Y., Yang H., Marrero L., Yavagal S., et al. (2007). CD1d degradation in Chlamydia trachomatis-infected epithelial cells is the result of both cellular and chlamydial proteasomal activity. J Biol Chem 282, 7368–7375. 10.1074/jbc.M610754200 [DOI] [PubMed] [Google Scholar]
- Klos A., Thalmann J., Peters J., Gérard H. C., Hudson A. P. (2009). The transcript profile of persistent Chlamydophila (Chlamydia) pneumoniae in vitro depends on the means by which persistence is induced. FEMS Microbiol Lett 291, 120–126. 10.1111/j.1574-6968.2008.01446.x [DOI] [PubMed] [Google Scholar]
- Lambden P. R., Pickett M. A., Clarke I. N. (2006). The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology 152, 2573–2578. 10.1099/mic.0.29032-0 [DOI] [PubMed] [Google Scholar]
- Loeb M. R. (1974). Bacteriophage T4-mediated release of envelope components from Escherichia coli. J Virol 13, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Manire G. P. (1970). Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J Bacteriol 101, 278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom A. J., Johnson A. P., Vemulapalli S., Kuehn M. J. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 188, 5385–5392. 10.1128/JB.00498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molano M., Meijer C. J., Weiderpass E., Arslan A., Posso H., Franceschi S., Ronderos M., Muñoz N., van den Brule A. J. (2005). The natural course of Chlamydia trachomatis infection in asymptomatic Colombian women: a 5-year follow-up study. J Infect Dis 191, 907–916. 10.1086/428287 [DOI] [PubMed] [Google Scholar]
- Moorman D. R., Sixbey J. W., Wyrick P. B. (1986). Interaction of Chlamydia trachomatis with human genital epithelium in culture. J Gen Microbiol 132, 1055–1067. [DOI] [PubMed] [Google Scholar]
- Moulder J. W. (1991). Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55, 143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette S. P., Hatch T. P., AbdelRahman Y. M., Rose L. A., Belland R. J., Byrne G. I. (2006). Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol Microbiol 62, 1387–1401. 10.1111/j.1365-2958.2006.05465.x [DOI] [PubMed] [Google Scholar]
- Rao X., Deighan P., Hua Z., Hu X., Wang J., Luo M., Wang J., Liang Y., Zhong G., et al. (2009). A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes Dev 23, 1818–1829. 10.1101/gad.1784009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. J., Eckmann L., Quayle A. J., Shen L., Zhang Y. X., Anderson D. J., Fierer J., Stephens R. S., Kagnoff M. F. (1997). Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest 99, 77–87. 10.1172/JCI119136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulston J. E. (1997). Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect Immun 65, 4539–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveneau N., Crane D. D., Fischer E., Caldwell H. D. (2005). Bactericidal activity of first-choice antibiotics against gamma interferon-induced persistent infection of human epithelial cells by Chlamydia trachomatis. Antimicrob Agents Chemother 49, 1787–1793. 10.1128/AAC.49.5.1787-1793.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey D. D., Scidmore M. A., Bannantine J. P., Brown W. J. (2002). Proteins in the chlamydial inclusion membrane. Microbes Infect 4, 333–340. 10.1016/S1286-4579(02)01546-0 [DOI] [PubMed] [Google Scholar]
- Roth A., König P., van Zandbergen G., Klinger M., Hellwig-Bürgel T., Däubener W., Bohlmann M. K., Rupp J. (2010). Hypoxia abrogates antichlamydial properties of IFN-γ in human fallopian tube cells in vitro and ex vivo. Proc Natl Acad Sci U S A 107, 19502–19507. 10.1073/pnas.1008178107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. C., Vandahl B. B., Larsen M. R., Roepstorff P., Gevaert K., Vandekerckhove J., Christiansen G., Birkelund S. (2002). Characterization of a secreted Chlamydia protease. Cell Microbiol 4, 411–424. 10.1046/j.1462-5822.2002.00200.x [DOI] [PubMed] [Google Scholar]
- Shen L., Shi Y., Douglas A. L., Hatch T. P., O’Connell C. M. C., Chen J.-M., Zhang Y.-X. (2000). Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Arch Biochem Biophys 379, 46–56. 10.1006/abbi.2000.1854 [DOI] [PubMed] [Google Scholar]
- Shen L., Li M., Zhang Y. X. (2004). Chlamydia trachomatis sigma28 recognizes the fliC promoter of Escherichia coli and responds to heat shock in chlamydiae. Microbiology 150, 205–215. 10.1099/mic.0.26734-0 [DOI] [PubMed] [Google Scholar]
- Stirling P., Richmond S. J. (1980). Production of outer membrane blebs during chlamydial replication. FEMS Microbiol Lett 9, 103–105. 10.1111/j.1574-6968.1980.tb05616.x [DOI] [Google Scholar]
- Ünal C. M., Schaar V., Riesbeck K. (2010). Bacterial outer membrane vesicles in disease and preventive medicine. Semin Immunopathol 1–14 10.1007/s00281-010-0231-y. [DOI] [PubMed] [Google Scholar]
- Wang Y., Berg E. A., Feng X., Shen L., Smith T., Costello C. E., Zhang Y.-X. (2006). Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci 15, 122–134. 10.1110/ps.051616206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Fischer E., Hackstadt T. (2000). Ultrastructural analysis of developmental events in Chlamydia pneumoniae-infected cells. Infect Immun 68, 2379–2385. 10.1128/IAI.68.4.2379-2385.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick P. (2006). Polarized epithelial cell culture for Chlamydia trachomatis. Wymondham, Norfolk, UK: Horizon Bioscience. [Google Scholar]
- Wyrick P. B. (2010). Chlamydia trachomatis persistence in vitro – an overview. J Infect Dis 201 (Suppl. 2), S88–S95. 10.1086/652394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick P. B., Knight S. T. (2004). Pre-exposure of infected human endometrial epithelial cells to penicillin in vitro renders Chlamydia trachomatis refractory to azithromycin. J Antimicrob Chemother 54, 79–85. 10.1093/jac/dkh283 [DOI] [PubMed] [Google Scholar]
- Zhong G., Fan T., Liu L. (1999). Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med 189, 1931–1938. 10.1084/jem.189.12.1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G., Liu L., Fan T., Fan P., Ji H. (2000). Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J Exp Med 191, 1525–1534. 10.1084/jem.191.9.1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G., Fan P., Ji H., Dong F., Huang Y. (2001). Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med 193, 935–942. 10.1084/jem.193.8.935 [DOI] [PMC free article] [PubMed] [Google Scholar]