Abstract
With rapid development of sequencing technologies such as deep sequencing and whole genome high-density tiling array, we now know that most of the “junk” genomic sequences are transcribed as non-coding RNAs (ncRNAs). A large number of long ncRNA transcripts (> 200bp) have been identified, and these long ncRNAs (LncRNAs) are found to be crucial regulators for epigenetic modulation, transcription, and translation. In this review, we briefly summarize the regulatory function of LncRNAs with a particular focus on the underlying mechanisms of LncRNAs in oncogenesis, tumor metastasis and suppression.
Keywords: Long non-coding RNA (LncRNA), epigenetic regulation, competitive endogenous RNA (ceRNA), oncogenic lncRNA, pseudogene transcript, natural antisense RNA (NAT)
Introduction
When the whole human genome was determined, it was a surprise that there are only about 20,000-25,000 protein-coding genes, representing less than 2% of whole human genome. It was hard to imagine that most of the genomic sequences are junk DNA since simple Drosophila melanogaster and Caenorhabditis elegans have a very close number of protein-coding genes. The limited number of protein-coding genes cannot explain the developmental and physiological complexity of humans. With the rapid development in high-throughput sequencing technologies such as deep sequencing and whole genome high-density tiling array, it is now known that about 98% of the “junk” DNAs are transcribed as non-coding RNAs (ncRNAs) including short and long ncRNAs [1,2].
A large variety of ncRNAs can be divided into two classes: structural and regulatory ncRNAs (Table 1). Structural ncRNAs include tRNA, rRNA, and snoRNA. Based on ncRNA length, regulatory ncRNA can be further divided into at least three groups: (1) Short ncRNA including MicroRNA (miRNA) (22-23 nts) and piwi-interacting RNA (piRNA) (26-31 nts); (2) medium ncRNA (50-200 nts); (3) long ncRNA (>200 nts). This review will focus on medium and long ncRNAs, which are collectively referred to as large or long ncRNAs (LncRNAs). Based on large-scale sequencing and prediction from chromatin-state maps of full length cDNA libraries in FANTOM2 and 3 as well as human transcriptomes, more than 4,600 LncRNAs in mouse and over 3,300 LncRNAs in human have been identified with a total of approximately 23,000 LncRNAs in a mammalian genome [3-7].
Table 1.
RNA Classifications
| Class | Functions | |
|---|---|---|
| mRNAs | Encoding proteins | |
| ncRNAs | ||
| 1. House-keeping or Structural ncRNAs | ||
| tRNA (transfer RNA) | mRNA translation | |
| rRNA (ribosomal RNA) | mRNA translation | |
| snoRNA (small nucleolar RNAs) | rRNA modification | |
| snRNA (small nuclear RNA including spliceosomal RNAs) | RNA splicing, polyadenylation | |
| 2. Regulatory ncRNAs | ||
| Short ncRNA | ||
| miRNA (22-23nt) | Degradation of mRNA or repression of translation | |
| piRNA (26-31nt, piwi-interacting RNA) | Silencing of transposons | |
| Medium ncRNA (50-200nt) | ||
| paRNA (promoter-associated ncRNA) | Gene repression in cis via interacting with PRC2 | |
| Long (large) ncRNA (>200nt) | ||
| Intergenic ncRNA | Epigenetic regulators of transcription in cis/in trans | |
| Intronic ncRNA | Ibid | |
| UTR LncRNA | Ibid | |
| Antisense transtcript | mRNA stability of its homologous coding gene | |
| Pseudogene transcript | Generation of NATs or ceRNAs, stabilization of its coding transcript by competitive binding miRNAs | |
| Enhancer-like ncRNA (eRNA) | Activation of promoter activity by unknown mechanism | |
| Mitochondrial ncRNA | Cell cycle regulation and more unknown functions | |
| Repeat-associated ncRNA | Regulation of repeat silencing | |
| Satellite ncRNA | Involvement of formation and function of centromere-associated complexes | |
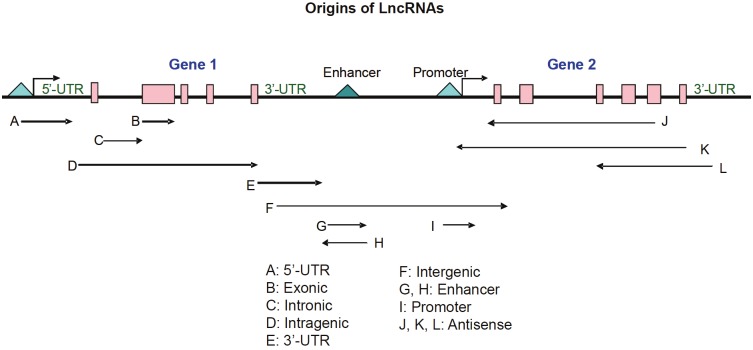
Biogenesis of LncRNAs is quite complicated. In general, LncRNA transcription and processing is very similar to protein-coding RNA. Most of LncRNAs are transcribed by RNA polymerase (RNAP) II, but some LncRNAs have been reported to be transcribed by RNAP III, and the majority of LncRNAs are spliced, polyadenylated and 5′-capped. Some LncRNAs are evolutionarily conserved and can be expressed at low level. A large proportion of LncRNAs has highly conserved proximal promoter sequence, exonic sequences, intronic sequences or secondary RNA structures. LncRNAs originate from intronic, exonic, intergenic, intragenic, promoter regions, 3′- and 5′-UTR, and enhancer sequences and are sometimes bidirectional transcripts (Figure 1). In particular, a large group of LncRNAs is antisense to known protein-coding transcripts that are also referred to as natural antisense transcripts (NATs) [6,8,9]. NATs are divided into two subtypes: cis-NATs, which are transcribed from opposite DNA strands at the same genomic loci; and trans-NATs, which are transcribed from distal loci (Figure 1). More recently, emerging experimental evidence revealed that NATs can also be generated from pseudogenes [10,11]. Notably, many cancerrelevant genes, particularly tumor suppressor genes, produce long antisense ncRNAs (Table 2).
Figure 1.
Origins of LncRNA. Arrows represent different types of LncRNA transcripts.
Table 2.
Identified tumor and disease-associated LncRNAs
| Name | Function/Characterization | References |
|---|---|---|
| H19 | Imprinted at the lgf2 locus; controls igf2 expression in cis, implicated in both tumor suppressors and oncogenes | Maher, ER. et al., 2000 [94]; Gaston, V. et al., 2001 [95] |
| HOTAIR | Intergenic transcript of HoxC locus, gene silencing in trans through interacting with PCR2 and LSD1 complex, involved in breast cancer metastasis | Tsai, MC. et al., 2010 [15]; Gupta, RA. et al., 2010 [16]; Rinn, JL. et al., 2007 [28] |
| AIR | Imprinted, monoallelically expressed from the paternal allele, interacts with histone methyltransferase G9a | Wutz, A. et al., 1997 [24]; Nagano, T. et al., 2008 [25]; Sleutels, F et al, 2002 [27] |
| ANRIL | Antisense transcript of INK4n/ARF/INK4a and p15/CDKN2B, required for the PRC2 recruitment to and silencing p15INK4b tumor suppressor gene | Pasmant, E. et al., 2007 [29]; Yap, KL. et al., 2010 [31]; Kotake, Y. et al., 2011 [32] |
| HOTAIRM1 | Antisense intergenic RNA myeloid 1, transcribed antisense to the HOXA genes, plays a role in the myelopoiesis through modulation of gene expression in the HOXA cluster | Zhang X., et al., 2009 [140] |
| KCNQ1OT1 | Tissue-specific imprinted genes within the Kcnq1 domain, interacting with both PRC2 and G9a leading to gene silencing in a lineage-specific manner | Kanduri, C. et al., 2006 [37]; Pandey, RR. et al., 2008 [33]; Lee, MP. et al., 1999 [97]; Nakano, S. et al., 2006 [98] |
| Evf-1 | Activates transcriptional activity by directly influencing Dlx-2 activity | Kohtz JD & Fishekk G., 2004 [141] |
| Evf-2 | An alternatively spliced form of Evf-1 activates transcriptional activity by directly influencing Dlx-2 activity | Feng, J. et al., 2006 [49] |
| P15AS | Antisense transcript of p15, highly expressed in leukemia, epigenetically silences the tumor suppressor gene p15 | Yu, W. et al., 2008 [107] |
| Xist | Mosaic expression, spreads on Xi in cis, interacts with BRCA1, correlated with breast cancer, cervical, ovarian, and testis tumors | Zhao, J. et al., 2008 [19]; Weakley SM et al. 2011 [80] |
| Tsix | Antisense transcript to Xist , prevents Xist stabilization and inhibits the interaction between Rep A and PRC2, silencing Xist expression | Zhao,J. et al., 2008 [19]; Navarro, P. et al., 2005 [22] |
| Zeb2NAT | Antisense to Zeb2, regulates splicing of the IRES-containing intron of Zeb 2, involved in EMT | Beltran, M. et al., 2008 [60] |
| MALAT-1/NEAT2 | Expressed in many cancers, regulates alternative splicing of pre-mRNA and promotes cell motility through transcriptional and post-transcriptional regulation of motility related gene expression | Tripathi, V., 2010 [66] |
| MEG3 | Imprinted transcripts, highly expressed in human pituitary, stimulates p53-mediated transactivation and suppresses tumor growth in the absence of p53 | Gejman, R. et al., 2008 [108]; Benetatos, L. et al., 2011 [110]; Zhou, Y. et al., 2007 [111] |
| GAS5 | Growth arrest-specific transcripts, controls apoptosis and cell cycle, down-regulated in breast cancers | Mourtada-Maarabouni, M. et al., 2008 [114]; Mour-tada-Maarabouni, M. et al., 2009 [115] |
| PCGEM1 | Prostate tissue-specific and prostate-associated, overexpressed in prostate cancers, regulates cell proliferation and apoptosis, promotes colony formation | Srikantan, V. et al., 2000 [76] |
| UCA1 | Urothelial carcinoma-associated transcript, upregulated in bladder carcinoma and embryo, influences cell growth and promotes invasion | Wang, F. et al., 2008 [142]; Wang, XS et al., 2006 [143] |
| SRA-1 | Alternative splicing of SRA-1, loss of coding frame, an increased expression is associated with tumor metastasis | Yao, H et al., 2010 [144] |
| CUDR | Upregulated in drug-resistant human squamous carcinoma, regulates drug sensitivity, cellular transformation and apoptosis | Tsang, WP et al., 2007 [145] |
| VL30-1 | A mouse noncoding retroelement RNA, binds and releases PSF from a proto-oncogene, thus activating Rab23 proto-oncogene transcription | Li, L et al., 2009 [146]; Wang, G. et al., 2009 [147] |
| Myc | Antisense transcript to myc gene, may be targeting the sense transcripts for immediate degradation | Yu, W. et al., 2008 [107] |
| p21NAT | Antisense to cdkn1a/p21, requires Ago1 for epigenetic silencing of Cdkn1a/p21 | Yu, W. et al., 2008 [107] |
| CCND1/Cyclin D1 | Transcribed from 5’ end of Cyclin D1 gene, induced by DNA damage and binding to TLS protein, leading to allosteric changes and repression of Cyclin D1 and anti-sense transcripts of tie-1 related to vascular malformation | Wang, X. et al., 2008 [52] |
| PTENP1 | Transcript of PTEN tumor suppressor pseudogene, PTENP1 3’-UTR exerts a tumor suppressive function by acting as a decoy for PTEN-targeting miRNAs | Poliseno, L et al, 2010 [67]; He, L. 2010 [68] |
| KRAS1P | Transcript of KRAS pseudogene, overexpression of KRAS1P 3’-UTR, increases KRAS mRNA abundance and accelerates cell growth | Poliseno, L et al, 2010 [67] |
| DHFR | Transcribed from upstream of DHFR gene, regulates DHFR expression by forming triple helix with the promoter and disassociating pre-initiation complex | Martianov, I. et al., 2007 [53] |
| TERRA | Telomeric UUAGG repeat-containing RNA, inhibits telomerase activity, also regulates Xist and HOTAIR | Azzalin, CM. et al., 2007 [42]; Schoeftner, S. and Blasco, MA., 2008 [44]; Luke, B. and Lingner, J., 2009 [120] |
| AK023948 | Antisense transcribed from the intron of SIR-like adaptor gene (SLA), significantly downregulated in most of papillary thyroid carcinoma | He, H et al., 2009 [148] |
| TUG1 | Ubiquitously expressed in human and mouse cell types and tissues, involves eye development, upregulated by p53, repressed cell proliferation via bound to PRC2 | Zhou, Y. et al., 2007 [111]; Huarte, M. et al., 2010 [113] |
| LincRNA ROR | Expressed in the induced pluripotent stem cells (iPSCs), involved in the conversion of lineage-committed cells by interacting with reprogramming complexes | Loewer, S. et al., 2010 [149] |
| HAR1 | REST target gene, decreased in the neurons of Huntington's disease | Tsai, MC. et al, 2010 [15] |
| Tie-1AS | Expressed temporally and spatially in vivo with its native gene tie-1, binds tie-1 mRNA, regulating tie-1 transcripts; imbalance of sense | Li, K. et al., 2010 [61] |
| BACE1AS | Antisense transcript for beta-secretase-1, directly implicated in the increased abundance of Abeta 1-42 in Alzheimer's disease | Faghihi, MA. et al., 2008 [62]; Jiang, Y. et al., 2010 [63] |
Unlike mRNA and structural ncRNAs, most LncRNAs are localized in the nucleus. Some of LncRNAs are found in both cytoplasm and nucleus. However, some LncRNAs are specifically distributed in cytoplasm [12,13]. Below, we will discuss various LncRNAs with respect to their roles in epigenetic regulation, transcriptional and posttranscriptional regulation as well as tumorigenesis.
Diverse regulatory functions of long non-coding RNAs (LncRNAs)
LncRNAs were initially thought to be spurious transcriptional noise due to low RNA polymerase fidelity. In recent years, it has become evident that LncRNAs are pervasively transcribed throughout eukaryotic genomes. The expression of many LncRNAs occurs in the developmental stage and is cell type-specific. Interestingly, a large number of LncRNAs are specifically expressed during embryonic stem cell differentiation, pathogenesis or tumorigenesis. Up to date, the functions of only a limited number of LncRNAs have been well characterized such as Xist/RepA, KCNQ1OT1, AIR, HOTAIR, Evf-2, H19, MALAT1, and some natural antisense LncRNAs (NATs). However, there is still a large number of LncRNAs for which their biological significance is not yet identified. Transcription of LncRNAs is now known to regulate the expression of protein-coding genes in close genomic proximity (in cis or cis-acting) and to target distant transcriptional complexes such as activator or repressors (in trans or trans-acting) via a variety of mechanisms. In this review we will highlight the role of LncRNAs in gene expression regulation at the level of chromatin modification and transcriptional and posttranscriptional modification. Also in this review, we will focus on the association of the identified LncRNAs with diseases, especially cancers.
Epigenetic regulation
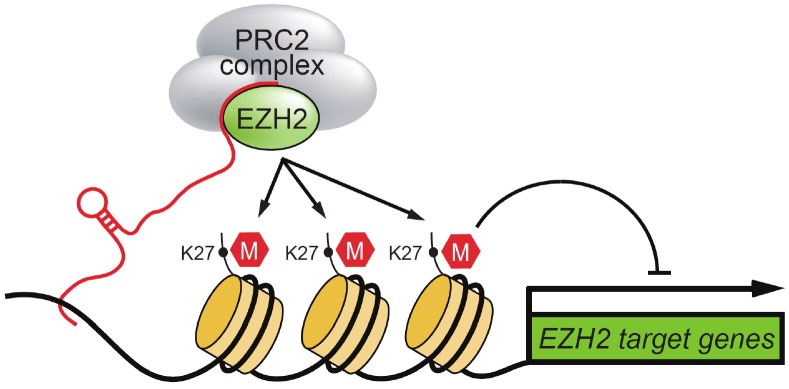
It has been demonstrated that LncRNAs can mediate epigenetic modification by recruiting chromatin remodeling complex to specific chromatin loci. A growing body of LncRNAs has been found to associate with chromatin-remodeling complexes and regulate gene expression. Several studies have shown that more than 20% of 3,300 human LncRNAs are bound by Polycomb Repressive Complexes (PRCs) or other types of chromatin remodeling complexes, e.g. SRA by SRC1, ANRIL by PRC1, Xist/RepA by PRC1 and PRC2 complexes, HOTAIR by PRC2 complex and/or LSD1, Air and Kcnq1ot1 by G9a, and CCND1/Cyclin D1 by translocated-in-liposarcoma (TLS)/histone acetyltransferase (HAT) complex (Figures 2 and 3) [14-20]. Accumulating data suggest that nuclear LncRNA molecules selectively interact with the components of chromatin-remodeling complexes directly or indirectly such as EZH2, SUZ12, CBX7, CoREST, and JARID1C/SMCX. Here, we will discuss how some of these LncRNAs mediate gene silencing or activation through their interaction with chromatin-remodeling complexes (Figure 2).
Figure 2.
LncRNA in chromatin-remodeling. LcnRNA such as Xist/RepA, Air, Hotair, Tsix, ANRIL and Kcnq1ot1 can recruit Polycomb Repressive Complex (PRC) via direct interaction with EZH2 or other components to the targeted locus where they promote trimethylation of H3K27, leading to silencing of the specific genes.
Xist/RepA
X-inactive specific transcript (Xist) is one of the first identified and best studied LncRNAs. It is a spliced and polyadenylated LncRNA with a size of 17 kb in mouse and 19 kb in human and only expressed in an inactive but not in the active X-chromosome of the female placental mammals [17,18]. Recent studies found that besides Xist, other LncRNAs including Tsix and Jpx are also involved in the X-chromosome inactivation (XCI) with Xist being the master regulator of the XCI [17-19,21-23]. In addition, a recent study identified a new 1.6 kb A-rich ncRNA transcript, termed repeat A (RepA), which is transcribed from Xist gene driven by an internal promoter and is comprised of the overlapping sequences of the 5′-end of Xist transcript [19]. RepA initially recruits PRC2 to the X-chromosome through interaction with EZH2 (Figure 2). Furthermore, depletion of RepA abolished full-length Xist induction and H3K27 trimethylation of X chromosome, and knock-down of PRC2 subunit Eed and EZH2 compromised Xist upregulation [19]. In addition, Kaneko et al. also showed that like HOTAIR, RepA binds to the N-terminus containing Thr 345 of EZH2 protein, which is defined as the ncRNA binding domain for the 5′-end of RepA/Xist and HOTAIR [21]. The promoter of Xist is repressed by a 40 kb antisense Tsix LncRNA, which also binds to PRC2, and thus inhibits the interaction between RepA and PRC by competing with RepA recruited to pre-XCI [21]. Tsix expression is turned on in both X chromosomes prior to initiation of XCI. At the onset of XCI, Tsix expression turns into monoallelic and is only associated with the future active X-chromosome until Xist expression is off. Tsix does not exist on the inactive X-chromosome once cells undergo the X-inactivation process [19,21]. Taken together, both RepA and PRC2 are required for the initiation and spread of XCI. The mechanism of how Xist is regulated remains unclear but likely involves negative and positive regulators. For the active X-chromosome, Tsix RNA functions as the established Xist repressor. Whereas for the inactive X-chromosome, another XCI-encoded LncRNA, Jpx, identified by Tian et al., functions as Xist activator [23]. Depletion of Jpx by either knockout or knockdown approach blocks Xist upregualtion, suggesting Jpx functions in trans and in cis [23]. In brief, the expression and function of Xist is controlled by two major ncRNA-based switches: Tsix for active X and Jpx for inactive X.
Air and HOTAIR
Antisense to Igf2r RNA (Air) is a 108 kb, polyadenylated, non-coding RNA that transcribed from an antisense promoter located in intron 2 of the Igf2r (insulin-like growth factor type 2 receptor) in the mouse chromosome 17 [20,24]. Igf2r gene cluster contains 3 imprinted genes: Igf2r, Slc22a2, and Slc22a3. Unlike Igf2r, Slc22a2, and Slc22a3 maternal transcription, the Air ncRNA is only expressed from the paternal allele [25]. Expression of the Air ncRNA results in a "cloud" nuclear pattern over the imprinted DNA locus during embryonic development in the placenta and the adult heart. The expression of unspliced Air is known to be unstable and exclusively localized to the nucleus while spliced isoforms are relatively stable and found in both nucleus and cytoplasm [25-27]. To date, the most characterized function of Air is regulation of genomic imprinting of the Igf2r gene cluster. This epigenetic process has recently been found to involve histone methylation to achieve monoallelic gene expression without altering the genetic sequence. Using the RNA TRAP (tagging and recovery of associated proteins) technique, Air was found to envelope the Slc22a3 loci in correlation with H3K9 histone methyltransferase (G9a) and transcriptional repression [24]. Genetic ablation of G9a further leads to nonimprinted and biallelic expression of Slc22a3, suggesting that Air recruits G9a to the Slc22a3 promoter, remodels the chromatin structure, and silences monoallelic expression [20,25,27]. Although the expression and function of Air LncRNA have been identified in the last decade, the underlying mechanism through which the chromatin remodel complex mediates a direct contact with the Air LncRNA remains unclear.
HOTAIR (HOX Antisense Intergenic RNA) is located at the boundary of two diametrical chromatin domains in the HOXC locus. HOTAIR is transcribed antisense to the canonical HOXC genes with a size of 2158-nts. This transcript is spliced and polyadenylated. HOTAIR distally regulates the chromosomal domain in trans on HOXD locus [28]. Recent study revealed that the 5′ domain of HOTAIR physically interacts with PRC2 methylase and increases its activity, which facilitates histone H3 lysine-27 trimethylation on the HOXD locus and results in silencing of the HOXD gene. RNAi-mediated depletion of HOTAIR dramatically induces the transcriptional activation in HOXD locus with the increased transcripts of HOXD8, HOXD9, HOXD10, and HOXD11 [28]. The 3′ domain of HOTAIR has recently been found to bind with demethylase LSD1 and is required for its occupancy on chromatin and normal function [15]. Therefore, HOTAIR provides a platform for at least two distinct histone modification complexes: a 5′-domain that binds PRC2 and a 3′-domain that interacts with the LSD1/CoREST/REST complex. Tethering two distinct chromatin-remodeling complexes by the HOTAIR scaffold enables RNA-mediated assembly of PRC2 and LSD1, and therefore, coordinates recruitments of both PRC2 and LSD1 to the chromatin for coupled histone H3K27 methylation and H3K4 demethylation. These results suggest that LncRNAs may function as scaffolds by providing binding surfaces to assemble selected histone modification enzymes, thereby specifying the patterns of histone modifications on target genes (Figure 2) [15].
ANRIL
ANRIL is an antisense non-coding RNA transcribed from the INK4 locus. Full-length of ANRIL is polyadenylated and spans 126.3 kb in genome. Within the INK4 locus is a p15/CDKN2B-p16/CDKN2A-p14/ARF gene cluster. The ANRIL gene is deleted along with CDKN2B and CDKN2A in the melanoma-neural system tumor syndrome family [31]. ANRIL is tightly associated with high-risk genetic markers of coronary artery disease (CAD) and is upregulated in prostate cancer [30]. Deletion of the posterior part of ANRIL in the mouse harboring CAD high-risk genetic markers gives rise to substantial suppression of both tumor suppressor genes CDKN2A and CDKN2B [30,31]. However, the mechanistic insight of the regulation and function of ANRIL is not yet fully understood. Experimental data also revealed that ANRIL is able to bind to chromobox 7 (CBX7) within the PRC1 complexes, leading to repression of the INK4b/ARF/INK4a gene for controlling cell senescence [29]. More recently, Kotake et al. demonstrated that expression of oncogenic Ras, which stimulates the expression of p15INK4B and p16INK4A, inhibits the expression of ANRIL [32]. The authors showed that the p15INK4B locus is bound by SUZ12, a component of PRC2, and is H3K27-trimethylated. Knock-down of ANRIL by shRNA disrupts binding of SUZ12 to the p15INK4B locus and increases the expression of p15INK4B but not p16INK4A or p14ARF, and thus inhibits cellular proliferation. In brief, these results present a working model in which ANRIL binds to and recruits PRC1 or PRC2 to repress the expression of tumor suppressor p15INK4B locus, facilitating oncogenesis.
KCNQ1OT1
Kcnq1ot1 is a 91 kb imprinting LncRNA [33] that is transcribed in the antisense direction to Kcnq1 gene from the paternal mouse chromosome 7 [34,35] or human chromosome 11. Kcnq1ot1 is an unspliced and polyadenylated transcript and localized exclusively in the nuclear compartment. Several groups [33,36-39] have demonstrated that Kcnq1ot1 RNA plays important roles in bidirectional silencing in controlling the expression in cis of neighboring genes, and such paternally gene repression of the Kcnq1-imprinted domain regulated by Kcnq1to1 is linked to the multilayered silencing pathways. In general, the Kcnq1ot1 RNA harbors a highly conserved domain at its 5′-end, which mediates transcriptional silencing through interaction with G9a/PRC2 complexes in a lineage and stage-specific manner (Figure 2). Moreover, studies further showed that EZH2 is required for Kcnq1ot1 association along the length of the Kcnq1 cluster, suggesting that EZH2 may mediate spreading of Kcnq1ot1 and/or play a role in higher-order genomic organization [40,41].
TERRA
Human telomeric DNA is transcribed into LncRNA, termed TERRA. TERRA is a heterogeneous ncRNA ranging in size from ~100 bases up to at least 9 kb that contains UUAGGG repeats and is complementary to the template sequence of telomerase RNA. Its 5′-end contains a 7-methylguanosine (m7G) cap structure [42-44]. TERRA is exclusively found in nuclear RNA fractions. TERRA can interact with several telomere-associated proteins such as telomere repeat factors 1 (TRF1) and 2 (TRF2), subunits of the Origin Recognition Complex (ORC), heterochromatin protein 1 (HP1), histone H3 trimethyl K9 (H3 K9me3), and some factors in the DNA damage sensing pathways [43-47]. It was shown recently that TERRA localization may be regulated developmentally, and in turn, might be important for orchestrating some aspects of the complex chromosome transactions that occur during cellular differentiation [44,45]. Several lines of evidence suggest that TERRA may act as a negative regulator of telomerase and thus establish telomere length homeostasis. Indeed, experimental evidence demonstrated that in addition to exerting an uncompetitive mode of inhibition, TERRA acts as a telomerase ligand and a natural potent competitive inhibitor for human telomerase [46]. TERRA contacts the telomerase reverse transcriptase (TERT) protein subunit independently of human telomerase RNA (hTR) [46]. Lieberman and other groups have shown that TERRA RNA can interact directly with the TRF2 amino-terminal basic glycine/arginine-rich (GAR) domain and ORC1 to form a stable ternary complex, facilitating TRF2 interaction with ORC, which has been implicated in transcriptional silencing, heterochromatin formation, centrosome function, and sister chromatid cohesion [43,47]. Taken together, TERRA plays a central role in telomere structural maintenance and heterochromatin formation.
Transcriptional regulation
Some ncRNAs were found to directly regulate transcription of their target genes. With advances in DNA sequencing technology, more and more widespread promoter- and enhancer-associated transcripts have been identified from yeast to human transcriptomes.
Promoter-associated ncRNAs (paRNAs)
paRNAs function mostly through RNAi-mediated pathways of gene repression; however, paRNA with a length of 50-200 nts, has been shown to be involved in epigenetic repression of Polycomb (PcG) target genes via an alternate mechanism. The mechanism of such paRNA gene regulation involves transcription of these RNA transcripts from proximal promoter regions that overlap with the genes targeted for regulation by these uncoding RNAs. Gene silencing occurs due to association of the paRNA with and the recruitment of the PRC2 component, SUZ12, to the target gene promoter regions, enhancing H3K27me3 at the target promoters, and thus repressing gene transcription in cis [48]. Another case of paRNA controlling transcriptional regulation involves an intergenic region between members of the Dlx/dll homeodomain-containing protein family, Dlx-5/6 [49]. Feng et al. [49] identifed a 3.8 Kb alternatively spliced form of Evf-1 from an ultra-conserved region in Dlx-5/6 and demonstrated that this paRNA Evf-2 is transcribed under Sonic hedgehog (Shh) treatment. Evf-2 associates with Dlx-2 to increase transcriptional activity of the Dlx-5/6 enhancer in a target- and homeodomain-specific manner in cis (Figure 3A). In these cases, transcription at the gene promoters, rather than the RNA product of transcription, mediates regulation of the overlapping genes in cis or distant enhancer/promoter regions in trans. The absence of these paRNAs from PcG target genes that are activated during differentiation in embryonic stem cells (ES) suggests the ncRNAs may maintain cells in an undifferentiated status. This principle may be present in other stem cell-related diseases involving PRC2 [50].
Figure 3.

LncRNA in transcriptional regulation. A. Transcriptional activation. LncRNA transcribed from the ultraconserved region of the gene Evf2 functions as coactivator of a homeodomain protein Dlx2, facilitating the Dlx6 gene transcription. B. Transcriptional suppression. Top, LncRNA CCND1 transcribed from the upstream region of the Cyclin D1 gene recruits the RNA-binding protein, termed translocated-in-liposarcoma (TLS), allosterically to modulate HAT activities of CREB-binding protein (CBP) and p300, resulting in inhibition of the gene expression. Bottom, LncRNA transcribed from the dihydrofolate reductase (DHFR) minor promoter can form triplex at its major promoter to prevent the binding of general transcription factors such as TFIID, and subsequently, silence the expression of DHFR.
Similarly, paRNA Evf-2 also plays a role in development and is essential for the development of interneurons that produce GABA, the major inhibitory neurotransmitter in the brain. The absence or reduction of GABA has been implicated in neurological disorders such as schizophrenia, Tourette's syndrome, epilepsy, and Rett syndrome, an autism spectrum disorder [49]. Taken together, a new mechanism of transcriptional regulation of gene discovered by both Kanhere et al. [48] and Feng et al. [49] in which the paRNA up-regulates enhancer activity of target genes through recruitment of transcriptional factor DLX-2 to form stable complex at the binding sites. Understanding the potential mechanisms that regulate the transcription and processing of these transcripts may provide therapeutic strategies by targeting these paRNA populations in different respective disease states [51].
In contrast, some paRNAs suppress transcription. For example, Wang et al. [52] demonstrated that DNA damage signals induce transcription of heterogeneous LncRNA CCND1/Cyclin D1 from upstream of the CCND1 promoter, and this low copy of LncRNA transcript is associated with the Cyclin D1 gene promoter and RNA-binding protein termed TLS in which they allosterically modulate the activity of RNA-binding protein, TLS. The modified TLS subsequently inhibits the HAT activities of CREB-binding protein (CBP) and p300, leading to silencing of Cyclin D1 expression (Figure 3B, top panel). In addition, some paRNAs can regulate RNAP II activity by competing with transcription factors. For example, in humans, the dihydrofolate reductase (DHFR) gene contains a minor and major promoter. The major promoter activity is suppressed in quiescent cells. A LncRNA DHFR transcribed from upstream of the minor promoter binds to both the major promoter of DHFR and the general transcription factor TFIIB, preventing the formation of pre-initiation complex at the major promoter site (Figure 3B, bottom panel). DHFR transcript and double-stranded DHFR promoter form a stable purine-purine-pyrimidine triple structure [53]. Triplex structure may be a common LncRNA mechanism for inhibition of targeted promoter activity.
Enhancer-like LncRNAs (eRNAs)
More recently, Ørom et al. [54,55] used a GENCODE annotation of the human genome to characterize over a thousand LncRNAs with sizes that ranged from 100 to 9100 nts that are expressed in multiple cell lines. Unexpectedly, in contrast to the well-characterized LncRNA such as XIST, they found that some LncRNAs have functional properties of enhancers in human cell lines. Depletion of this type of LncRNAs (eRNAs) led to potent decreased expression of their neighboring protein-coding genes, including the master transcription factors TAL1/SCL, Snail (Snai1) and Slug (Snai2). For example, depletion of ncRNA-a3 significantly decreased the expression of its flanking gene Tal1/SCL, which is a key regulator in hematopoiesis. In addition, knock-down of ncRNA-a7 reduced the expression of its neighboring gene Snai1, which plays a crucial role in cell adhesion, migration, and epithelial-messenchymal transition [54]. These findings demonstrate that such enhancer-like eRNAs display a transcriptional activator function for their neighboring genes, suggesting that an unanticipated role for this class of LncRNAs in activation of critical regulators involved in development and differentiation [54,55]. Similarly, using the active chromatin marker H3K4 monomethylation, Kim et al. [56] found that enhancer domains can be transcribed by RNAPII bi-directionally to a novel class of eRNAs. Moreover, the eRNA expression level at neuronal enhancers positively correlates with the level of mRNA synthesis at nearby genes. Their solid data demonstrate that eRNA synthesis occurs specifically at the enhancers that are actively involved in promoting mRNA synthesis [56]. The reported evidence indicates that LncRNAs that associates with enhancer regions are important regulators for modulating their neighboring gene expression [54-56].
Post-transcriptional regulation
In addition to the above mechanisms, LncRNAs are also involved in posttranslational processing of mRNAs, including splicing, editing, trafficking, translation, and degradation. They are summarized below:
Naturally occurring antisense transcripts (NATs)
Of the most particular types of LncRNAs, NATs play a very crucial role in regulating mRNA dynamics. It is estimated in human and mouse that 61-72% of all transcribed regions posses LncRNAs in antisense orientation (NATs) [8,57-59]. Unlike classical NAT LncRNAs to the imprinting genes such as Tsix, Air, HOTAIR, and Evf-2, which guide the epigenetic silencing complexes to the targeted loci, some NATs can form RNA duplexes to mask key cis-regulatory elements in the mRNA of overlapping gene, leading to alternative splicing pattern of the paired gene. For example, the Zeb2/Sip1 NAT is complementary to the 5′ splice site of an intron in the 5′-UTR of the zinc finger Hox mRNA Zeb2, which is involved in epithelial-mesenchymal transition (EMT). Expression of the Zeb2 NAT upon EMT can mask the splice site, preventing splicesome from functioning. As a result, the translation machinery can then recognize and bind to an internal ribosome entry site (IRES) in the retained intron, resulting in more efficient Zeb2 translation (Figure 4A) [60]. More recently, Li et al. [61] identified a NAT for tyrosine kinase containing immunoglobin and epidermal growth factor homology domain-1 (Tie-1AS) in zebrafish, mouse, and human. The tie-1 NAT selectively binds tie-1 mRNA in vivo to form RNA-RNA duplex, resulting in downregulation of the Tie-1 protein and thus specific defects in endothelial cell contact junctions in vivo and in vitro. In contrast, the expression of antisense transcript (BACE-1AS) of a crucial enzyme in Alzeimers’s disease pathophysiology, Beta-secretase-1 (BACE-1), increases BACE-1 mRNA stability and generates more Abeta (amyloid-beta) 1-42 through a post-transcriptional feed-forward mechanism [62,63].
Figure 4.
LncRNA in posttranscriptional regulation. A. Increase in translational efficiency. Transcripts of the antisense LncRNA, in this case, NAT Zeb2 can mask the 5′ splice site of an intron in the 5′-UTR of the zinc finger Hox mRNA Zeb2 by complementary interaction, preventing spliceosome binding to generate an inhibitory sequences for ribosome scanning. This results in binding of the translational machinery to an internal ribosome entry site (IRES), leading to more efficient translation of Zeb2. B. Stabilization of mRNA. Transcripts of pseudogenes and its counterpart gene (in this example, PTENP1 and PTEN) are highly conserved at 3’-UTR, such transcripts act as competing endogenous RNA (ceRNA) and thereby the PTEN mRNA is protected from common miRNA (A, B) targeting for degradation by binding with PTENP1 transcripts competitively, as a result, increasing the PTEN mRNA abundance and protein level.
LncRNAs affecting pre-mRNA splicing
Higher eukaryotes are frequently observed to use a well-known mechanism termed alternative splicing of pre-mRNA to diversify their transcriptomes and to increase their proteomic complexities. The serine/arginine (SR) splicing factors have been demonstrated to be engaged in regulating tissue- or cell-type-specific alternative splicing in a concentration- and phosphorylation-dependent manner [64-66]. However, the underlying mechanisms that modulate the cellular levels of active SR proteins remain unclear. Recent studies present evidence showing that long nuclear-retained regulatory RNA (nrRNA), referred to as metastasis-associated lung carcinoma transcript 1 (MALAT1), plays a crucial role in alternative splicing regulation [64,66]. MALAT1, also known as NEAT2 (noncoding nuclear-enriched abundant transcript 2), is about 7 kb long and localized in nuclear speckles and interacts with SR splicing factor, SRSF1, which affects the distribution of these and other splicing factors in nuclear speckle domains. Depletion of MALAT1 with antisense oligonucleotides or transient overexpression of SRSF1 changes the alternative splicing of the endogenous pre-mRNAs. Importantly, MALAT1 controls cellular phosphorylation status of SR proteins, thereby regulating cellular ratio of phosphorylated vs. dephosphorylated form of SR proteins [66], suggesting that MALAT1 regulates pre-mRNA processing via modulating the levels of active SR proteins.
Pseudogene transcripts
Recently, over 10,000 pseudogene mRNAs have been identified, accounting for 10% of the FANTOM3 transcriptome [10]. The biological importance of pseudogene mRNAs, however, is less understood. There is a growing body of evidence to demonstrate that pseudogene transcripts play an important role in regulating mRNA stability of its coding paralogue. Interestingly, recent studies revealed that pseudogene transcripts of tumor suppressor gene PTEN (PTENP1) and oncogenic KRAS (KRASP) are biologically active as they can specifically regulate cellular levels of their counterpart genes, PTEN and KRAS [67,68]. Emerging evidence demonstrated the functional importance of PTEN dosage during tumor development. Among the key regulators of PTEN dosage are a number of noncoding RNAs, including miRNAs and pseudogene transcripts, which regulate PTEN abundance at the posttranscriptional level. Recent investigations unveiled that PTEN and PTENP1 3′-UTR are highly conserved, and PTEN mRNA is protected from common miRNA binding at the microRNA response elements in 3'-UTR region by PTENP1 RNA, which is also called competing endogenous RNA (ceRNA). The competitive binding of ceRNAs to the common miRNAs results in the increase of PTEN mRNA abundance and protein level (Figure 4B) [67-72]. Similarly, KRAS mRNA level is also increased by expression of ceRNAs of KRASP [67-69]. Besides pseudogene transcripts, it should be noted that emerging evidence supports the notion that some protein-coding mRNAs also function as ceRNAs. By a combined computational and experimental approach researchers have identified and validated more endogenous ceRNAs [69-72]. Interestingly, Pandolfi’s group expanded ceRNAs to include some protein-coding mRNA transcripts such as ZEB2, VAPA, CNOT6L. Like pseudogene transcripts, these types of endogenouse ceRNAs also modulate PTEN protein levels in a microRNA-dependent, protein coding-independent manner [69-70,72].
Trafficking regulator
LncRNAs have been shown to modulate the activity of proteins by regulating their subcellular localization exemplified by the transcription factor NFAT (nuclear factor of activated T cells). NFAT transcript is localized in the cytoplasm until calcium-dependent signals cause it to be imported into the nucleus, where it activates transcription of target genes. One of the key regulators of NFAT trafficking is NRON (noncoding repressor of NFAT). The NRON gene contains three exons, and it can be alternatively spliced to produce variant transcripts ranging in size from 0.8 to 3.7 kb. By directly binding to importin-beta 1, one of the components of the nucleocytoplasmic trafficking machinery, NRON specifically inhibits the nuclear accumulation of NFAT but not that of other transcription factors such as p53, AP1, the fork-head FOXO1 and NFκB that also translocate from the cytoplasm to nucleus [73].
LncRNAs and cancer
The relevance of LncRNAs in normal physiological processes and pathogenesis, especially in cancer is increasingly recognized. In recent years, different new approaches such as genome-wide gene expression screen, genomewide association studies, region-targeted association assay and conventional linkage screen, designed LncRNA array, RIP-RNA sequencing as well as transgenic expression and gene knockdown/knockout have all been successfully used to identify the functions of LncRNAs in cancer. Accumulating data shows that many identified LncRNAs are crucial players in a variety of tissue carcinogenesis, invasion, and metastasis [74,75]. Based on their functions, LncRNAs can be roughly classified into oncogenic and tumor-suppressor groups (Table 2).
Oncogenic LncRNAs
It is now known that similar to protein-coding oncogenes, some LncRNAs can also regulate cellular pathways that lead to oncogenesis or metastasis. These types of LncRNAs are referred to as oncogenic transcripts. The first identified oncogenic or pro-oncogenic LncRNAs are PCGEM1 and PINC, which are highly expressed in prostate and breast cancer [76-79]. Recently, more and more putative onco-LncRNAs have been discovered such as KRASP, HULC, HOTAIR, MALAT1/NEAT1, p15AS, ANRIL, H19, SRA1, p21NAT, and RICTOR (Table 2). It is worth noting that some LncRNAs may have oncogenic and/or tumor suppressive effect, depending on the cellular context. For example, XIST transcript is upregulated in some male cancers but downregulated in female cancers, respectively [80]. Here, we will discuss some examples of such LncRNAs.
MALAT-1 LncRNA
Using subtractive hybridization approach, one LncRNA, referred to as cancer metastasis-associated lung adenocarcinoma transcript, MALAT-1, was originally identified in non-small-cell lung cancer [81]. MALAT-1 is an abundant LncRNA of ~8.7 kb and plays a pivotal role in cell proliferation, migration, and invasion. As mentioned above in this review, MALAT-1 predominantly localizes to nucleus, especially in nuclear speckles in a transcription dependent manner to regulate post-transcriptional processing events such as alternative splicing of mRNAs [64-65]. This LncRNA was significantly upregulated in many cancers such as in the lung, breast, prostate, liver, and colon [81-86]. Investigations have provided evidence that much higher expression of MALAT-1 was found in the metastatic tumors than in non-metastatic tumors, and its expression level in early stage of non-small cell lung cancer is closely correlated with poor prognosis [81]. Lin et al. showed that in all of nodules of procarcinogen-induced murine hepatocellular carcinomas (HCCs) and human HCCs, MALAT-1 expression was markedly elevated compared with the normal liver tissues [83]. In addition, the study also found a significant increase in MALAT1 expression in breast, pancreatic, colon and lung cancers compared with the surrounding normal tissues [83]. Therefore, MALAT1 could be a prognostic marker for metastasis and survival of human cancers [81,83,85]. Recent study also demonstrated that MALAT-1 is involved in regulating cell mobility as knockdown of MALAT-1 impaired the in vitro migration of the lung cancer cells [86]. Furthermore, the researchers also measured the premRNA and mature mRNA levels of some MALAT1 target genes involved in migration. Their results suggested that MALAT1 regulates its target gene expression at both transcriptional and post-transcriptional level [86]. However, the underlying mechanism of MALAT-1 contributing to tumor metastatic process remains unclear.
Tumor marker LncRNA
Recent genome-wide association study (GWAS) unveiled strong and reproducible associations of multiple genetic variants in a large “gene-desert” region of chromosome 8q24 with susceptibility to prostate cancer (PC). Using fine mapping and re-sequencing approaches combined with single-nucleotide polymorphism (SNP) analysis, a 13 kb intron-less LncRNA, termed PRNCR1 (prostate cancer non-coding RNA1) was identified [87]. PRNCR1 expression was upregulated in some of the PC cells as well as precursor lesion prostatic intraepithelial neoplasia and considered as one of tumor markers. Knock-down of PRNCR1 by siRNA attenuated the viability of PC cells and the transactivation activity of androgen receptor, which indicates that PRNCR1 could be involved in prostate carcinogenesis possibly through androgen receptor activity. These findings could provide a new insight in understanding the pathogenesis of genetic factors for PC susceptibility and prostate carcinogenesis [87].
The experiments performed by Silva JM et al. [88] revealed that many of the Long Stress-Induced Non-coding Transcripts (LSINCTs) were overexpressed in a number of lung and some breast cancer cell lines compared with normal human bronchial epithelial cells or normal breast epithelial cell line and immortalized MCF10 cells. LSINCT transcripts are intergenic and intragenic LncRNAs, ranging from 2 kb to 4 kb in length. Interestingly, the study indicates that some LSINCTs are overexpressed in the HER2- and TP53+ breast cell lines [88]. By RNA Amplification of cDNA Ends (RACE) and Northern blots, the authors validated these LSINCTs overexpressed in a panel of breast cancer cells. The larger transcript of LSINCT5 is about 2.5 kb in length. LSINCT5 is polyadenylated and transcribed in trans. Increased expression of LSINCT5 has been observed in a panel of breast cancer cells. Moreover, several other cancers including cervix and ovary cancers also have increased LSINCT5 expression [88].
Intergenic LncRNA (LincRNA)
LincRNA HOTAIR is expressed mainly in posterior and distal sites and highly conserved during vertebrate evolution [28], and the latest study showed that HOTAIR is also involved in the metastasis of breast cancer [16]. HOTAIR is overexpressed in metastatic breast cancers and predicts poor prognosis. Dysregulation of HOTAIR represses the expression of a set of cell-cell interaction promoting genes including JAM2, PCDH10, PCDHB5, and EPHA1 and conversely enhances the expression of many metastasis-facilitating genes, including ABL2, SNAIL, LAMB3, and LAMC2. Moreover, the function of HOTAIR is counteracted by knock-down of PRC2 [16]. Therefore, the interaction of HOTAIR and PRC2, leading to increased H3K27 trimethylation and silencing of metastasis suppressor genes, is responsible for the HOTAIR-mediated tumor cell invasion and subsequent metastasis. In addition, recent study showed that an increased expression level of HOTAIR transcript in hepatocellular carcinoma (HCC) patients correlates with a significantly shorter recurrence-free survival, suggesting that HOTAIR transcripts in HCC could be a candidate biomarker for predicting tumor recurrence in HCC patients who underwent liver transplant therapy [89]. Moreover, in vitro assays demonstrated that knock-down of HOTAIR in liver cancer cell line reduced cell viability and invasion, and sensitized TNF-α-induced apoptosis, resulting in increase of chemotherapeutic sensitivities of the cancer cells to cisplatin and doxorubicin [89]. These findings indicate that lincRNAs have active roles in modulating the cancer epigenome and may be an important predictor for cancer outcome and novel targets for caner therapy.
Imprinted LncRNAs
Abnormal genomic imprinting, in particular, loss of imprinting (LOI) is involved in a number of human hereditary diseases and cancers [90]. Studies have demonstrated that a disruption in the expression of imprinted genes such as H19, p57kip2, IGF2, and KvLQT1 in chromosome band 11p15.5 results in approximately 80% of Beckwith-Wiedemann syndrome (BWS). About 5-10% of BWS patients are predisposed to a variety of childhood tumors, including hepatoblastoma, rhabdomyosarcoma, neuroblastoma, adrenal carcinoma, and Wilms tumor [91-95]. Kcnq1ot1 (also called LIT1) is an imprinted antisense LncRNA within the human KvLQT1 locus, which is about 60 kb long and has a silencing domain at its 5′-end [40]. Kcnq1ot1 transcript is associated with multiple balanced chromosomal rearrangements in BWS and additional breakpoint in embryonal rhabdoid tumors [96]. Kcnq1ot1 is expressed normally from the paternal allele, from which the KvLQT1 transcription is silent. Abnormal expression of Kcnq1ot1 in both the paternal and maternal allele, e.g. LOI, was found in 50% BWS patients and 53% colorectal cancer [96-98]. LOI of Kcnq1ot1 transcript is closely accompanied by loss of methylation (LOM) of maternal allele of the differentially methylated regions (DMR) located in intron 10 of KvLQT1 gene (KvDMR1). Moreover, LOM of the control element CpG island, namely KvDMR1, strongly correlates with loss of H3K9 dimethylation in both of BWS and cancer [96-100]. KvDMR contains the promoter for the paternally expressed Kcnq1ot1. Disruption of the promoter abolishes Kcnq1ot1 transcripts, leading to activation of neighbor silencing imprinted genes, such as CDKN1C, a tumor repressor [36,37,99]. It has been found that silencing of the imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at KvDMR in esophageal cancers [100]. Taken together, these data suggest that abnormal expression of Kcnq1ot1 contributes to carcinogenesis.
Antisense LncRNA
Recently, many studies described consistent and significant differences in the distribution of sense and antisense transcripts between normal and neoplastic tissues. Many of the differentially expressed antisense transcripts likely represent LncRNAs [57-60]. A subset of genes that mainly generate NATs in normal but not cancer cells is involved in essential metabolic processes, and a large body of evidence shows that altered ratio of sense and antisense transcripts contributes to tumorigenesis and cancer progression [101-106]. For instance, Yu et al. found that leukemia cells had larger amounts of p15 NAT (p15AS) and smaller amounts of its partner p15 mRNA than normal lymphocytes [107]. The length of the p15AS transcript is around 3.5 kb. Furthermore, they demonstrated that ectopic expression of p15AS induces p15 silencing in cis and in trans through heterochromatin formation but not DNA methylation [107]. It is noteworthy to mention that this group also found that many NAT LncRNAs to the cancer-relevant genes exist such as those encoding p21, p53, E-cadherin, myc, Tie-1, p27KIP1, APC, RB1, NF1, PTEN, CDKN1A, CDKN2A, CDKN2B, BRCA1, BRCA2, VHL, TP63, TP73, ARF, WT1, and MYC [107]. Thus, tumorigenic NATs may be a trigger for heterochromatin formation and DNA methylation in tumor suppressor silencing in tumorigenesis. Interestingly, experimental evidence also demonstrated that NATs are involved in epithelial-mesenchymal transition (EMT) process exemplified by Zeb2 NAT [60]. Beltran et al. showed that the expression of a transcriptional repressor of E-cadherin Zeb2 (also called Sip1, Smad-interacting protein 1) is upregulated after Snail1-induced EMT [60]. Snail1 does not change Zeb2 mRNA level, but alters the processing of a large intron located within its 5′-UTR (Figure 4A). Ectopic expression of Zeb2 NAT in epithelial cells prevents splicing of the Zeb2 5′-UTR and increases translation efficiency of Zeb2, resulting in downregulation of E-cadherin. Therefore, Zeb2 NAT plays a crucial role in regulating E-cadherin expression during EMT [60].
Tumor suppressor LncRNAs
Like protein-coding tumor suppressors, some LncRNAs are found to function as tumor suppressors, including MEG3, GAS5, LincRNA-p21, PTENP1, TERRA, CCND1/Cyclin D1, and TUG1.
p53-related LncRNAs
The LncRNA MEG3 is a transcript of a maternally imprinted RNA gene. All normal human pituitary cell types express MEG3, but the loss of MEG3 expression occurs in pituitary adenomas of a gonadotroph origin [108]. In addition, loss of MEG3 expression was also found in the majority of human meningiomas or the human meningioma cell lines [109]. MEG3 was found to be a positive regulator of p53, a tumor suppressor protein. Ectopic expression of MEG3 significantly increases p53 protein level and dramatically stimulates p53-dependent transcription from p53-responsive promoter [110,111]. Coactivation of p53 is MEG3-transcription-dependent and requires intact secondary structure of the LncRNA. Furthermore, MEG3 selectively enhances p53 binding to its target promoter such as GDF15 but not p21. MEG3 is also able to inhibit cell proliferation in the absence of p53 [108-112]. These data suggest that MEG3 function as a tumor suppressor in both p53-dependent and p53-independent manner.
Another example of tumor suppressor LncRNA is LincRNA-p21, whose expression is directly induced by the p53 signaling pathway. LincRNA-p21 is required for global repression of genes that interfere with p53 function to regulate cellular apoptosis. LincRNA-p21-mediated gene repression occurs through physical interaction with RNA-binding protein hnRNP-K, leading to its localization to the promoters of genes to be repressed in a p53-dependent manner [113].
Receptor-binding LncRNA
Growth arrest-specific 5 or GAS5 is another example of tumor-suppressor LncRNA which plays an essential role in normal growth arrest in lymphocytes. Overexpression of GAS5 causes both an increase in apoptosis and a reduction in the rate of progression through the cell cycle while downregulation of endogenous GAS5 inhibits apoptosis and maintains a more rapid cell cycle, indicating that GAS5 expression is both necessary and sufficient for normal growth arrest in lymphocytes [114]. GAS5 LncRNA regulates the expression of a critical group of genes with tumor suppressive consequences [115]. GAS5 encodes two canonical splice variants. In addition, multiple snoRNAs are solely transcribed from the introns of GAS5. Under starvation conditions, GAS5 is induced and directly interacts with the DNA binding domain of glucocorticoid receptor (GR) through an RNA-sequence mimic of the GR response element (GRE) DNA, leading to inhibition of GR binding at its target gene promoters to induce their transcription. Repression of the transcriptional activity by GRE-mimic binding of GAS5 is not unique to GR, and other members of nuclear receptors are also affected [116]. In breast cancer, GAS5 was found to be significantly downregulated [115].
Promoter-associated LncRNA
In human cell lines, CCND/Cyclin D1, a cis-acting heterogeneous LncRNA that is about 200 bp and >330 bp transcripts. These LncRNAs are originated from the promoter region of the CCND1/Cyclin D1 gene. CCND1/Cyclin D1 transcripts have been shown to bind TLS (also termed FU, Fused in Ewing's Sarcoma). The binding of ncRNA to TLS allosterically activates the repressor and tethers the LncRNA to the CCND1 promoter to inhibit CCND1 expression (Figure 3B) [52]. Cyclin D1 encoded by CCND1 gene is frequently overexpressed in human tumors, and nuclear Cyclin D1 is oncogenic driver in human cancers [117]. Therefore, CCND1/ Cyclin D1 transcript functions as tumor suppressor to repress tumorigenesis.
Pseudogene LncRNA
Some pseudogene transcripts of tumor suppressors possess tumor suppressive effect [67-68]. An exceptional example is PTENP1, a coding-independent pseudogene of the PTEN mRNA. PTENP1 is biologically active and functions as ceRNA, which can positively regulate PTEN protein level, exerting a growth-suppressive role via competitively binding of interfering miRNAs to the 3′-UTR of PTEN for degradation (Figure 4B) [67,69-72]. These findings may lead to development of a novel approach for cancer therapy.
Telomere-related LncRNAs
TERRA expression is highly dependent on developmental status, including nuclear reprogramming, telomere length, cellular stresses, tumor stage, and chromatin structure [43-47]. Mis-regulated TERRA is responsible for many of the abnormal telomere phenotypes seen in aging and cancer cells [118-120]. Interestingly, TERRAs are significantly downregulated in advanced stages of different types of human cancers compared with normal tissues [44]. Consistently, low levels of TERRA have been observed in the tumor-derived and in vitro-immortalized cell lines [121]. These findings suggest that regulation of TERRA level and TERRA-regulated telomere length play important roles in tumor development that involve telomerase misregulation.
Opportunities and challenges
The rapid advances in biotechnology combined with bioinformatics accelerate the progression of genome-wide studies of LncRNA. It has been known to date that a high percentage of the genomic DNA of mammalian cells is transcribed into RNAs, of which most are non-protein coding transcripts. In particular, around 50% of protein-coding RNAs have their paired long antisense ncRNAs [57-59]. These LncRNAs function as key regulators implicated in numerous functions in regulating gene expression pattern, modulating protein activity, altering the RNA processing, serving as structural components and precursors for small RNAs, and influencing mRNA stability. Up to date, only a small portion of LncRNAs has been identified. Like miRNAs and pRNAs, LncRNAs have emerged as important regulatory molecules in developmental, tumor-suppressor and oncogenic pathways and other diseases [69-71,122-128]. Some of the LncRNAs have become diagnostic markers and potential therapeutic targets [123-125]. Currently, the study of LncRNAs is becoming one of the most popular fields in the biological and medical sciences.
However, an existing challenge is to understand how the molecular functions of LncRNAs affect the phenotypes of the organisms. Limited by technological advances in the identification of the biological function of LncRNAs, siRNA knock-down is currently the only effective approach, and ectopic expression of LncRNA in trans does not always recapitulate the function in cis. Moreover, some of the LncRNAs are very large in size and can sometimes exceed 8 kb in length with many secondary structures, which present challenges for cloning and ectopic expression. Future work will focus on whether expression of LncRNA itself is enough to produce observable functional effects.
Characterization of interacting domains such as the stem-loop structure of LncRNAs is also imperative for deciphering their biological functions that will provide effective basis for further validation by genetic approaches and for novel drug development. Generation of knockout and/or transgenic mouse model can determine those LncRNAs, which are not transcriptional noise or junk but are required for normal development and pathogenesis. Recently, two LncRNA mouse models have been reported [126-132]. When maintained in a semi-natural environment, knockout mice of BC1, a rodent-specific LncRNA, display global phenotypes such as reduced exploration, increased anxiety, and mortality compared with wild-type [126-128]. The neural phenotype is consistent with the functional absence of a BCL-interacting protein, FMRP, which is the product of the fragile X mental retardation gene. Another special transgenic mouse model of human spinocerebellar ataxia type 8 (SCA8) demonstrated that antisense RNA of the Kelch-like 1 gene (KLHL1) is associated with a dominantly inherited, slowly progressive neurodegeneration disorder SCA8 [129]. Patients with SCA8 show a trinucleotide (CUG) expansion in a noncoding RNA termed ataxin 8 opposite strand (ATXN8OS), an antisense transcript to the KLHL1 gene [130]. Transgenic mice containing this pathogenic CTG expansion in their DNA, like human patient, exhibit a progressive neurological phenotype in a dose-dependent manner [129]. Tri- or tetranucleotide repeat expansions within ATXN8OS are proposed to form hairpin structures that drive splicing regulators away from their normal pre-mRNA targets [129,131,132].
From these studies, it is now clear that LncRNAs are linked to many diseases such as cancer and neuropathy [74,75,127,128]. The implication of LncRNAs in tumorigenesis, metastasis, and progression remain to be further investigated. Unlike protein-coding RNAs, mutations and polymorphisms in LncRNAs are not frequently reported and genome-wide mutagenesis and association studies have yet completed to define the causative changes in LncRNA sequences. So far, only a few studies pinpoint to high-risk alleles within LncRNA genes such as prostate cancers [123]. Expression profiling of LncRNAs during brain development, iPS cell generation, ES cells and lymphocyte differentiation as well as breast cancers have only been recently documented [12-14,113,132-139]. Therefore, characterization of oncogenic and tumor suppressor LncRNAs is an attractive field, which will lead to new markers of cancer diagnosis and identification of novel therapeutic targets.
Acknowledgments
We are grateful to Dr. Stephanie Sellers for her assistance in the preparation of this review manuscript.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 4.Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, Bult CJ, Fletcher CF, Forrest AR, Furuno M, Hill D, Itoh M, Kanamori-Katayama M, Katayama S, Katoh M, Kawashima T, Quackenbush J, Ravasi T, Ring BZ, Shibata K, Sugiura K, Takenaka Y, Teasdale RD, Wells CA, Zhu Y, Kai C, Kawai J, Hume DA, Carninci P, Hayashizaki Y. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muro EM, Andrade-Navarro MA. Pseudogenes as an alternative source of natural antisense transcripts. BMC Evol Biol. 2010;10:338. doi: 10.1186/1471-2148-10-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frith MC, Wilming LG, Forrest A, Kawaji H, Tan SL, Wahlestedt C, Bajic VB, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Huminiecki L. Pseudo-messenger RNA: phantoms of the transcriptome. PLoS Genet. 2006;2:e23. doi: 10.1371/journal.pgen.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;92:18–25. doi: 10.1016/j.ygeno.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 18.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X -specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yotova IY, Vlatkovic IM, Pauler FM, Warczok KE, Ambros PF, Oshimura M, Theussl HC, Gessler M, Wagner EF, Barlow DP. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics. 2008;92:464–473. doi: 10.1016/j.ygeno.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 25.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 26.Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 27.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 28.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 31.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey RR, Ceribelli M, Singh PB, Ericsson J, Mantovani R, Kanduri C. NF-Y regulates the antisense promoter, bidirectional silencing, and differential epigenetic marks of the Kcnq1 imprinting control region. J Biol Chem. 2004;279:52685–52693. doi: 10.1074/jbc.M408084200. [DOI] [PubMed] [Google Scholar]
- 36.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanduri C, Thakur N, Pandey RR. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. Embo J. 2006;25:2096–2106. doi: 10.1038/sj.emboj.7601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis A, Green K, Dawson C, Redrup L, Huynh KD, Lee JT, Hemberger M, Reik W. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- 39.Kanduri C. Functional insights into long antisense noncoding RNA Kcnq1ot1 mediated bidirectional silencing. RNA Biol. 2008;5:208–211. doi: 10.4161/rna.7113. [DOI] [PubMed] [Google Scholar]
- 40.Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 42.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 43.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 45.Schoeftner S, Blasco MA. Chromatin regulation and non-coding RNAs at mammalian telomeres. Semin Cell Dev Biol. 2010;21:186–193. doi: 10.1016/j.semcdb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Silanes IL, d'Alcontres MS, Blasco MA. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat Commun. 2010;1:33. doi: 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- 48.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Carninci P. Molecular biology: The long and short of RNAs. Nature. 2009;457:974–975. doi: 10.1038/457974b. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 54.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 59.Galante PA, Vidal DO, de Souza JE, Camargo AA, de Souza SJ. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, Horswill MA, Stanhope SA, North PE, Miao RQ, Wilkinson GA, Affolter M, Ramchandran R. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang Y, Mullaney KA, Peterhoff CM, Che S, Schmidt SD, Boyer-Boiteau A, Ginsberg SD, Cataldo AM, Mathews PM, Nixon RA. Alzheimer's-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci USA. 2010;107:1630–1635. doi: 10.1073/pnas.0908953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He L. Posttranscriptional regulation of PTEN dosage by noncoding RNAs. Sci Signal. 2010;3:pe39. doi: 10.1126/scisignal.3146pe39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, Krauthammer M, Halaban R, Provero P, Adams DJ, Tuveson DA, Pandolfi PP. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 74.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino K, Buzard GS, Mostofi FK, McLeod DG, Moul JW, Srivastava S. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW, Srivastava S. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 78.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate- specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 79.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci USA. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 82.Guffanti A, Iacono M, Pelucchi P, Kim N, Soldà G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, Callari M, Mignone F, Pesole G, Bertalot G, Bernardi LR, Albertini A, Lee C, Mattick JS, Zucchi I, De Bellis G. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 84.Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, Ranganathan S, Michalopoulos GK. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmidt LH, Spieker T, Koschmieder S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 86.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4780. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 87.Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono T, Tsunoda T, Daigo Y, Matsuda K, Kamatani N, Nakamura Y, Kubo M. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 88.Silva JM, Perez DS, Pritchett JR, Halling ML, Tang H, Smith DI. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95:355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 89.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of Long non-coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 90.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 91.Ping AJ, Reeve AE, Law DJ, Young MR, Boehnke M, Feinberg AP. Genetic linkage of Beckwith-Wiedemann syndrome to 11p15. Am J Hum Genet. 1989;44:720–723. [PMC free article] [PubMed] [Google Scholar]
- 92.Koufos A, Grundy P, Morgan K, Aleck KA, Hadro T, Lampkin BC, Kalbakji A, Cavenee WK. Familial Wiedemann-Beckwith syndrome and a second Wilms tumor locus both map to 11p15.5. Am J Hum Genet. 1989;44:711–719. [PMC free article] [PubMed] [Google Scholar]
- 93.Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C, Steele L, Cameron J, Smith A, Ambus I, Li M, Ray PN, Sadowski P, Squire J. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet. 2001;10:2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 94.Maher ER, Reik W. Beckwith-Wiedemann syndrome: imprinting in clusters revisited. J Clin Invest. 2000;105:247–252. doi: 10.1172/JCI9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gaston V, Le Bouc Y, Soupre V, Burglen L, Donadieu J, Oro H, Audry G, Vazquez MP, Gicquel C. Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2001;9:409–418. doi: 10.1038/sj.ejhg.5200649. [DOI] [PubMed] [Google Scholar]
- 96.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 97.Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, Kugoh H, Mukai T, Ikeguchi M, Oshimura M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006;97:1147–1154. doi: 10.1111/j.1349-7006.2006.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Horike S, Mitsuya K, Meguro M, Kotobuki N, Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y, Oshimura M. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum Mol Genet. 2000;9:2075–2083. doi: 10.1093/hmg/9.14.2075. [DOI] [PubMed] [Google Scholar]
- 100.Soejima H, Nakagawachi T, Zhao W, Higashimoto K, Urano T, Matsukura S, Kitajima Y, Takeuchi M, Nakayama M, Oshimura M, Miyazaki K, Joh K, Mukai T. Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene. 2004;23:4380–4388. doi: 10.1038/sj.onc.1207576. [DOI] [PubMed] [Google Scholar]
- 101.Maruyama R, Shipitsin M, Choudhury S, Wu Z, Protopopov A, Yao J, Lo PK, Bessarabova M, Ishkin A, Nikolsky Y, Liu XS, Sukumar S, Polyak K. Breast Cancer Special Feature: Altered antisense-to-sense transcript ratios in breast cancer. Proc Natl Acad Sci USA. 2012;109:2820–2824. doi: 10.1073/pnas.1010559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grigoriadis A, Oliver GR, Tanney A, Kendrick H, Smalley MJ, Jat P, Neville AM. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics. 2009;10:324. doi: 10.1186/1471-2164-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Numata K, Osada Y, Okada Y, Saito R, Hiraiwa N, Nakaoka H, Yamamoto N, Watanabe K, Okubo K, Kohama C, Kanai A, Abe K, Kiyosawa H. Identification of novel endogenous antisense transcripts by DNA microarray analysis targeting complementary strand of annotated genes. BMC Genomics. 2009;10:392. doi: 10.1186/1471-2164-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kohno K, Chiba M, Murata S, Pak S, Nagai K, Yamamoto M, Yanagisawa K, Kobayashi A, Yasue H, Ohkohchi N. Identification of natural antisense transcripts involved in human colorectal cancer development. Int J Oncol. 2010;37:1425–1432. doi: 10.3892/ijo_00000794. [DOI] [PubMed] [Google Scholar]
- 105.Monti L, Cinquetti R, Guffanti A, Nicassio F, Cremona M, Lavorgna G, Bianchi F, Vignati F, Cittaro D, Taramelli R, Acquati F. In silico prediction and experimental validation of natural antisense transcripts in two cancer-associated regions of human chromosome 6. Int J Oncol. 2009;34:1099–1108. doi: 10.3892/ijo_00000237. [DOI] [PubMed] [Google Scholar]
- 106.Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, Yamamoto D, Alvarenga R, da Silva CC, Brito GC, Simon SD, Moreira-Filho CA, Leite KR, Camara-Lopes LH, Campos FS, Gimba E, Vignal GM, El-Dorry H, Sogayar MC, Barcinski MA, da Silva AM, Verjovski-Almeida S. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 107.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2008;93:4119–4125. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 111.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 112.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic non-coding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 115.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a nonprotein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 116.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest-and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 119.Caslini C. Transcriptional regulation of telomeric non-coding RNA: implications on telomere biology, replicative senescence and cancer. RNA Biol. 2010;7:18–22. doi: 10.4161/rna.7.1.10257. [DOI] [PubMed] [Google Scholar]
- 120.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. Embo J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–1659. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 125.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lewejohann L, Skryabin BV, Sachser N, Prehn C, Heiduschka P, Thanos S, Jordan U, Dell'Omo G, Vyssotski AL, Pleskacheva MG, Lipp HP, Tiedge H, Brosius J, Prior H. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res. 2004;154:273–289. doi: 10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 127.Zhong J, Chuang SC, Bianchi R, Zhao W, Paul G, Thakkar P, Liu D, Fenton AA, Wong RK, Tiedge H. Regulatory BC1 RNA and the fragile X mental retardation protein: convergent functionality in brain. PLoS One. 2010;5:e15509. doi: 10.1371/journal.pone.0015509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, Brosius J, Denman RB, Khandjian EW, Kindler S, Tiedge H. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, Ranum LP. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 130.He Y, Zu T, Benzow KA, Orr HT, Clark HB, Koob MD. Targeted deletion of a single Sca8 ataxia locus allele in mice causes abnormal gait, progressive loss of motor coordination, and Purkinje cell dendritic deficits. J Neurosci. 2006;26:9975–9982. doi: 10.1523/JNEUROSCI.2595-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koob MD, Moseley ML, Schut LJ, Benzow KA, Bird TD, Day JW, Ranum LP. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat Genet. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 132.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim DH, Jeon Y, Anguera MC, Lee JT. X-chromosome epigenetic reprogramming in pluripotent stem cells via noncoding genes. Semin Cell Dev Biol. 2011;22:336–342. doi: 10.1016/j.semcdb.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Do JT, Han DW, Gentile L, Sobek-Klocke I, Stehling M, Scholer HR. Enhanced reprogramming of Xist by induced upregulation of Tsix and Dnmt3a. Stem Cells. 2008;26:2821–2831. doi: 10.1634/stemcells.2008-0482. [DOI] [PubMed] [Google Scholar]
- 138.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin Immunol. 2010;22:353–361. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 139.Navarro P, Oldfield A, Legoupi J, Festuccia N, Dubois A, Attia M, Schoorlemmer J, Rougeulle C, Chambers I, Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 140.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kohtz JD, Fishell G. Developmental regulation of EVF-1, a novel non-coding RNA transcribed upstream of the mouse Dlx6 gene. Gene Expr Patterns. 2004;4:407–412. doi: 10.1016/j.modgep.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 142.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 143.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, Zhang Y, Xin DQ, Na YQ, Chen WF. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 144.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. Rna. 2007;13:890–898. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang G, Cui Y, Zhang G, Garen A, Song X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci USA. 2009;106:16794–16798. doi: 10.1073/pnas.0909022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69:625–631. doi: 10.1158/0008-5472.CAN-08-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]