Abstract
The conversion of nicotinamide to nicotinic acid by nicotinamidase enzymes is a critical step in maintaining NAD+ homeostasis and contributes to numerous important biological processes in diverse organisms. In Borrelia burgdorferi, the nicotinamidase enzyme, PncA, is required for spirochaete survival throughout the infectious cycle. Mammals lack nicotinamidases and therefore PncA may serve as a therapeutic target for Lyme disease. Contrary to the in vivo importance of PncA, the current annotation for the pncA ORF suggests that the encoded protein may be inactive due to the absence of an N-terminal aspartic acid residue that is a conserved member of the catalytic triad of characterized PncA proteins. Herein, we have used genetic and biochemical strategies to determine the N-terminal sequence of B. burgdorferi PncA. Our data demonstrate that the PncA protein is 24 aa longer than the currently annotated sequence and that pncA translation is initiated from the rare, non-canonical initiation codon AUU. These findings are an important first step in understanding the catalytic function of this in vivo-essential protein.
Introduction
Human infection with the pathogenic spirochaete Borrelia burgdorferi following the bite of an infected tick can result in a multi-stage, inflammatory disorder called Lyme disease. Humans are incidental hosts for B. burgdorferi and, in nature, the spirochaete cycles between Ixodes sp. ticks and small rodents and birds. In order to survive in these diverse host environments, B. burgdorferi must employ molecular mechanisms for persistence and transmission (Rosa et al., 2005; Tilly et al., 2008).
The genome of B. burgdorferi is complex and segmented, consisting of a small, linear chromosome and, in the characterized type strain B31, at least 21 linear and circular plasmids (Casjens et al., 2000; Fraser et al., 1997). In general, the genes present on the chromosome encode proteins found in other bacterial species, whereas approximately 60 % of the genes found on the plasmids appear to encode proteins unique to Borrelia (Casjens et al., 2000; Fraser et al., 1997). Consistent with its reduced genome size, B. burgdorferi lacks many of the genes encoding metabolic enzymes required for de novo synthesis of nucleotides, amino acids, fatty acids and enzyme cofactors, all of which are essential for growth and survival of the spirochaete throughout its infectious cycle. Consequently, B. burgdorferi must scavenge required nutrients from its host environments (Bono et al., 1998; Byram et al., 2004; Jewett et al., 2007b, 2009; Lawrence et al., 2009; Purser et al., 2003; Tilly et al., 2004).
The pncA gene (BBE22) present on linear plasmid 25 (lp25) encodes a nicotinamidase that hydrolyses nicotinamide to nicotinic acid for production of NAD+ (Purser et al., 2003). B. burgdorferi does not appear to harbour genes encoding enzymes required for de novo NAD+ biosynthesis (Casjens et al., 2000; Fraser et al., 1997) and must therefore rely on salvage and interconversion of nicotinamide from the host for NAD+ homeostasis. The pncA gene is dispensable for spirochaete growth in nutrient-rich in vitro culture medium. Spirochaetes lacking the entire lp25 plasmid are non-infectious in the mouse, but reintroduction of the pncA gene alone to spirochaetes lacking lp25 is sufficient to restore mouse infectivity to near-wild-type levels, demonstrating that the nicotinamidase activity encoded by pncA is essential for survival of B. burgdorferi in the mammalian environment (Purser et al., 2003). Furthermore, pncA is critical for B. burgdorferi survival and replication in the tick (Grimm et al., 2005; Strother & de Silva, 2005). However, the current annotation for the 5′ end of the pncA ORF does not include an obvious ribosome-binding site proximal to the start codon and the predicted PncA protein is 20 aa shorter than Escherichia coli PncA. In addition, B. burgdorferi PncA lacks an N-terminal aspartic acid residue that completes the catalytic triad conserved among nicotinamidase enzymes of other bacterial species, suggesting that the encoded protein may be inactive (French et al., 2010; Purser et al., 2003). These observations are inconsistent with the known critical importance of PncA in the infectious cycle of B. burgdorferi. Therefore, herein we have characterized this in vivo-essential protein through genetic and biochemical identification of the N-terminal sequence of B. burgdorferi PncA.
Methods
Bacterial clones and growth conditions.
All low-passage B. burgdorferi clones are listed in Table 1 and were derived from strain B31 clone A3. B. burgdorferi was grown in liquid Barbour–Stoenner–Kelly (BSK) II medium supplemented with gelatin and 6 % rabbit serum (Barbour, 1984) and plated in solid BSK medium as described previously (Rosa & Hogan, 1992; Samuels, 1995). All spirochaete cultures were grown at 35 °C supplemented with 2.5 % CO2. Kanamycin was used at 200 µg ml−1.
Table 1. Bacterial clones used in this study.
| Species/clone | Description | Reference |
| B. burgdorferi | ||
| A3 68-1 | Low-passage clone derived from B31 (lp25−, lp56−) | Lawrence et al. (2009) |
| +/pBSV2*TT pncA-FLAG A | Harbours the shuttle vector carrying promoterless pncA-FLAG A | This work |
| +/pBSV2*TT flaBp-pncA-FLAG A | Harbours the shuttle vector carrying pncA-FLAG A under the control of the flaB promoter | This work |
| +/pBSV2*TT pncA-FLAG N | Harbours the shuttle vector carrying pncA-FLAG N under the control of the endogenous pncA promoter | This work |
| S. typhimurium | ||
| pncAnadB mutant | LT2 pncA278 : : Tn10d-cam nadB215 : : Tn10 | Purser et al. (2003); Zhu et al. (1991) |
| pncA-FLAG A | Harbours the shuttle vector carrying promoterless pncA-FLAG A | This work |
| pncA-FLAG B | Harbours the shuttle vector carrying promoterless pncA-FLAG B | This work |
| pncA-FLAG N | Harbours the shuttle vector carrying pncA-FLAG N under the control of the endogenous pncA promoter | This work |
| flaBp-pncA-FLAG A | Harbours the shuttle vector carrying pncA-FLAG A under the control of the flaB promoter | This work |
| flaBp-pncA-FLAG B | Harbours the shuttle vector carrying pncA-FLAG B under the control of the flaB promoter | This work |
Salmonella typhimurium wild-type strain LT2 and the isogenic pncA−nadB− mutant (TT400 sty LT2 pncA278 : : Tn10d-cam nadB215 : : Tn10) (Zhu et al., 1991) were kindly provided by S. J. Norris, University of Texas Health Science Center at Houston, TX, USA. All S. typhimurium strains were grown in Luria–Bertani (LB) medium, on LB agar or on M9 glucose agar (Sambrook & Russell, 2000). Strains transformed with pBSV2*TT constructs were supplemented with 50 µg kanamycin ml−1.
All cloning and plasmid propagation was conducted in either E. coli Top10 (Invitrogen) or E. coli DH5α cells grown in LB medium or on LB agar supplemented with 50 µg kanamycin ml−1.
Construction of pBSV2*TT vector.
Vector pBSV2* (Bestor et al., 2010), a derivative of pBSV2 (Stewart et al., 2001), was generated by removal of a second EcoRI site present outside the multiple cloning site of pBSV2 by using targeted PCR mutagenesis (Bestor et al., 2010). In order to prevent any potential readthrough transcription that might originate from the sequence upstream of the multiple cloning site, the Rho-independent transcription-terminator sequence for bmpB (Ramamoorthy et al., 2005) repeated three times in tandem was cloned into the EcoRI site within the multiple cloning site of pBSV2*. Briefly, the forward and reverse sequences of the bmpB terminator repeated three times in tandem, each harbouring a 5′ MfeI site and a 3′ EcoRI site, were synthesized as ssDNA oligos (Table 2, primers 1 and 2). To anneal the two ssDNA oligos, 2 nmol of each oligo was combined, heated to 95 °C for 10 min and cooled to room temperature over 90 min. The double-stranded terminator fragment was digested with MfeI and EcoRI and ligated into EcoRI-cut pBSV2*, creating pBSV2*TT. Sequence analysis confirmed retention of a single EcoRI restriction site 3′ of the terminator sequence, which was used for subsequent cloning as described below.
Table 2. Oligonucleotides used in this study.
Functional restriction-enzyme sites added to the primer are underlined. The sequence for one copy of the bmpB terminator is indicated in bold. The sequence for the B. burgdorferi codon usage-optimized FLAG epitope is indicated in italics. The engineered ATG sequence in primer 5 is indicated in lower-case letters.
| Number | Name | Sequence 5′–3′ |
| 1 | bmpB termX3 FWD | CCCCCCCAATTGCTTTTTTGGCTGGCTATATTGCAGCCAAAAAAAGCTTTTCGCTTTTTTGGCTGGCTATATTGCAGCCAAAAAAAGCTTTTCGCTTTTTTGGCTGGCTATATTGCAGCCAAAAAAAGCTTTTCGGAATTCCG |
| 2 | bmpB termX3 REV | CGGAATTCCGAAAAGCTTTTTTTGGCTGCAATATAGCCAGCCAAAAAAGCGAAAAGCTTTTTTTGGCTGCAATATAGCCAGCCAAAAAAGCGAAAAGCTTTTTTTGGCTGCAATATAGCCAGCCAAAAAAGCAATTGGGGGGG |
| 3 | pncA 5′ EcoRI N | CGGAATTCCTATGCTATCCCCTTGTTCAAGTC |
| 4 | pncA FLAG XbaI 3′ | GCTCTAGATTATTTATCATCATCATCTTTATAATCTATATTAAGCTTACTTTGGCTG |
| 5 | pncA 5′ EcoRI A | CGGAATTCatgGCACTTATTTTAATAGATATAC |
| 6 | pncA 5′ EcoRI B | CGGAATTCATGCCAGTATCTAACAGTAATG |
| 7 | flaB prom 5′ EcoRI | CGGAATTCTGTCTGTCGCCTCTTGTGGCTTCC |
| 8 | flaB prom 3′ EcoRI | CGGAATTCGATTGATAATCATATATCATTCC |
Cloning of pncA fragments.
A DNA fragment containing approximately 1000 bp upstream of pncA and the annotated PncA ORF followed by a C-terminal FLAG-epitope tag with EcoRI and XbaI sites was PCR-amplified from B. burgdorferi genomic DNA using primers 3 and 4 (Table 2). The resulting DNA fragment was cloned into EcoRI/XbaI-cut pBSV2*TT, yielding pBSV2*TT pncA-FLAG N (native). Positive clones were analysed and verified by restriction digest and DNA sequence analysis, respectively. Two fragments of the pncA gene, differing in the extent of their 5′ sequence (pncA-FLAG A and pncA-FLAG B) and with EcoRI/XbaI ends, were generated by PCR from B. burgdorferi genomic DNA using 5′ primers 5 and 6, respectively, and 3′ primer 4 (Table 2). Individual PCR fragments were cloned into EcoRI/XbaI-cut pBSV2*TT, resulting in plasmids pBSV2*TT pncA-FLAG A and pBSV2*TT pncA-FLAG B (Fig. 2). Plasmids were analysed and verified by restriction digest and sequence analysis. The constitutive flaB promoter including a ribosome-binding site with EcoRI ends was amplified from B. burgdorferi genomic DNA using primers 7 and 8 and cloned into the EcoRI-cut plasmids pBSV2*TT pncA-FLAG A and pBSV2*TT pncA-FLAG B in front of the promoterless pncA genes, resulting in plasmids pBSV2*TT flaBp-pncA-FLAG A and pBSV2*TT flaBp-pncA-FLAG B. Plasmids were analysed and verified by restriction digest and sequence analysis.
Nicotinamidase functional-complementation assay.
The S. typhimurium pncAnadB mutant was made chemically competent and transformed by heat shock with plasmids pBSV2*TT pncA-FLAG A, B and N and plasmids pBSV2*TT flaBp-pncA-FLAG A and B. Transformants were selected on LB plates containing kanamycin. Individual transformants were tested for nicotinamidase activity (resulting from synthesis of a functional B. burgdorferi PncA protein) by growth on M9 glucose agar supplemented with 1 mM MgSO4, 0.0005 % thiamine, 0.1 % Casamino acids, 50 µg kanamycin ml−1 and 0.1 mM either nicotinamide or nicotinic acid. Growth on LB agar plus kanamycin was used as a positive control. The production of PncA–FLAG was determined by immunoblot analysis of total protein lysates using an anti-FLAG M2 monoclonal antibody (Sigma).
Functional analysis of pBSV2*TT pncA-FLAG A in mice.
The pBSV2*TT pncA-FLAG A and pBSV2*TT flaBp-pncA-FLAG A plasmids were each transformed by electroporation into A3 68-1 (Lawrence et al., 2009) as described previously (Jewett et al., 2007) and transformants were selected in solid BSK medium containing kanamycin and confirmed by PCR using primers 4 and 5 (Table 2). Total genomic DNA was prepared from PCR-positive clones and screened for the presence of B. burgdorferi plasmid content (Elias et al., 2002). Clones that retained the plasmid content of the parent clone were used in further experiments. The University of Central Florida (UCF) is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for all animal experiments were prepared according to the guidelines of the NIH and approved by the UCF’s Institutional Animal Care and Use Committee. Groups of six 6–8-week-old C3H/HeN female mice were needle-inoculated as described previously (Jewett et al., 2009) with 1×104 spirochaetes. Mouse infection was assessed 3 weeks post-inoculation by reisolation of spirochaetes from ear, bladder and joint tissues as described previously (Elias et al., 2002; Grimm et al., 2004).
Purification and N-terminal sequencing of PncA–FLAG.
In order to purify the PncA protein from B. burgdorferi, the pBSV2*TT pncA-FLAG N plasmid was transformed into A3 68-1 (Lawrence et al., 2009), as described above. The production of PncA–FLAG in in vitro-grown B. burgdorferi was confirmed by immunoblot analysis of total protein lysate from clone A3 68-1/pBSV2*TT pncA-FLAG N using anti-FLAG M2 monoclonal antibody (Sigma). Clone A3 68-1/pBSV2*TT pncA-FLAG N was grown to stationary phase (>1×108 spirochaetes ml−1) in 250 ml BSK II medium. Spirochaetes were harvested by centrifugation at 8000 r.p.m. (JA-14 rotor) for 10 min and washed in 10 ml ice-cold HN buffer (50 mM HEPES, 50 mM NaCl; pH 7.6). The washed cells were resuspended and lysed in 10 ml Cell Lytic B (Sigma) and 1 ml protease inhibitor cocktail (Sigma). The cell lysate was centrifuged for 15 min at 21 000 g and 4 °C to remove the cellular debris and the supernatant was stored on ice. A 3 ml column containing 500 µl anti-FLAG M2 affinity resin (Sigma) was prepared according to the manufacturer’s recommendations. The soluble protein lysate was loaded onto the column and the flow-through was collected. The flow-through was reloaded and collected three times. The column was washed with 20 column volumes of TBS (50 mM Tris, 150 mM NaCl; pH 7.6). Protein was eluted from the column by the addition of 6 ml 0.1 M glycine–HCl, pH 3.5. Six 1 ml fractions were collected into 1.5 ml microfuge tubes containing 25 µl 1 M Tris, pH 8.0. Protein samples were resolved by electrophoresis on an SDS-PAGE gel (12.5 % polyacrylamide) and blotted onto nitrocellulose. Immunoblots were hybridized with anti-FLAG M2 monoclonal antibody. PncA–FLAG protein from elution 1 was resolved by electrophoresis on an SDS-PAGE gel (12.5 % polyacrylamide) and blotted onto a PVDF membrane. The PVDF membrane was stained with Coomassie brilliant blue to visualize the PncA–FLAG protein. The PncA–FLAG protein fragment was cut from the PVDF membrane and sent to the Protein Sequencing Unit of the Protein Chemistry Section of the Research Technologies Branch, NIAID, NIH, Rockville, MD, USA, for N-terminal sequencing. The nucleotide and amino acid sequences for pncA have been submitted to GenBank under accession no. JN223414.
Results
Additional upstream coding sequence is necessary for the nicotinamidase activity of B. burgdorferi PncA
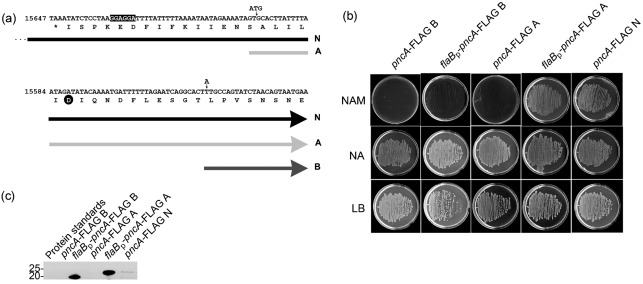
The pncA gene of B. burgdorferi encodes a functional nicotinamidase enzyme that is critical for B. burgdorferi survival in mice and ticks (Grimm et al., 2005; Purser et al., 2003). The current annotation of the pncA ORF (GenBank accession no. NP_045429) (Casjens et al., 2000; Fraser et al., 1997) does not have an obvious proximal ribosome-binding site, encodes a protein that is 20 aa shorter than homologues in other bacteria and lacks an N-terminal aspartic acid residue found in the catalytic triad of well-characterized nicotinamidase enzymes (Purser et al., 2003). In order to explore the hypothesis that the pncA ORF contains additional 5′ coding sequence that is necessary for its enzymic activity, we took advantage of a prediction made by Purser et al. (2003), who suggested that the actual translational start site of the B. burgdorferi PncA protein may be 48 nt upstream of the current annotation (Purser et al., 2003). This sequence does not contain an in-frame start codon; therefore, we engineered an in-frame ATG start codon upstream of the 48 nt flanking sequence preceding the pncA ORF, as well as a 3′ FLAG-epitope tag. This fragment was cloned into the B. burgdorferi shuttle vector pBSV2*TT, creating plasmid pBSV2*TT pncA-FLAG A (Fig. 1a). Although this pncA gene probably did not use the endogenous mechanism of translational regulation because of the introduction of the engineered ATG start codon, it allowed us to test the function of the newly included upstream sequence in producing an active B. burgdorferi PncA enzyme. In addition, we cloned two other pncA genes into pBSV2*TT. The pncA ORF as currently annotated, along with a single nucleotide point mutation (T to A) to create an ATG start codon [the start codon for the pncA ORF is currently proposed to be TTG (Casjens et al., 2000; Fraser et al., 1997)] and including a 3′ FLAG-epitope tag sequence, was cloned to create pBSV2*TT pncA-FLAG B (Fig. 1a). The 1000 bp pncA upstream sequence, believed to carry the endogenous pncA promoter sequence, followed by the pncA ORF including a 3′ FLAG-epitope tag sequence were cloned to create pBSV2*TT pncA-FLAG N (native) (Fig. 1a). Plasmids pBSV2*TT pncA-FLAG A and pBSV2*TT pncA-FLAG B both lacked promoter sequences; therefore, the B. burgdorferi flaB promoter including a ribosome-binding site sequence, which has been shown to function well in E. coli (Elias et al., 2003; Stewart et al., 2001), was added in order to drive transcription of the pncA-FLAG gene, creating expression plasmids pBSV2*TT flaBp-pncA-FLAG A and pBSV2*TT flaBp-pncA-FLAG B.
Fig. 1.
Identification of the B. burgdorferi pncA sequence sufficient to encode a functional nicotinamidase enzyme. (a) Schematic representation of the three pncA-FLAG constructs used for functional complementation of an S. typhimurium pncAnadB mutant for growth on minimal medium containing 0.1 mM nicotinamide. The reverse complement of nt 15678–15558 on lp25 is shown, with the translated single-letter amino acids indicated below. The 5′ sequences of each pncA construct are represented by block arrows: pncA-FLAG N (N, black arrow; note that the 5′ end of this construct continues approximately 900 bp upstream of the sequence that is shown, as indicated by three dots), pncA-FLAG A (A, light-grey arrow) and pncA-FLAG B (B, dark-grey arrow). The putative ribosome-binding site sequence is highlighted in black. The aspartic acid residue conserved among the catalytic triad of PncA homologues is indicated by a black circle. The locations of the engineered ATG start codon for construct pncA-FLAG A, and the nucleotide point mutation (T to A) to create an ATG start codon for construct pncA-FLAG B, are indicated by small vertical arrows. (b) Functional-complementation assay for nicotinamidase activity of pncA-FLAG constructs. Representative data are shown for the overnight growth at 37 °C of the S. typhimurium pncAnadB mutant harbouring each pncA-FLAG construct (indicated above each column) on minimal agar containing 0.1 mM nicotinamide (NAM) or 0.1 mM nicotinic acid (NA). Rich agar (LB) was used as a positive control. All agar plates contained 50 µg kanamycin ml−1. (c) Immunoblot analysis of PncA–FLAG production in the S. typhimurium pncAnadB mutant harbouring each pncA-FLAG gene. Total protein lysates were resolved by SDS-PAGE, transferred to a nitrocellulose membrane and probed for PncA–FLAG using an anti-FLAG M2 monoclonal antibody. Sizes of the protein standards are indicated in kDa. The predicted molecular masses for PncA–FLAG B and PncA–FLAG A are 21 and 24 kDa, respectively.
A genetic approach involving functional complementation of a Salmonella mutant lacking nicotinamidase activity was employed to investigate the sequence necessary to produce an enzymically active B. burgdorferi PncA protein. This approach was used successfully for the initial characterization of BBE22 (PncA) as a nicotinamidase (Purser et al., 2003). The plasmids carrying the different B. burgdorferi pncA constructs were transformed into an S. typhimurium pncAnadB mutant that is unable to grow on minimal medium supplemented with nicotinamide, due to a lack of nicotinamidase activity (Purser et al., 2003). Salmonella mutants harbouring the promoterless pncA-FLAG A and pncA-FLAG B genes were unable to grow in the presence of nicotinamide alone and did not produce detectable PncA–FLAG protein (Fig. 1b, c). However, upon introduction of the flaB promoter, flaBp-pncA-FLAG A allowed growth of the Salmonella mutant on nicotinamide and production of an estimated 24 kDa PncA–FLAG protein (Fig. 1b, c), indicating that this construct harboured sufficient coding sequence to produce an active nicotinamidase enzyme. In contrast, despite the presence of a promoter to drive transcription of the pncA gene, flaBp-pncA-FLAG B did not allow growth of the Salmonella mutant on nicotinamide (Fig. 1b), although a protein corresponding to the predicted size of the current annotation for PncA (21 kDa) was detected by immunoblot analysis using anti-FLAG antibodies (Fig. 1c). The pncA sequence of flaBp-pncA-FLAG B, which follows the current annotation of the pncA ORF, starts 30 bp downstream of the GAT codon encoding a putative catalytic aspartic acid residue (Fig. 1a), without which a functional PncA enzyme may not be produced, even in the presence of a constitutive promoter. The plasmid pBSV2*TT pncA-FLAG N, presumed to harbour the endogenous pncA promoter region, allowed growth of the Salmonella nicotinamidase mutant on nicotinamide (Fig. 1b). Interestingly, Salmonella mutants expressing pncA-FLAG N produced a PncA–FLAG protein of similar size to that produced by Salmonella mutants expressing flaBp-pncA-FLAG A, albeit at a lower level, perhaps due to a difference between the strengths of the pncA and flaB promoters in Salmonella. These data suggested that pncA promoter-driven expression of pncA-FLAG N in Salmonella was sufficient to produce a functional B. burgdorferi PncA enzyme to allow growth of the Salmonella mutant on nicotinamide alone. In addition, these data indicate that native B. burgdorferi PncA may be 3 kDa larger than the protein as currently annotated. All Salmonella transformants were able to grow on minimal medium supplemented with nicotinic acid, which bypasses the requirement for nicotinamidase activity, as well as rich medium (LB) (Fig. 1b). Together, these data demonstrate that the pncA ORF that includes an additional 48 nt of upstream coding sequence, when driven by the flaB promoter, is sufficient to produce a PncA protein with functional nicotinamidase activity and suggest that the native PncA protein harbours additional upstream sequence relative to the current annotation for the pncA ORF.
Expression of pncA–FLAG A restores mouse infectivity to spirochaetes lacking lp25
Our data indicated that a pncA gene including an engineered ATG start codon and 48 nt of additional sequence upstream of the annotated start of translation of the pncA ORF, when driven by the flaB promoter, produced nicotinamidase activity in a Salmonella functional-complementation assay (Fig. 1). The nicotinamidase activity of PncA is critical for B. burgdorferi mouse infectivity (Purser et al., 2003). The pncA gene alone, under the control of its endogenous promoter, has been shown to restore near-wild-type levels of mouse infectivity to spirochaetes lacking the entire lp25 plasmid (Purser et al., 2003). Therefore, we sought to determine the ability of pBSV2*TT flaBp-pncA-FLAG A to complement attenuated lp25− spirochaetes in vivo. B. burgdorferi clone A3 68-1, lacking lp25 and lp56, was transformed with either plasmid pBSV2*TT pncA-FLAG A (promoter-negative) or pBSV2*TT flaBp-pncA-FLAG A (promoter-positive) and groups of mice were inoculated with 1×104 spirochaetes carrying either plasmid. Three weeks post-inoculation, the number of mice infected in each group was determined by serology and reisolation of spirochaetes from tissues. Introduction of pBSV2*TT flaBp-pncA-FLAG A into spirochaetes lacking the entire lp25 plasmid was sufficient to restore mouse infectivity, as five of six mice inoculated with these spirochaetes became infected. These data suggest that spirochaetes harbouring pBSV2*TT flaBp-pncA-FLAG A were able to produce active nicotinamidase enzyme in vivo. Those spirochaetes harbouring pBSV2*TT pncA-FLAG A, which lacks a promoter to drive pncA expression, remained non-infectious, as none of six mice became infected. These data indicate that expression of an additional 48 nt of upstream coding sequence along with the pncA ORF resulted in in vivo production of a PncA protein that harbours sufficient activity to restore infectivity to spirochaetes lacking lp25. In addition, these data, along with the functional-complementation data in the Salmonella pncAnadB mutant, demonstrated that the presence of the FLAG-epitope tag on the C terminus of the PncA proteins did not affect the nicotinamidase activity of the functional enzymes.
The PncA protein of B. burgdorferi uses the non-canonical initiation codon AUU: expression and sequencing of the native PncA protein
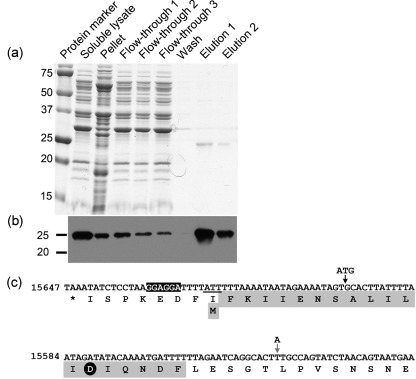
Our data demonstrate that the addition of 48 nt of upstream coding sequence to the pncA ORF resulted in production of a B. burgdorferi PncA protein with functional nicotinamidase activity in a Salmonella mutant functional-complementation assay (Fig. 1b), as well as during an active B. burgdorferi mouse infection (see above). However, the pncA construct used in these experiments harboured an engineered ATG start codon and was under the control of the constitutive flaB promoter. Therefore, we sought to define experimentally the N-terminal amino acid sequence of the PncA protein expressed in B. burgdorferi under the control of its endogenous promoter. To this end, plasmids pBSV2*TT pncA-FLAG N and pBSV2*TT alone were each transformed into B. burgdorferi clone A3 68-1. Positive transformants with the plasmid content of the parent were identified and used for further studies. Native PncA–FLAG protein was produced in and purified to homogeneity from B. burgdorferi using anti-FLAG agarose beads (Fig. 2a, b). No FLAG epitope-tagged protein was detected in or purified from B. burgdorferi carrying the Borrelia shuttle vector alone (data not shown). Purified PncA–FLAG was separated by SDS-PAGE, transferred to a PVDF membrane and N-terminally sequenced using Edman degradation. A single peptide was detected with the amino acid sequence MFKIIENSALILIDIQNDF (Fig. 2c). This N-terminal amino acid sequence demonstrates that the PncA protein is 24 aa longer than currently predicted and includes the critical aspartic acid residue (Fig. 3), thereby completing the catalytic triad that is conserved among nicotinamidase enzymes. Furthermore, these data revealed that the B. burgdorferi PncA protein uses the non-canonical initiation codon AUU, which is located 5 bp downstream of a putative ribosome-binding sequence (Fig. 2c). Together, these data identify the translational start site of the B. burgdorferi PncA protein.
Fig. 2.
Purification and N-terminal sequence analysis of PncA–FLAG protein from B. burgdorferi. (a) Coomassie-stained SDS-PAGE gel of protein fractions collected during purification of the PncA–FLAG protein from B. burgdorferi. (b) Immunoblot of the protein fractions collected during purification of the PncA–FLAG protein from B. burgdorferi using antibodies against the FLAG-epitope tag. Numbers to the left of panels (a) and (b) indicate the masses in kDa of the protein standards. (c) Reverse complement of nt 15647–15525 from lp25. The corresponding translated single-letter amino acid sequence is shown below the nucleotide sequence. The sequence of the first 20 aa of native purified PncA–FLAG as determined by Edman degradation and N-terminal sequencing is highlighted in grey. The putative ribosome-binding site sequence is highlighted in black. The non-canonical translational start codon of native purified PncA–FLAG (ATT) is underlined. The aspartic acid residue conserved among the catalytic triad of PncA homologues is indicated by a black circle. The locations of the engineered ATG start codon and translational start site for construct pncA-FLAG A, and the nucleotide point mutation (T to A) to create an ATG start codon and translational start site for construct pncA-FLAG B (which reflects the current proposed translational start site for the pncA ORF), are indicated by vertical arrows (black and grey, respectively).
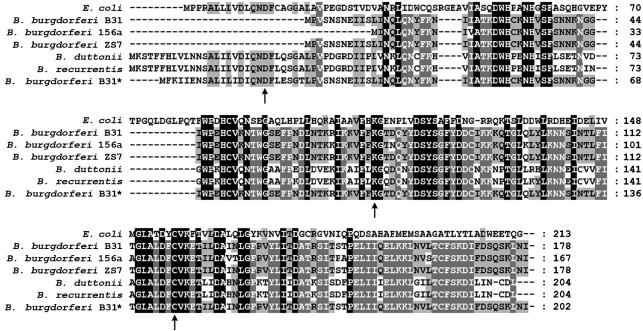
Fig. 3.
Alignment of the PncA homologues from E. coli and Borrelia spp. Shown is an amino acid alignment of the PncA proteins, according to current annotation, from E. coli (GenBank accession no. AAC74838), Lyme disease spirochaetes B. burgdorferi B31 (NP_045429), B. burgdorferi 156a (YP_002477119.1) and B. burgdorferi ZS7 (YP_002455325.1), relapsing fever spirochaetes B. duttonii (YP_002223541.1) and B. recurrentis (YP_002223408.1), and the PncA protein sequence for B. burgdorferi B31 defined herein (*) (JN223414). Amino acids identical to the consensus sequence are shaded. Amino acid residues conserved in the catalytic triad of nicotinamidase enzymes are indicated by arrows. Dashes represent spaces introduced for optimal sequence alignment. Amino acid sequences were aligned using the clustal w algorithm in the megalign program from the DNASTAR Lasergene suite.
Discussion
Nicotinamidase enzymes play a critical role in maintaining NAD+ homeostasis and have been found to contribute to important biological processes in both prokaryotes and eukaryotes, including bacterial pathogenesis (Anderson et al., 2003; Balan et al., 2008; Evans et al., 2010; Frothingham et al., 1996; Ghislain et al., 2002; Hunt et al., 2007; Kim et al., 2004; Minard & McAlister-Henn, 2010; Purser et al., 2003; Raynaud et al., 1998; Silva et al., 2009; van der Horst et al., 2007; Vrablik et al., 2009; Wang & Pichersky, 2007). Because of the absence of nicotinamidases in mammals, these enzymes may be potential drug targets for mammalian pathogens (Sauve, 2008), including the Lyme disease spirochaete B. burgdorferi.
B. burgdorferi lacks the enzymes for de novo biosynthesis of NAD+ and must therefore rely on salvage of nicotinamide from the host for survival (Casjens et al., 2000; Fraser et al., 1997; Purser et al., 2003). The B. burgdorferi nicotinamidase, PncA, serves an essential physiological function required for survival of B. burgdorferi throughout its infectious cycle (Grimm et al., 2005; Purser et al., 2003). However, in conflict with the essential in vivo role of PncA, the current annotation of the pncA ORF suggests that the encoded protein may lack catalytic activity due to the absence of an N-terminal aspartic acid that is conserved in the catalytic triad of nicotinamidase enzymes from other species (French et al., 2010; Purser et al., 2003). Using a genetic approach involving functional complementation of a Salmonella mutant lacking nicotinamidase activity, we have found that addition of 48 nt of upstream flanking sequence to the annotated pncA ORF results in production of an active nicotinamidase enzyme when expressed under the control of the flaB promoter (Fig. 1). In contrast, the pncA ORF as currently annotated, similarly expressed alone under the control of the flaB promoter, did not yield nicotinamidase activity (Fig. 1). Furthermore, the plasmid carrying the pncA ORF along with 48 nt of additional upstream sequence under the control of the flaB promoter was capable of restoring mouse infectivity to spirochaetes lacking the entire lp25 plasmid. Recently, recombinant B. burgdorferi PncA, expressed in and purified from E. coli, was found to have poor catalytic activity (French et al., 2010). Although the authors acknowledged that the current annotation of the pncA ORF may be inaccurate and stated that additional upstream sequence was added to the pncA gene in their expression plasmid, it remains unclear whether sufficient sequence was used to express enzymically active PncA (French et al., 2010). We have identified a pncA sequence that is sufficient to encode an active nicotinamidase enzyme, which supports B. burgdorferi survival in the mouse. These data, as well as the in vivo requirement for pncA, raise the possibility of using exogenous expression of pncA in spirochaetes lacking lp25 as an in vivo genetic selection mechanism for shuttle-vector maintenance without the need for antibiotic selection. The requirement for pncA in vivo is absolute, rather than the conditional requirement for antibiotic resistance, making flaBp-pncA a novel selection cassette with application in genetic studies in vivo.
Purification of native PncA from B. burgdorferi using a C-terminal FLAG-epitope tag followed by N-terminal sequencing determined that translation of pncA initiates from the rare, non-canonical initiation codon AUU, which is 72 nt upstream of the current annotation for the start codon of the PncA protein (Fig. 2). The identified PncA start codon is 5 nt downstream of the putative ribosome-binding site and produces a 203 aa protein that harbours the N-terminal aspartic acid residue conserved in the catalytic triad of PncA homologues (Fig. 3). This is the first report of a B. burgdorferi gene using AUU as the initiation codon. Bioinformatic analysis suggests that AUU is the initiation codon for the pncA ORFs of all Lyme disease spirochaetes and that these ORFs are currently annotated incorrectly (Fig. 3). In contrast, the relapsing fever spirochaetes Borrelia recurrentis and Borrelia duttonii appear to initiate translation of pncA using a canonical AUG initiation codon and are predicted to produce 204 aa PncA proteins, which include the conserved N-terminal aspartic acid residue (Lescot et al., 2008) (Fig. 3). The few genes demonstrated to use an AUU initiation codon to initiate protein synthesis are bacterial infC, which encodes initiation factor 3 (IF-3) (Hu et al., 1993; Liveris et al., 1993; Pon & Gualerzi, 1986; Sacerdot et al., 1996), E. coli pcnB, which encodes poly(A) polymerase (PAPI) (Binns & Masters, 2002), orfB of bacterial transposable IS3 element family member IS911 (Prère et al., 2011), gp26* of bacteriophage T4 (Nivinskas et al., 1992) and a small number of eukaryotic genes (Ivanov et al., 2008). Genes infC and pcnB are the only two examples in which AUU is the definitive initiation codon; the other examples use AUU as an alternative initiation codon in addition to a more conventional initiation site (Binns & Masters, 2002).
Given the few characterized examples of AUU-initiated proteins, the biological significance of having an AUU start codon is unclear. Characterization of AUU-initiated translation of the E. coli pcnB-encoded PAPI suggests a possible role for the non-canonical start codon in the control of protein production (Binns & Masters, 2002). PAPI catalyses the template-independent addition of adenosine monophosphate to the 3′ ends of RNA species. Overproduction of E. coli PAPI is deleterious for cell growth (Binns & Masters, 2002; Cao & Sarkar, 1992a, b). Using experimentally modified genes, the efficiency of translation with AUU as an initiation codon has been found to be 1–13 % of that of AUG-initiated translation due to IF-3 initiation codon-mismatch discrimination and destabilization (Sacerdot et al., 1996; Sussman et al., 1996). Because of the reduced level of translation resulting from AUU initiation, the AUU initiation codon of pcnB is believed to be the mechanism that limits PAPI synthesis (Binns & Masters, 2002). We have not found overproduction of B. burgdorferi PncA to be cytotoxic, nor has this been reported in the literature. No increase in B. burgdorferi PncA production or change in spirochaete growth rate was observed when the AUU sequence was mutated to a canonical AUG (A. K. Linowski & M. W. Jewett, unpublished data). Elucidation of the role, if any, of the AUU initiation codon in translational control of pncA is the focus of future studies.
B. burgdorferi is an A+T-rich organism with a G+C content of only 30 mol% (Casjens et al., 2000; Fraser et al., 1997). The codon-usage frequency of AUU in the B. burgdorferi B31 genome is reported to be 55.7 %, compared with 18.1 % for AUG (http://www.kazusa.or.jp/codon/). Further bioinformatics analysis is required to determine the frequency of AUU codons as putative initiation codons, rather than encoding an isoleucine residue. Our findings stress the importance of critical evaluation of genome annotation of ORFs of interest and highlight the possibility that additional AUU-initiated ORFs in the B. burgdorferi genome may remain to be identified.
Together, our data have defined experimentally the complete amino acid sequence of the B. burgdorferi PncA protein. These findings provide a foundation for determination of the structure and catalytic function of this in vivo-essential protein that may contribute to rational design of nicotinamidase enzyme inhibitors that could serve as novel antimicrobials for the treatment of Lyme disease.
Acknowledgements
Many thanks to S. J. Norris, University of Texas Health Science Center at Houston, TX, USA, for giving us the Salmonella strains and to M. K. Garfield in the Protein Chemistry Section of the Research Technologies Branch, NIAID, NIH, Rockville, MD, USA, for N-terminal sequence analysis. Thank you to T. J. Jewett for critical reading of this manuscript. This research was supported, in part, by NIH, NIAID grant 5K22AI081730 to M. W. J. and the Intramural Research Program of the NIH, NIAID.
Abbreviations:
- IF-3
initiation factor 3
- lp
linear plasmid
- PAPI
poly(A) polymerase
References
- Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003). Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423, 181–185. 10.1038/nature01578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan V., Miller G. S., Kaplun L., Balan K., Chong Z. Z., Li F., Kaplun A., VanBerkum M. F., Arking R., et al. (2008). Life span extension and neuronal cell protection by Drosophila nicotinamidase. J Biol Chem 283, 27810–27819. 10.1074/jbc.M804681200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A. G. (1984). Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57, 521–525. [PMC free article] [PubMed] [Google Scholar]
- Bestor A., Stewart P. E., Jewett M. W., Sarkar A., Tilly K., Rosa P. A. (2010). Use of the Cre-lox recombination system to investigate the lp54 gene requirement in the infectious cycle of Borrelia burgdorferi. Infect Immun 78, 2397–2407. 10.1128/IAI.01059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns N., Masters M. (2002). Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol Microbiol 44, 1287–1298. 10.1046/j.1365-2958.2002.02945.x [DOI] [PubMed] [Google Scholar]
- Bono J. L., Tilly K., Stevenson B., Hogan D., Rosa P. (1998). Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144, 1033–1044. 10.1099/00221287-144-4-1033 [DOI] [PubMed] [Google Scholar]
- Byram R., Stewart P. E., Rosa P. (2004). The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J Bacteriol 186, 3561–3569. 10.1128/JB.186.11.3561-3569.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. J., Sarkar N. (1992a). Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci U S A 89, 10380–10384. 10.1073/pnas.89.21.10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. J., Sarkar N. (1992b). Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc Natl Acad Sci U S A 89, 7546–7550. 10.1073/pnas.89.16.7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S., Palmer N., van Vugt R., Huang W. M., Stevenson B., Rosa P., Lathigra R., Sutton G., Peterson J., et al. (2000). A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35, 490–516. 10.1046/j.1365-2958.2000.01698.x [DOI] [PubMed] [Google Scholar]
- Elias A. F., Stewart P. E., Grimm D., Caimano M. J., Eggers C. H., Tilly K., Bono J. L., Akins D. R., Radolf J. D., et al. (2002). Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun 70, 2139–2150. 10.1128/IAI.70.4.2139-2150.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias A. F., Bono J. L., Kupko J. J., III, Stewart P. E., Krum J. G., Rosa P. A. (2003). New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol 6, 29–40. 10.1159/000073406 [DOI] [PubMed] [Google Scholar]
- Evans C., Bogan K. L., Song P., Burant C. F., Kennedy R. T., Brenner C. (2010). NAD+ metabolite levels as a function of vitamins and calorie restriction: evidence for different mechanisms of longevity. BMC Chem Biol 10, 2. 10.1186/1472-6769-10-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C. M., Casjens S., Huang W. M., Sutton G. G., Clayton R., Lathigra R., White O., Ketchum K. A., Dodson R., et al. (1997). Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390, 580–586. 10.1038/37551 [DOI] [PubMed] [Google Scholar]
- French J. B., Cen Y., Vrablik T. L., Xu P., Allen E., Hanna-Rose W., Sauve A. A. (2010). Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry 49, 10421–10439. 10.1021/bi1012518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frothingham R., Meeker-O’Connell W. A., Talbot E. A., George J. W., Kreuzer K. N. (1996). Identification, cloning, and expression of the Escherichia coli pyrazinamidase and nicotinamidase gene, pncA. Antimicrob Agents Chemother 40, 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Talla E., François J. M. (2002). Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19, 215–224. 10.1002/yea.810 [DOI] [PubMed] [Google Scholar]
- Grimm D., Tilly K., Byram R., Stewart P. E., Krum J. G., Bueschel D. M., Schwan T. G., Policastro P. F., Elias A. F., Rosa P. A. (2004). Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci U S A 101, 3142–3147. 10.1073/pnas.0306845101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Tilly K., Bueschel D. M., Fisher M. A., Policastro P. F., Gherardini F. C., Schwan T. G., Rosa P. A. (2005). Defining plasmids required by Borrelia burgdorferi for colonization of tick vector Ixodes scapularis (Acari: Ixodidae). J Med Entomol 42, 676–684. 10.1603/0022-2585(2005)042[0676:DPRBBB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hu W. S., Wang R. Y., Shih J. W., Lo S. C. (1993). Identification of a putative infC-rpmI-rplT operon flanked by long inverted repeats in Mycoplasma fermentans (incognitus strain). Gene 127, 79–85. 10.1016/0378-1119(93)90619-E [DOI] [PubMed] [Google Scholar]
- Hunt L., Holdsworth M. J., Gray J. E. (2007). Nicotinamidase activity is important for germination. Plant J 51, 341–351. 10.1111/j.1365-313X.2007.03151.x [DOI] [PubMed] [Google Scholar]
- Ivanov I. P., Loughran G., Atkins J. F. (2008). uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci U S A 105, 10079–10084. 10.1073/pnas.0801590105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett M. W., Lawrence K., Bestor A. C., Tilly K., Grimm D., Shaw P., VanRaden M., Gherardini F., Rosa P. A. (2007). The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol 64, 1358–1374. 10.1111/j.1365-2958.2007.05746.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett M. W., Lawrence K. A., Bestor A., Byram R., Gherardini F., Rosa P. A. (2009). GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J Bacteriol 191, 6231–6241. 10.1128/JB.00450-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kurokawa D., Watanabe K., Makino S., Shirahata T., Watarai M. (2004). Brucella abortus nicotinamidase (PncA) contributes to its intracellular replication and infectivity in mice. FEMS Microbiol Lett 234, 289–295. 10.1111/j.1574-6968.2004.tb09546.x [DOI] [PubMed] [Google Scholar]
- Lawrence K. A., Jewett M. W., Rosa P. A., Gherardini F. C. (2009). Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol Microbiol 72, 1517–1529. 10.1111/j.1365-2958.2009.06740.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Audic S., Robert C., Nguyen T. T., Blanc G., Cutler S. J., Wincker P., Couloux A., Claverie J. M., et al. (2008). The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet 4, e1000185. 10.1371/journal.pgen.1000185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liveris D., Schwartz J. J., Geertman R., Schwartz I. (1993). Molecular cloning and sequencing of infC, the gene encoding translation initiation factor IF3, from four enterobacterial species. FEMS Microbiol Lett 112, 211–216. 10.1111/j.1574-6968.1993.tb06450.x [DOI] [PubMed] [Google Scholar]
- Minard K. I., McAlister-Henn L. (2010). Pnc1p supports increases in cellular NAD(H) levels in response to internal or external oxidative stress. Biochemistry 49, 6299–6301. 10.1021/bi100825z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivinskas R., Vaiskunaite R., Raudonikiene A. (1992). An internal AUU codon initiates a smaller peptide encoded by bacteriophage T4 baseplate gene 26. Mol Gen Genet 232, 257–261. [DOI] [PubMed] [Google Scholar]
- Pon C. L., Gualerzi C. O. (1986). Mechanism of translational initiation in prokaryotes. IF3 is released from ribosomes during and not before 70 S initiation complex formation. FEBS Lett 195, 215–219. 10.1016/0014-5793(86)80163-6 [DOI] [PubMed] [Google Scholar]
- Prère M. F., Canal I., Wills N. M., Atkins J. F., Fayet O. (2011). The interplay of mRNA stimulatory signals required for AUU-mediated initiation and programmed −1 ribosomal frameshifting in decoding of transposable element IS911. J Bacteriol 193, 2735–2744. 10.1128/JB.00115-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser J. E., Lawrenz M. B., Caimano M. J., Howell J. K., Radolf J. D., Norris S. J. (2003). A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol 48, 753–764. 10.1046/j.1365-2958.2003.03452.x [DOI] [PubMed] [Google Scholar]
- Ramamoorthy R., McClain N. A., Gautam A., Scholl-Meeker D. (2005). Expression of the bmpB gene of Borrelia burgdorferi is modulated by two distinct transcription termination events. J Bacteriol 187, 2592–2600. 10.1128/JB.187.8.2592-2600.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Etienne G., Peyron P., Lanéelle M. A., Daffé M. (1998). Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144, 577–587. 10.1099/00221287-144-2-577 [DOI] [PubMed] [Google Scholar]
- Rosa P. A., Hogan D., (1992). Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants. In Proceedings of the First International Conference on Tick-Borne Pathogens at the Host–Vector Interface, pp. 95–103. Edited by Munderloh U. G., Kurtti T. J.St Paul, MN: University of Minnesota. [Google Scholar]
- Rosa P. A., Tilly K., Stewart P. E. (2005). The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol 3, 129–143. 10.1038/nrmicro1086 [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Chiaruttini C., Engst K., Graffe M., Milet M., Mathy N., Dondon J., Springer M. (1996). The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol Microbiol 21, 331–346. 10.1046/j.1365-2958.1996.6361359.x [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2000). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Samuels D. S. (1995). Electrotransformation of the spirochete Borrelia burgdorferi. In Methods in Molecular Biology, pp. 253–259. Edited by Nickoloff J. A. Totowa, NJ: Humana Press, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve A. A. (2008). NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther 324, 883–893. 10.1124/jpet.107.120758 [DOI] [PubMed] [Google Scholar]
- Silva R. M., Duarte I. C., Paredes J. A., Lima-Costa T., Perrot M., Boucherie H., Goodfellow B. J., Gomes A. C., Mateus D. D., et al. (2009). The yeast PNC1 longevity gene is up-regulated by mRNA mistranslation. PLoS One 4, e5212. 10.1371/journal.pone.0005212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P. E., Thalken R., Bono J. L., Rosa P. (2001). Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol 39, 714–721. 10.1046/j.1365-2958.2001.02256.x [DOI] [PubMed] [Google Scholar]
- Strother K. O., de Silva A. (2005). Role of Borrelia burgdorferi linear plasmid 25 in infection of Ixodes scapularis ticks. J Bacteriol 187, 5776–5781. 10.1128/JB.187.16.5776-5781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. K., Simons E. L., Simons R. W. (1996). Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol Microbiol 21, 347–360. 10.1046/j.1365-2958.1996.6371354.x [DOI] [PubMed] [Google Scholar]
- Tilly K., Grimm D., Bueschel D. M., Krum J. G., Rosa P. (2004). Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis 4, 159–168. 10.1089/1530366041210738 [DOI] [PubMed] [Google Scholar]
- Tilly K., Rosa P. A., Stewart P. E. (2008). Biology of infection with Borrelia burgdorferi. Infect Dis Clin North Am 22, 217–234, v. 10.1016/j.idc.2007.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A., Schavemaker J. M., Pellis-van Berkel W., Burgering B. M. (2007). The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev 128, 346–349. 10.1016/j.mad.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Vrablik T. L., Huang L., Lange S. E., Hanna-Rose W. (2009). Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development 136, 3637–3646. 10.1242/dev.028431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Pichersky E. (2007). Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J 49, 1020–1029. 10.1111/j.1365-313X.2006.03013.x [DOI] [PubMed] [Google Scholar]
- Zhu N., Olivera B. M., Roth J. R. (1991). Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J Bacteriol 173, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]