Abstract
SLCO1B1 gene variants are associated with severe statin-induced myopathy. We examined whether these variants are also associated with general statin intolerance in a large population of patients with type 2 diabetes prescribed statins as part of routine clinical care.
4196 individuals were genotyped for rs4149056 (Val174Ala) and rs2306283 (Asp130Asn). Intolerance was defined by serum biochemistry and statin discontinuation, switching or dose reduction. Ala174 was associated with higher intolerance (OR=2.05, p=0.043), while Asp130 was associated with lower intolerance (OR=0.71, p= 0.026). Ala174 was associated with a reduced LDLc response (p=0.01) and 130D was associated with a greater LDLc response to statins (p=0.048) as previously reported, however this association was not present when intolerant individuals were removed.
This study suggests that common genetic variants, selected for an extreme phenotype of statin-induced myopathy also predispose to more common milder statin intolerance and may, for this reason, impact on lipid-lowering efficacy.
Introduction
The HMG-CoA Reductase Inhibitors, or statins, are very widely prescribed for the primary and secondary prevention of cardiovascular disease (1). Statins are generally well tolerated being associated with very few serious side effects, the most important of these being myopathy which occurs only rarely (2). Early studies have however indicated that around 12% of individuals develop symptoms relating to statin use with small but significant accompanying changes in serum creatinine kinase (CK) and liver enzymes (3). It is well known that for many drugs that are taken chronically only about 50% of patients will remain on the drug after a year, and this is also true of statins (4-6). While for antihypertensive drugs side effects can play a role in adherence (7), the reasons for discontinuation of statins appears to have been less researched, although 7% of discontinuation of statins may be due to side effects (4). A common non-synonymous coding variant in the solute carrier organic anion transporter gene SLCO1B1 has been demonstrated to associate strongly with the risk of development of simvastatin induced myopathy in a whole genome association analysis of the SEARCH study (8). This variant involves a valine to alanine substitution at position 174 in the protein (V174A, rs4149056), organic anion transporting polypeptide OATP1B1. The alanine allele gives rise to a less functional form of OATP1B1 (9), with reduced maximal transport activity (10-12), possibly as a result of intracellular sequestration and reduced surface activity. This may give rise to higher blood concentrations of statin (13), driving the higher risk of stain-induced myopathy.
Another common variant in SLCO1B1 involving the substitution of asparagine to aspartic acid at amino acid 130 (N130D, rs2306283) may also have functional consequences (12). This variant has been associated with a reduced area under the curve for plasma pravastatin concentration, suggesting a gain of function effect of this allele, however this has not been confirmed at the biochemical level (14).
As these variants appear to influence plasma concentrations and hepatic uptake of statins through the OAT1B1 transporter, we hypothesized that they may be having an impact upon general side effects experienced by patients being prescribed statins, which may in turn influence how these drugs are taken. We therefore sought to investigate the impact of the V174A and N130D variants of the SLCO1B1 gene on statin tolerance and lipid-lowering response in a large population of statin taking individuals with type 2 diabetes in Tayside.
Results
There were a total of 4196 patients genotyped for V174A and N130D. The 174A allele frequency was 0.162 (SE 0.004) and for 130D it was 0.382 (SE 0.005).
Characteristics of study population prior to commencement of statin treatment
The baseline characteristics of the study population according to genotype are shown in supplementary table 1. There was no difference by 174A genotype in mean age, gender ratio or BMI. Individuals homozygous for 130D allele were on average 1.2 years older than other individuals (p=0.0085, recessive model). The 130D allele was associated with a slightly lower mean baseline total cholesterol (mean difference 0.1mmol/l p=0.034, recessive model). The 174A allele was associated with a higher HDLc (mean difference 0.1 mmol/l p=0.0295, recessive model). There was no difference by 174A genotype in initial statin dose, however there was a significant linear trend by 130D genotype with homozygous individuals more likely to start on a lower dose (p=0.0025, additive model). There was no difference in prior CK or ALT tests by genotype. In addition, there was no difference in frequency of fibrate prescribing at baseline.
Characteristics of study population after commencement of statin treatment
The characteristics of the population during the statin exposed period according to genotype are shown in table 1. There was no difference in study duration, maximum statin dose prescribed, adherence to statins, statin switching or discontinuation rate by genotype. There was also no genotypic differences in the type of statin used at the beginning or at the end of treatment period (supplementary table 2). However, there was a difference in percentage of patients who switched to a lower dose observed in both genotypes, with the 174A homozgotes more likely to drop dose (p=0.05, recessive model) and the 130D homozygotes less likely (p=0.0333, recessive model). There was no difference in CK or ALT tests by genotype.
Table 1. Characteristics of study population after commencement of statin exposure.
Data are Means (SD) or individual counts (%)
| V174A | N130D | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All | VV | VA | AA | P(add) | P(rec) | NN | ND | DD | P(add) | P(rec) | |
| Study duration (yrs) | 5.5 | 5.4 | 5.7 | 5.0 | 0.3094 | 0.5978 | 5.4 | 5.4 | 5.6 | 0.9387 | 0.9712 |
|
| |||||||||||
| Maximum Dose (mg): | |||||||||||
| ≤10 | 261(6.2) | 178(6.1) | 75(6.4) | 8(8.3) | 97(6.0) | 113(5.8) | 51(8.2) | ||||
| >10 - <40 | 1364(32.5) | 960(32.8) | 379(32.5) | 25(25.8) | 522(32.3) | 634(32.5) | 208(33.3) | ||||
| ≥40 - <80 | 1757(41.9) | 1223(41.7) | 490(42.0) | 44(45.4) | 689(42.6) | 812(41.6) | 256(41.0) | ||||
| ≥80 - <160 | 601(14.3) | 422(14.4) | 165(14.1) | 14(14.4) | 242(15.0) | 279(14.3) | 80(12.8) | ||||
| ≥160 | 213(5.1) | 148(5.1) | 59(5.1) | 6(6.2) | 0.9280 | 0.6319 | 68(4.2) | 116(5.9) | 29(4.7) | 0.3719 | 0.1851 |
|
| |||||||||||
| Statin Adherence (%) | 77.5(19.1) | 77.4(18.9) | 78.1(19.3) | 74.6(22.0) | 0.8348 | 0.1289 | 77.9(18.6) | 77.0(19.6) | 78.2(18.8) | 0.8063 | 0.3286 |
| Dropped dose | 1107(26.4) | 772(26.3) | 301(25.8) | 34(35.1) | 0.5108 | 0.0500 | 447(27.6) | 517(26.5) | 143(22.9) | 0.0344 | 0.0333 |
| Discontinued | 283(6.7) | 201(6.9) | 74(6.3) | 8(8.3) | 0.8241 | 0.5504 | 109(6.7) | 130(6.7) | 44(7.1) | 0.8559 | 0.6982 |
|
| |||||||||||
| CK & ALT tests: | |||||||||||
| CK test | 2511(59.8) | 1741(59.4) | 712(61.0) | 58(59.8) | 0.4290 | 0.9921 | 956(59.1) | 1188(60.8) | 367(58.8) | 0.7904 | 0.5700 |
| Abnormal CK test (X1) | 729(17.4) | 501(17.1) | 207(17.7) | 21(21.7) | 0.3280 | 0.2608 | 263(16.3) | 365(18.7) | 101(16.2) | 0.5287 | 0.3960 |
| ALT test | 4063(96.8) | 2839(96.9) | 1132(96.9) | 92(94.9) | 0.6286 | 0.2589 | 1565(96.7) | 1894(96.9) | 604(96.8) | 0.8491 | 0.9563 |
| Abnormal ALT test (X1) | 1764(42.0) | 1233(42.1) | 488(41.8) | 43(44.3) | 0.9316 | 0.6439 | 691(42.7) | 826(42.3) | 247(39.6) | 0.2375 | 0.1778 |
Table 2 shows the frequencies of combined genotypes and their categorical coding (genotype scores) based on predicted functional status. A174 was predominantly found in association with 130D as the *15 haplotype (D’=0.83).
Table 2.
Cross-tabulation of SLCO1B1 genotypes
| V174A genotype | ||||
|---|---|---|---|---|
|
| ||||
| VV | VA | AA | ||
| N130D genotype (%) | NN | 1423(33.9)** | 186(4.4)*** | 9(0.2)**** |
| ND | 1221(29.1)* | 698(16.6)*** | 35(0.8)**** | |
| DD | 287(6.8) | 284(6.8)** | 53(1.3)**** | |
SLCO1B1 Genotype score 0; *1;**2; ***3; ****4. Where 0 is homozygous for haplotype *1B (DV) predicted to have the lowest AUC for simvastatin, and 4 is homozygous for *5/*15 (NA or DA) predicted to have the highest AUC for simvastatin. The intermediate scores assume a dose dependency of D and A separately with A174 being dominant over D130 as shown in previous pharmacokinetic studies.
Statin intolerance
Only 55 individuals had evidence of significant CK rise of over 3 times the normal range and these were removed from the analysis (supplementary table 3, supplementary figure 1). Based on our definition of tolerance, we were able to classify 1275 (30.4%) as tolerant and 816 (19.4%) as intolerant which gave a total of 2091 in the study. The remaining 2050 individuals were coded as missing as they were not classifiable as tolerant or intolerant. This may have been due to the occurrence of a prescribing change without a relevant biochemical measure or vice versa, or due to no indication of intolerance while being exposed to only a very low dose of statin.
Table 3 shows the breakdown of intolerance by genotype and genotype score. There was an insignificant association of the 174A allele with intolerance (p=0.0645, recessive model) whereas the 130D allele was associated with tolerance (p=0.0446, recessive model). These associations gave rise to a significant trend (p=0.0093) by genotype score.
Table 3.
Tolerant/Intolerant frequencies by genotype and combined genotype score.
| V174A | N130D | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | VV | VA | AA | P(add) | P(rec) | NN | ND | DD | P(add) | P(rec) | |
| Intolerant (%) |
816 | 565(69.2) | 227(27.8) | 24(2.9) | 316(38.7) | 399(48.9) | 101(12.4) | ||||
| Tolerant (%) | 1275 | 905(71.0) | 348(27.3) | 22(1.7) | 0.1972 | 0.0645 | 471(36.9) | 606(47.5) | 198(15.5) | 0.1064 | 0.0446 |
| SLCO1B1 genotype score | |||||||||||
| 0 | 1 | 2 | 3 | 4 | P(add) | ||||||
|
|
|||||||||||
| Intolerant (%) | 48(5.9) | 237(29.0) | 321(39.3) | 186(22.8) | 24(2.9) | ||||||
| Tolerant (%) | 100(7.8) | 396(31.1) | 497(39.0) | 260(20.4) | 22(1.7) | 0.0093 | |||||
In a logistic regression model accounting for both genotypes, statin adherence, study duration, maximum dose, prescription for other lipid-regulating drugs, CYP3A4 inhibiting drugs and age, the association of 174A with intolerance to statins (2.05, CI 1.02-4.09, p=0.0427) and 130D with tolerance (0.71, 0.52-0.96, p=0.0257) (table 4) was confirmed. This was also observed by genotype score, with an odds ratio of 1.14 (CI 1.02-1.28, p=0.0200) for each genotypic step. Similar results were seen when using only CK or ALT results to determine intolerance (supplementary tables 4 and 5 respectively).
Table 4.
Logistic regression results for Intolerance
| Single genotypes | Combined genotype | |||
|---|---|---|---|---|
|
| ||||
| Odds Ratio (95% CI) | P | Odds Ratio (95% CI) | P | |
| V174A Recessive | 2.05(1.02-4.09) | 0.0427 | 1.14(1.02-1.28) | 0.0200 |
| N130D Recessive | 0.71(0.52-0.96) | 0.0257 | ||
| Dose (MG): | ||||
| <40 | 1.00 | 1.00 | ||
| ≥40 - <80 | 1.59(1.25-2.01) | 0.0001 | 1.60(1.26-2.03) | 0.0001 |
| ≥80 - <160 | 3.89(2.83-5.34) | <.0001 | 3.90(2.84-5.36) | <.0001 |
| ≥160 | 5.21(3.10-8.75) | <.0001 | 5.24(3.12-8.82) | <.0001 |
| Other lipid-regulating drug: | ||||
| No | 1.00 | 1.00 | ||
| Yes | 2.27(1.43-3.60) | 0.0005 | 2.24(1.41-3.56) | 0.0006 |
| CYP3A4 inhibitor drugs: | ||||
| No | 1.00 | 1.00 | ||
| Yes | 1.58(1.28-1.95) | <.0001 | 1.58(1.28-1.95) | <.0001 |
| Statin Adherence (per 10%) | 0.85(0.81-0.90) | <.0001 | 0.85(0.81-0.90) | <.0001 |
| Statin Duration (per 1 yr) | 1.26(1.22-1.31) | <.0001 | 1.26(1.22-1.31) | <.0001 |
| Age at baseline (per 10 yrs) | 1.06(0.96-1.18) | 0.2436 | 1.06(0.96-1.17) | 0.2645 |
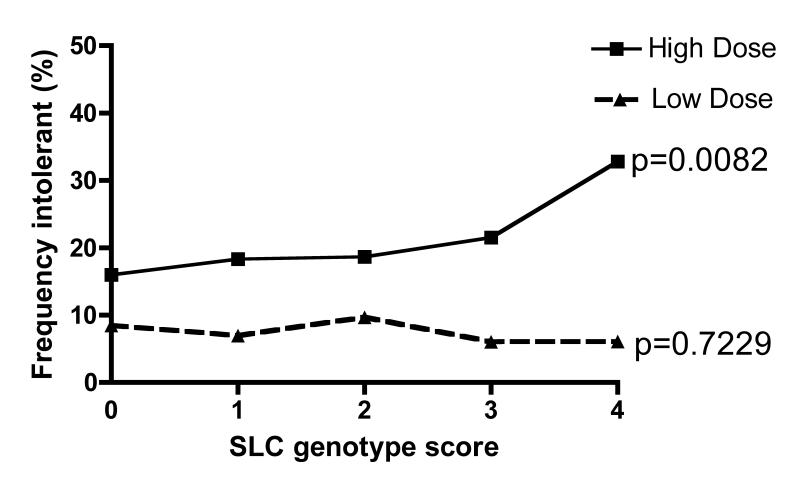
When stratified by statin dose we found a significant linear trend (p=0.0023) for an increased frequency of intolerant individuals by genotype category in the individuals prescribed statins at higher equivalent doses (≥40mg) with no evidence for this trend at lower doses (table 5). Furthermore, when the frequency of intolerant individuals was determined as a proportion of the total population (rather than just compared to the definitely tolerant group, total n=4141), this association remained (p for trend across groups=0.0434), with 36% of patients with the *15/*15 or *15/*5 genotypes (group 4) on a statin equivalent dose of ≥40mg classifiable as intolerant compared with only 21% in group 0 (figure 1). This suggests an overall number needed to harm in the entire population on ≥40mg statin equivalent for this genotype of approximately nine (15). As a large fraction of the population displayed some aspect of intolerance, but were not classed as intolerant by our criteria, this number needed to harm may represent an underestimate.
Table 5.
Frequency of tolerant/intolerant by dose group.
| SLCO1B1 genotype score | ||||||||
|---|---|---|---|---|---|---|---|---|
| All | 0 | 1 | 2 | 3 | 4 | P(trend) | ||
| Low dose (<40mg) | Intolerant (%) | 178 (24.2) | 13 (22.8) | 51 (23.8) | 79(26.0) | 33(21.9) | 2(18.2) | |
| Tolerant | 559 (75.9) | 44 (77.2) | 163 (76.2) | 225 (74.0) | 118 (78.2) | 9 (81.8) | 0.8076 | |
|
| ||||||||
| High dose (≥40mg) | Intolerant | 638 (47.1) | 35 (38.5) | 186(44.4) | 242(47.1) | 153(51.9) | 22 (62.9) | |
| Tolerant | 716 (52.9) | 56 (61.5) | 233 (55.6) | 272(52.9) | 142 (48.1) | 13 (37.1) | 0.0023 | |
Figure 1. Frequency of intolerance in whole population stratified by dose.
In order to contextualize the putative risk burden for intolerance in the entire statin treated population conferred by this variation, we coded the individuals who we could positively identify as intolerant as 1, and coded all other statin treated individuals as 0 regardless of the classification for tolerance. The data was then stratified by dose group as those treated with less than 40mg statin (simvastatin equivalent, Low Dose) and those treated with 40mg statin or above (simvastatin equivalent, High Dose). P values shown are for Chi squared tests for trend (1df.) across the SLCO1B1 genotype score.
Lipid-Lowering Efficacy
Previous studies have suggested a difference in statin efficacy in lipid-lowering may be modulated by these gene variants (8). We investigated the impact of these variants on maximum LDLc response. Individuals recessive for 174A (*5/*15) had a lower response (p=0.01) and individuals recessive for 130D variant had a greater response (p=0.0483) (Table 6). After accounting for variables that are affected by intolerance (statin adherence, study duration, maximum dose) in a multiple regression model, the association with LDLc response was attenuated (p=0.0534 and p=0.0850 for 174A and N130D respectively)(table 6). Furthermore, once we removed the group of patients defined as intolerant from the analysis, to see how the genotypic effect on lipid-lowering efficacy may be confounded by the effect of intolerance, the trend was completely eliminated (p=0.4962, 0.1758 for 174A and N130D respectively).
Table 6.
Multiple regression for maximum LDLc response
| Whole population (n=3710) |
Intolerant patients removed (n=2978) |
|||||
|---|---|---|---|---|---|---|
|
| ||||||
| Adjusted for covariates unrelated to intolerance |
Adjusted for intolerance related covariates |
|||||
|
| ||||||
| Parameter Estimate (SE) |
P | Parameter Estimate (SE) |
P | Parameter Estimate (SE) |
P | |
| V174A Recessive | 0.1822(0.0707) | 0.0100 | 0.1252(0.0648) | 0.0534 | 0.0507(0.0745) | 0.4962 |
| N130D Recessive | −0.0592(0.0300) | 0.0483 | −0.0488(0.0274) | 0.0750 | −0.0396(0.0293) | 0.1758 |
| Baseline LDLc (per 1mmol/L) | 0.2855(0.0110) | <.0001 | 0.3444(0.0105) | <.0001 | 0.3385(0.0117) | <.0001 |
| Age at baseline (per 10 yrs) | −0.0647(0.0102) | <.0001 | −0.0690(0.0095) | <.0001 | −0.0612(0.0101) | <.0001 |
| Dose (MG): | ||||||
| <20 | - | 0 | 0 | |||
| ≥20 - <40 | - | −0.1856(0.0389) | <.0001 | −0.1574(0.0402) | <.0001 | |
| ≥40 | - | −0.2777(0.0378) | <.0001 | −0.2868(0.0395) | <.0001 | |
| Statin Adherence (per 10%) | - | −0.0848(0.0051) | <.0001 | −0.0937(0.0057) | <.0001 | |
| Statin Duration (per 1 yr) | - | −0.0657(0.0032) | <.0001 | −0.0661(0.0037) | <.0001 | |
Discussion
We have demonstrated that the two functional variants, V174A and N130D of the SLCO1B1 gene encoding OATP-C/OATP1B1 are associated with intolerance of statins in this large observational study of statin prescribing in patients with type 2 diabetes. The direction of effect across haplotypes observed in our study is consistent with previous observations regarding the biochemistry and/or pharmacokinetics of these variants (16). These results suggest that these variants not only modulate susceptibility to the rare event of severe muscle toxicity during statin treatment (8), but are also responsible for more common and mild manifestations of intolerance. Our definition of intolerance was derived from statin discontinuation, switching or dose reduction as a surrogate indicator of intolerance, and as such may incorporate a range of more subtle side effects (not necessarily related to myopathy or myalgia).
It has been estimated that around 12% of individuals taking statins experience symptoms related to their use, resulting in 7% discontinuation due to these side effects (3, 4). Recently a large observational study, which only considered myopathic symptoms (17), found that in patients with diabetes statins were associated with a doubling of the risk of myopathy with 7.9% of statin users developing myopathy compared to 5.5% in the non statin users. However, this singular approach is likely to have underestimated the actual overall burden of side effects related to statins in this population, when compared to our, more general, approach.
The validity of the Go-DARTS study database for examining the role of pharmacogenetic variants that have been discovered in clinical trials is underscored by previous studies of genes such as APOE and HMGCR that influence lipid lowering efficacy in this population (18, 19), and further confirms the utility of this population-based approach to assess the external validity and generalisability of observations made in a clinical trial setting.
Real world validation of statin intolerance is of vital importance as trials can underestimate the true incidence of such problems due to their study design (20). Trial subjects are seldom representative of the subsequent target population, and often exclude groups of individuals who may be at particular risk. For example, the Heart Protection Study (21), screened a total of 63,603 individuals of whom only approximately 50% (32,145) went forward into the pre-randomisation phase of statin therapy. Of these, 9% reported a problem with the run in medication, with 65% choosing not to continue for unspecified reasons. As the major expressed role for the run in phase was to ensure compliance and minimal drop out it is not surprising therefore that in the final 20,000 randomised the side effect rates between placebo and treatment groups were very similar.
Clearly one of the weaknesses of our study is the derivation of a necessarily imprecise surrogate index of statin intolerance based on statin prescribing changes and routinely-measured biochemical assays. We have included discontinuation as a component of our definition of intolerance and it is clear that individuals may stop statins in a real life setting for reasons that are unrelated to statin intolerance. It is also clear that individuals may present with abnormal CK levels for reasons unrelated to the statin treatment, therefore we derived our definition using the co-existence of both CK testing and a relevant prescribing change. Regression modelling confirmed that this combined definition is highly related to the dose of statin used and indicates that this is a suitably statin-related measure. Furthermore, it is clear that our approach has been successful in revealing a genetic signal of the impact of these variants on statin tolerability regardless of the nature of this intolerance definition. The vast majority of our subjects were treated with simvastatin (supplementary table 2) and it is likely that this genotype may be selective for certain statins, indeed our data shows that the signal obtained with the SLCO1B1 genotypes is stronger when limited to those initially treated with simvastatin (supplementary table 6). A recent clinical trial with pravastatin and simvastatin reported that SLCO1B1 genotypes may predict intolerance to simvastatin but not pravastatin consistent with a lack of correlation between genotype and serum pravastatin statin levels in this study (22). Previous work has however demonstrated a robust role for these SLCO1B1 variants in pravastatin disposition. Our data are not powered to confirm or refute the role of pravastatin in this context, but it does appear that the effect of the genotype is consistent in the case of simvastatin, with the original myopathy findings being made in two simvastatin trials (8). It is therefore possible that individuals with the susceptible genotypes of SLCO1B1 may find other forms of statin more tolerable, and further research should be undertaken to examine this hypothesis.
The observation that the genotype effect is more apparent at higher doses is potentially clinically relevant as more recent clinical trial evidence has indicated the importance of the use of higher doses of statins to obtain maximal efficacy, and, as a result, there is an increasing tendency for patients to receive higher and higher doses with the associated increased risk of side effects. Our study suggests that at least one third of individuals with the *5/*15 haplotypes of SLCO1B1 are likely to suffer side effects at such high doses of statins. While this points to a small group of individuals who are at high risk of intolerance, our data would suggest that in a population treated with ≥40mg of simvastatin only nine people would need to be treated to result in one individual being intolerant due to this genotype. Indeed it has recently been suggested that doses of statins should be tailored to an individual’s SLCO1B1 genotype to avoid the risk of side effects (23). Side effects may give rise to discontinuation which would clearly lead to reduced lipid-lowering response to statins, which we have shown in our population. This would be predicted to result in turn to an increased susceptibility to cardiovascular events highlighting the potential importance of adjusting statin therapy to an individual’s SLCO1B1 genotype.
Methods
We performed an observational incident cohort study using data from the Genetics of Diabetes Audit and Research (Go-DARTS) database. This includes detailed information on people with diabetes in Tayside Scotland (population 400,000) including all prescriptions dispensed from Tayside pharmacies, all biochemistry from the region-wide clinical laboratory database and other clinical data relating to diabetes care from 1990 to present (24), and this is record-linked to consented genetic information.
Study Population
All patients taking part in Go-DARTS who were resident in Tayside during the study period 1st January 1990 to 30th September 2008, who were genotyped for both SLCO1B1 variants and with a record of at least two dispensed prescriptions for a statin were included in the study. The study period for each individual was defined as time from first to last statin prescription (encashed before 30th September 2008).
Definition of intolerance
A composite measure of intolerance was defined using both biochemical abnormalities and relevant prescribing changes within individuals. Biochemical abnormalities were defined as those exceeding the upper limit of the normal ranges for both alanine aminotransferase (ALT) and creatinine kinase (CK), as determined for the age and sex of each individual using routine clinical biochemistry classifications. Relevant prescribing changes were defined as switching statin to equivalent or lower dose, dose reduction of same statin, or discontinuation of statin prescribing.
Indicators of intolerance were either (i.) abnormal CK measure 1 to 3 times over upper limit of normal (>3 were excluded from analyses), with no abnormal CK recorded before statin commencement; or (ii) an abnormal ALT measure, with no abnormal ALT before statin commencement (≥50% increase in ALT from baseline during the study period was also considered abnormal). These abnormalities had to be accompanied by evidence of a relevant change in prescribing in order for an individual to be defined as intolerant. In addition, individuals with normal CK measured directly prior to a prescribing change were also classified as intolerant. Individuals with abnormal CK or ALT values but no change in prescribing were defined as neither tolerant nor intolerant, as were individuals with a prescribing change without a relevant biochemical test. The tolerant group were defined as individuals with no recorded abnormal CK or ALT measures recorded during the period of exposure to statin treatment, with no prescribing changes suggestive of intolerance and a consistent statin dose equivalent to 10mg of simvastatin or greater.
Definition of lipid-lowering response
To measure response to statins we considered maximum response. This was defined as the lowest LDLc measured while on statins. An LDLc measurement before each individual’s first statin prescription was determined (baseline measurement). Where this was not available values were estimated using multiple imputation methods as previously described (18, 19, 25). Patients were required to have at least one post-treatment LDLc for their lipid-lowering response to be measured. All lipid measurements recorded between first, and last prescription of statins were considered, and the maximum response observed. Any measurements taken after patients reached a simvastatin (or equivalent) dose of 80mg, or started on another lipid-regulating drug (BNF chapter 2.12) were not included.
Drug adherence and dose
Detailed drug dispensing records were used to calculate the percentage maximum possible adherence to statins for each patient using previously validated methods (25, 26).
Dose was calculated as the maximum dose prescribed during the study period, and where other statins were prescribed expressed as the equivalent dose of simvastatin (19).
Genotyping
Genotyping for rs4149056 (V174A) and rs2306283 (N130D) was performed using TAQMAN assays. Both variants were in Hardy-Weinberg equilibrium and were in linkage disequilibrium (D’=0.83), however the higher frequency of 130D resulted in a low correlation coefficient (R2=0.2). Genotyping success rate was >98%.
SLCO1B1 analysis
The V174A and N130D variants were analysed separately as recessive models. In addition, the genotypes were combined to make genotype scores based on the combinations of the 4 haplotypes and their putative functions based on the body of evidence summarized on the Pharmacogenomics Knowledge Base, which includes pharmacokinetic and biochemical data (16), indicating the likely incremental functional implications of the groups. The Ala174 variant was coded as 4 in the homozygote form regardless of N130D genotype, as both the *5 and the *15 haplotypes are associated with increased serum levels of statins, and the A174 appears to lead to cytoplasmic retention of the protein. In contrast the D130 homozygote was coded 0 only in the absence of A174 as this haplotype (*1B) has been shown to be associated with decreased serum levels of statins. The *1a haplotype acted as the reference (N130,V174).
Statistical methods
Quantitative variables are presented as means (±SD) and categorical variables as percentages. For testing significance of the additive model, comparison between groups of continuous variables utilized ANOVA test for trend and for categorical data the chi-square test for trend was used. For the recessive model the t-test was used for continuous data and chi-square for categorical variables.
The primary outcome of intolerance was analysed by logistic regression and maximum LDLc response to statins by multiple linear regression. Covariates selected for the model were maximum statin dose, adherence to statins, age at baseline and study duration. For the intolerance model, whether patients were on concurrent lipid-regulating medication, or on any medication known to inhibit CYP3A4 (16), were also included as a covariate, and a baseline LDLc measurement was added to the lipid-lowering response model.
Supplementary Material
Acknowledgements
This work was funded by The Wellcome Trust (Award 072960 and 084726) and the UK Medical Research Council (Award G0601261). The genotyping was facilitated by a capital funding from the CSO Generation Scotland initiative.
Footnotes
Disclosures: The authors have no conflicts of interest to disclose.
References
- (1).Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–78. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- (2).Armitage J. The safety of statins in clinical practice. Lancet. 2007;370:1781–90. doi: 10.1016/S0140-6736(07)60716-8. [DOI] [PubMed] [Google Scholar]
- (3).Lansberg PJ, Mitchel YB, Shapiro D, Kastelein JJ, Altman R, Jerums G, et al. Long-term efficacy and tolerability of simvastatin in a large cohort of elderly hypercholesterolemic patients. Atherosclerosis. 1995;116:153–62. doi: 10.1016/0021-9150(95)05523-y. [DOI] [PubMed] [Google Scholar]
- (4).Simons LA, Levis G, Simons J. Apparent discontinuation rates in patients prescribed lipid-lowering drugs. Med J Aust. 1996;164:208–11. doi: 10.5694/j.1326-5377.1996.tb94138.x. [DOI] [PubMed] [Google Scholar]
- (5).Pedan A, Varasteh L, Schneeweiss S. Analysis of factors associated with statin adherence in a hierarchical model considering physician, pharmacy, patient, and prescription characteristics. J Manag Care Pharm. 2007;13:487–96. doi: 10.18553/jmcp.2007.13.6.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Donnelly LA, Doney AS, Morris AD, Palmer CN, Donnan PT. Long-term adherence to statin treatment in diabetes. Diabet Med. 2008;25:850–5. doi: 10.1111/j.1464-5491.2008.02476.x. [DOI] [PubMed] [Google Scholar]
- (7).After the diagnosis: adherence and persistence with hypertension therapy. Am J Manag Care. 2005;11:S395–9. [PubMed] [Google Scholar]
- (8).Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- (9).Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem. 2001;276:35669–75. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- (10).Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005;15:513–22. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI] [PubMed] [Google Scholar]
- (11).Iwai M, Suzuki H, Ieiri I, Otsubo K, Sugiyama Y. Functional analysis of single nucleotide polymorphisms of hepatic organic anion transporter OATP1B1 (OATP-C) Pharmacogenetics. 2004;14:749–57. doi: 10.1097/00008571-200411000-00006. [DOI] [PubMed] [Google Scholar]
- (12).Nozawa T, Nakajima M, Tamai I, Noda K, Nezu J, Sai Y, et al. Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J Pharmacol Exp Ther. 2002;302:804–13. doi: 10.1124/jpet.302.2.804. [DOI] [PubMed] [Google Scholar]
- (13).Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–9. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- (14).Niemi M, Schaeffeler E, Lang T, Fromm MF, Neuvonen M, Kyrklund C, et al. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1) Pharmacogenetics. 2004;14:429–40. doi: 10.1097/01.fpc.0000114750.08559.32. [DOI] [PubMed] [Google Scholar]
- (15).Altman DG. Confidence intervals for the number needed to treat. Bmj. 1998;317:1309–12. doi: 10.1136/bmj.317.7168.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–70. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- (17).Nichols GA, Koro CE. Does statin therapy initiation increase the risk for myopathy? An observational study of 32,225 diabetic and nondiabetic patients. Clin Ther. 2007;29:1761–70. doi: 10.1016/j.clinthera.2007.08.022. [DOI] [PubMed] [Google Scholar]
- (18).Donnelly LA, Doney AS, Dannfald J, Whitley AL, Lang CC, Morris AD, et al. A paucimorphic variant in the HMG-CoA reductase gene is associated with lipid-lowering response to statin treatment in diabetes: a GoDARTS study. Pharmacogenet Genomics. 2008;18:1021–6. doi: 10.1097/FPC.0b013e3283106071. [DOI] [PubMed] [Google Scholar]
- (19).Donnelly LA, Palmer CN, Whitley AL, Lang CC, Doney AS, Morris AD, et al. Apolipoprotein E genotypes are associated with lipid-lowering responses to statin treatment in diabetes: a Go-DARTS study. Pharmacogenet Genomics. 2008;18:279–87. doi: 10.1097/FPC.0b013e3282f60aad. [DOI] [PubMed] [Google Scholar]
- (20).Thompson PD, Clarkson P, Karas RH. Statin-associated myopathy. Jama. 2003;289:1681–90. doi: 10.1001/jama.289.13.1681. [DOI] [PubMed] [Google Scholar]
- (21).MRC/BHF Heart Protection Study of cholesterol-lowering therapy and of antioxidant vitamin supplementation in a wide range of patients at increased risk of coronary heart disease death: early safety and efficacy experience. Eur Heart J. 1999;20:725–41. doi: 10.1053/euhj.1998.1350. [DOI] [PubMed] [Google Scholar]
- (22).Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54:1609–16. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Niemi M. Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther. 87:130–3. doi: 10.1038/clpt.2009.197. [DOI] [PubMed] [Google Scholar]
- (24).Morris AD, Boyle DI, McMahon AD, Greene SA, MacDonald TM, Newton RW. Adherence to insulin treatment, glycaemic control, and ketoacidosis in insulin-dependent diabetes mellitus. The DARTS/MEMO Collaboration. Diabetes Audit and Research in Tayside Scotland. Medicines Monitoring Unit. Lancet. 1997;350:1505–10. doi: 10.1016/s0140-6736(97)06234-x. [DOI] [PubMed] [Google Scholar]
- (25).Wei L, Wang J, Thompson P, Wong S, Struthers AD, MacDonald TM. Adherence to statin treatment and readmission of patients after myocardial infarction: a six year follow up study. Heart. 2002;88:229–33. doi: 10.1136/heart.88.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Donnan PT, MacDonald TM, Morris AD. Adherence to prescribed oral hypoglycaemic medication in a population of patients with Type 2 diabetes: a retrospective cohort study. Diabet Med. 2002;19:279–84. doi: 10.1046/j.1464-5491.2002.00689.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.