The extracellular matrix (ECM) provides vital structure and organization to all metazoans, but it is far from an inanimate, unchanging assemblage of collagens, proteoglycans, and glycoproteins. ECM undergoes constant remodeling, most obviously during development, wound healing, and other repair processes. Defects in regulation of the deposition and turnover of ECM play important roles in pathological processes such as osteoporosis, atherosclerosis, arthritis, and fibrotic diseases. The deposition and remodeling of ECM is accomplished by cells, and the paper of Ohashi et al. in a recent issue of the Proceedings (1) provides some of the first observations of these processes in real time in living cell cultures. Ohashi et al. transfected cells with a chimera of green fluorescent protein and fibronectin (FN). FN is a prominent constituent of ECM around and beneath many cells, and FN-rich matrices provide substrates for cell adhesion and migration during development, wound healing, and other situations, as well as affecting many cellular functions including proliferation, survival, and differentiation (2). Ohashi et al. show that the FN–GFP chimera assembles into the FN-rich matrix surrounding their transfected cells, and they then use fluorescence microscopy to observe alterations in that matrix over time. This approach offers many opportunities for future research, but the observations they report already reveal some intriguing features that have implications not only for ECM structure and function but more widely in cell adhesion and beyond.
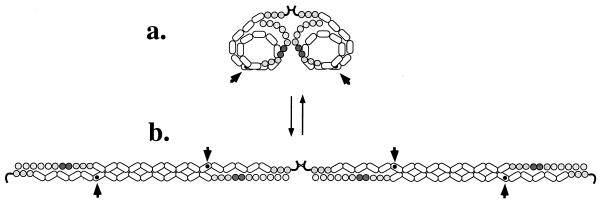
Like most ECM proteins, FN is a large molecule; a dimer of subunits of around 250 kDa, each composed of a series of independently folding modular domains known as FN repeats I, II, and III; (12, 2, and 15–17 copies, respectively). Soluble dimeric FN molecules are loosely folded, rather like a string of beads clumped into a loose ball (Fig. 1a). In the ECM, these strings of beads are extended and laterally associated, probably in antiparallel fashion (Fig. 1b), although the exact arrangement is still unclear (3). This fibrillogenesis is not spontaneous and generally occurs only in the presence of cells; the cells perform a “spider” or “knitting” function to assemble the FN-rich ECM around themselves. It has been known for 20 years that the arrangement of FN fibrils relates to the arrangement of actin filaments inside the cells (and vice versa) and that cells appear to pull on the matrix, generating tension (4, Fig. 2). It also is clear that integrin receptors are involved (5–7) and that cell-derived tension is necessary (5, 8, 9) for FN matrix assembly. So it was expected that FN-matrix fibrils would move in response to cellular traction forces, and indeed, Ohashi et al. observed just that. They describe extensions, deformations, and fusions of ECM fibrils over time intervals ranging from a few minutes to hours. More interestingly, they observed that the fibrils were elastic, appearing to stretch and retract in response to cellular movements. Extended fibrils were observed to detach at one end and retract into much shorter fibrils and small clumps of FN. The contractions could be to <25% of the original fibril length and occur in less than 5 min, indicating that the fibrils were extensively stretched, presumably via integrin-mediated attachments to cells.
Figure 1.
Assembly of fibronectin (FN) (a) In solution, FN is a globular protein consisting of a linear array of independently folded globular domains coiled into a relatively compact conformation. The elongated modules are Fn3 repeats (2.5 × 4nm). (b) When assembled into filaments, the FN molecules are extended and laterally associated, probably in an antiparallel fashion. The details of the intersubunit interactions are not known. The RGD-integrin-binding site in each subunit (arrowhead) is shown partially occluded in the soluble molecule and exposed in the filament as an example of cryptic sites exposed during FN fibrillogenesis.
Figure 2.
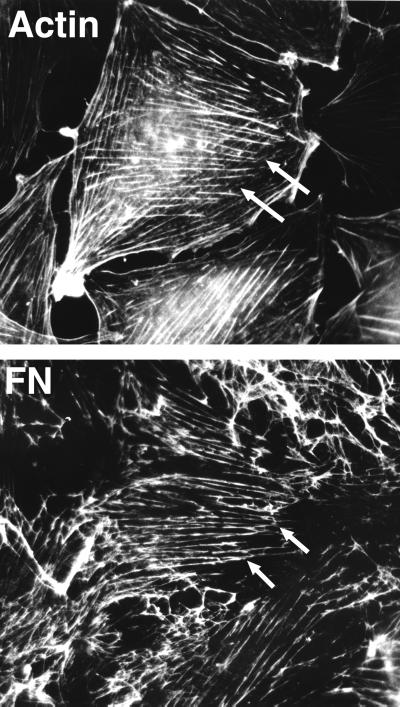
Coalignment of actin microfilament bundles and extracellular FN fibrils. Double-label immunofluorescence of a well spread, nonmotile cell showing extensive codistribution of the two sets of fibrils. Note the extension beyond the cell boundary of FN fibrils linked to stress fibers within the cells (arrows). This demonstrates the tension applied by cells on the ECM. Reproduced with permission from Hynes and Destree, 1978 (4).
To understand the implications of this stretched state of FN fibrils, we must return to the molecular structure of FN, about which a fair amount is known (2, 10). Three-dimensional structures are available for each of the three types of modules; type I and II modules are compact disulfide-bonded modules, whereas the type III repeats (Fn3), which comprise more than half the protein, are β-sandwiches rather similar in overall structure to Ig domains, although Fn3 repeats lack S–S bonds. Fn3 repeats are among the most widespread protein modules in the genome (11), occurring in other ECM proteins in addition to FN, in many cell adhesion receptors and cytokine receptors, and in intracellular muscle proteins such as titin and twitchin. Each repeat is around 2.5 nm wide and 4 nm long, and FN has 15–17 of them (depending on alternative splicing) arranged in tandem. Atomic resolution structures of tandem arrays of Fn3 repeats show them to be elongated arrays with fairly small interdomain interfaces; the angle between consecutive repeats varies (12). Another matrix molecule, tenascin, has a similar tandem array of Fn3 repeats (13).
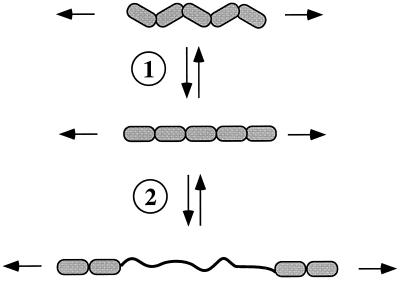
So, what is the relevance of these facts to the elasticity of FN matrix fibrils? The presumed antiparallel organization of individual FN molecules (see Fig. 1) is consonant with the smallest diameter filaments being ≈5 nm in diameter, as observed (2). However, many matrix fibrils are much thicker than this and presumably comprise lateral associations of filaments. So, extension of fibrils under tension could arise from sliding of filaments within the fibrils, although this is likely restricted by covalent crosslinking, known to occur in the fibrils (2). However, that would not readily explain the elasticity and rapid recoil of fibrils when detached. Other work suggests that the elasticity arises from intrinsic features of the tandem array of Fn3 repeats. Erickson (14) proposed that Fn3 repeats could unfold under tension, and Soteriou et al. (15) proposed the same for the multiple, tandem Fn3 repeats and Ig domains of titin, an enormous elastic protein of skeletal muscle. Estimates of the free energy required to unfold an Fn3 domain (7–14 kcal/mol; 1 cal = 4.18 J) and of the force necessary to extend a folded Fn3 domain (2.5 × 4 nm) into an extended polypeptide (≈29 nm) showed that the required tensile forces are comparable to those generated by relatively few myosin or kinesin molecules (14). Thus, it is entirely reasonable to propose that molecules such as FN and tenascin in the ECM, or titin and twitchin inside muscle cells, could extend under the influence of forces generated by the cytoskeleton. Tests of this hypothesis using optical tweezers or atomic force microscopy to stretch individual molecules (16–19) have shown that both the Fn3 repeats of tenascin and the intracellular Ig repeats of titin (which also lack S–S bonds) can be extended by pulling on them. The extensions observed were up to 20–25 nm per repeat, and when the force was removed, the repeats refolded (see Fig. 3). The results (16–19) conform well with the predictions (14), except that the forces required to unravel the Fn3 and Ig domains are larger than had been calculated. Nonetheless, they are well within the range of forces that cells could exert. At even lower forces, comparable to those exerted by single motor proteins, significant elastic extension of these tandem arrays of repeats can be obtained simply by extending the zigzag array of domains (see Fig. 3). Whether unfolding of individual domains or merely extension of tandem repeating arrays occurs in the in vitro situation remains uncertain, although the 4-fold contraction observed by Ohashi et al. (1) would be hard to explain solely on the basis of alignment of the Fn3 repeats. Whichever mechanism(s) apply, it is clear that FN, tenascin, and titin are extensible and elastic proteins (Fig. 3).
Figure 3.
Elastic extension in response to applied force. The diagram depicts two steps in unfolding of a series of Fn3 repeats (five are shown). In the first step (①), the zigzag array of repeats is extended into a linear array. In the second step (②), which requires greater force, individual Fn3 repeats unfold to an extended polypeptide chain with a 5- to 6-fold increase in length. Both steps are reversible giving elastic recoil.
This elasticity of matrix molecules and titin arises from their repeating arrays of Fn3 and Ig repeats and may also occur in other important proteins. This is particularly the case in cell-adhesion receptors, many of which are also built of tandem arrays of Ig and Fn3 repeats. Indeed, evolutionary considerations suggest that such molecules represent some of the earliest uses of these modules; these cell-adhesion molecules arose before the divergence of vertebrates, arthropods, and nematodes, whereas fibronectin and Igs, although they have given their names to the domains, both appear to be vertebrate inventions. Given what we now know about the behavior of tandem arrays of Fn3 and Ig domains, it is reasonable to propose that cell-adhesion receptors may also exhibit elastic extensibility. Such elasticity could extend to other repeating modules found in cell-adhesion molecules, including cadherin repeats, which, like Fn3 repeats, are β-sandwiches and lack disulfide bonds.
What could be other implications of this elastic stretching of FN? One intriguing possibility is that cell-derived extension of the FN molecule could expose cryptic sites (refs. 5 and 9, and refs. therein). FN contains many binding sites for cells and for other molecules, including itself. In the soluble molecule, folded on itself, many of these are not accessible (Fig. 1a). Unfolding of the extended FN subunits as they assemble into filaments and further unfolding under tension could well expose additional binding sites. Indeed, certain segments of FN that are involved in the assembly of FN fibrils need to be exposed by interactions with integrins (this is believed to be why cells are needed for matrix assembly—the “spider” function), and a key FN self-assembly site has been shown to be exposed by cell contractility, pulling on the FN fibrils (9). Thus, cell-surface FN receptors and their attachments to the cytoskeleton extend the FN molecule, allowing it to display a fuller repertoire of binding sites. It will not be surprising if additional functional sites remain to be discovered by analyses of extended matrix fibrils that have not been revealed by studies of unextended matrix molecules such as FN attached to glass or plastic. Matrix glycoproteins are large and conserved in sequence, but many domains have no assigned functions—these may yet be revealed by using dynamic tension. Another example of this phenomenon of reciprocal interactions between cells and matrix proteins comes from von Willebrand factor, a large glycoprotein of the vascular endothelial matrix. von Willebrand factor only normally binds its receptors on platelets when the molecule is perturbed by binding to ECM or by extreme shear forces arising from blood flow (20, 21). Furthermore, the initial binding to one platelet receptor, glycoprotein Ib, activates a second receptor, αIIbβ3, which then binds to a second site in von Willebrand factor.
The ECM is not just a static scaffold; it is a dynamic, information-rich source of inputs for cells, and some of those inputs are only revealed after cellular deformation of the matrix molecules; the discourse between cells and matrix is a dialogue. The ability to observe these two-way interactions as they are happening, by using approaches such as those pioneered by Ohashi et al. (1), promises to reveal many more secrets of this two-way conversation.
Footnotes
The companion to this commentary begins on page 2153 in issue 5 of volume 96.
References
- 1.Ohashi T, Kiehart D P, Erickson H P. Proc Natl Acad Sci USA. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes R O. Fibronectins. New York: Springer; 1990. [Google Scholar]
- 3.Mosher D F. Curr Opin Struct Biol. 1993;3:214–222. [Google Scholar]
- 4.Hynes R O, Destree A T. Cell. 1978;15:875–886. doi: 10.1016/0092-8674(78)90272-6. [DOI] [PubMed] [Google Scholar]
- 5.Wu C, Keivens V M, O’Toole T E, McDonald J A, Ginsberg M H. Cell. 1995;83:715–724. doi: 10.1016/0092-8674(95)90184-1. [DOI] [PubMed] [Google Scholar]
- 6.Yang J T, Hynes R O. Mol Cell Biol. 1996;7:1737–1748. doi: 10.1091/mbc.7.11.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wennerberg K, Lohikangas L, Gullberg D, Pfaff M, Johansson S, Fassler R. J Cell Biol. 1996;132:227–238. doi: 10.1083/jcb.132.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliday N L, Tomasek J J. Exp Cell Res. 1995;217:109–117. doi: 10.1006/excr.1995.1069. [DOI] [PubMed] [Google Scholar]
- 9.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin A M, Burridge K. J Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potts J R, Campbell I D. Curr Opin Cell Biol. 1994;6:648–655. doi: 10.1016/0955-0674(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 11.Bork P, Doolittle R F. Proc Natl Acad Sci USA. 1992;89:8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leahy D J, Aukhil I, Erickson H P. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 13.Erickson H P. Curr Opin Cell Biol. 1993;5:869–876. doi: 10.1016/0955-0674(93)90037-q. [DOI] [PubMed] [Google Scholar]
- 14.Erickson H P. Proc Natl Acad Sci USA. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soteriou A, Clarke A, Martin S, Trinick J. Proc R Soc London Ser B. 1993;254:83–86. doi: 10.1098/rspb.1993.0130. [DOI] [PubMed] [Google Scholar]
- 16.Tskhovrebova L, Trinick J, Sleep J A, Simmons R M. Nature (London) 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 17.Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 18.Kellermayer M S, Smith S B, Granzier H L, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 19.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 20.Wagner D D. Annu Rev Cell Biol. 1990;6:217–246. doi: 10.1146/annurev.cb.06.110190.001245. [DOI] [PubMed] [Google Scholar]
- 21.Savage B, Saldivar E, Ruggeri Z M. Cell. 1996;84:289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]