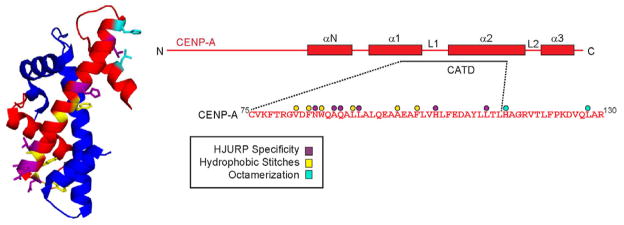
Summary
Centromeres are defined by the presence of chromatin containing the histone H3 variant, CENP-A, whose assembly into nucleosomes requires the chromatin assembly factor HJURP. We find that while surface-exposed residues in the CENP-A targeting domain (CATD) are the primary sequence determinants for HJURP recognition, buried CATD residues that generate rigidity with H4 are also required for efficient incorporation into centromeres. HJURP contact points adjacent to the CATD on the CENP-A surface are not used for binding specificity but rather to transmit stability broadly throughout the histone fold domains of both CENP-A and H4. Further, an intact CENP-A/CENP-A interface is a requirement for stable chromatin incorporation immediately upon HJURP-mediated assembly. These data offer insight into the mechanism by which HJURP discriminates CENP-A from bulk histone complexes and chaperones CENP-A/H4 for a substantial portion of the cell cycle prior to mediating chromatin assembly at the centromere.
Introduction
The centromere is the site of mitotic kinetochore assembly, spindle microtubule attachment, and final metaphase sister chromatid cohesion on each chromosome (Cheeseman and Desai, 2008). Centromeres are specified epigenetically (Allshire and Karpen, 2008), except for in some budding yeasts (Clarke and Carbon, 1980), and CENP-A (Cse4 in budding yeast, CID in flies, CenH3 in plants) is a histone H3 variant that has emerged as the best candidate to carry the centromere-specifying epigenetic mark (Black and Bassett, 2008). The underlying centromeric DNA is highly divergent through eukaryotic evolution. In humans, megabase stretches of highly repetitive α-satellite DNA are typically found at centromeres (Willard, 1990). CENP-A has been shown to direct de novo centromere activity to sites that lack centromeric repeats, by a pulse of overexpression in fruit fly cells (Heun et al., 2006; Olszak et al., 2011), initial targeting of CENP-A to an ectopic locus directly (Mendiburo et al., 2011), or via its dedicated chromatin assembly factor, HJURP (Barnhart et al., 2011), and by tethering recombinant CENP-A-containing nucleosomal arrays to a solid support in frog egg extracts (Guse et al., 2011). These artificial conditions generate CENP-A accumulation in the absence of any repetitive centromere DNA, perhaps recapitulating some aspects of the establishment steps of the naturally-occurring neocentromeres present at low frequency in the human population (Warburton, 2004). Once established, newly arising centromeres are propagated in perpetuity and are thought to recruit constitutive centromere components, as well as inner centromere and kinetochore components recruited at mitosis (Bassett et al., 2010; Amor et al., 2004).
There are diverse proposals under consideration for how CENP-A physically marks centromere location. These proposals include radical models where CENP-A directs the alteration of nucleosomal histone stoichiometry (Dalal et al., 2007; Williams et al., 2009), the recruitment of non-histone proteins to a nucleosome-like particle (Mizuguchi et al., 2007), or the handedness of DNA wrapping (Furuyama and Henikoff, 2009). Other proposals include alterations of nucleosome structure and/or dynamics from within a conventional octameric nucleosome that wraps DNA in a left-handed manner (Black et al., 2004; Black et al., 2007a; Dechassa et al., 2011; Sekulic et al., 2010; Tachiwana et al., 2011; Panchenko et al., 2011). Perhaps the most conservative proposal comes from recent experiments in frog egg extracts in which the unstructured C-terminal four to six a.a. residues are sufficient to impart conventional H3 with the ability to form nucleosomal arrays that recruit a functional kinetochore (Guse et al., 2011).
The faithful delivery of CENP-A to centromeres requires a conserved chromatin assembly factor termed HJURP in animals (Dunleavy et al., 2009; Foltz et al., 2009) and Scm3 in yeast (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007). How histone variants, in general, are sorted into specific chromatin assembly pathways is key to understanding how they may transmit epigenetic information. However, relatively little is known of the molecular recognition between histone variants and their dedicated assembly proteins (De Koning et al., 2007). CENP-A provides a unique challenge to the cell, since it is present at a remarkably low stoichiometry to bulk H3 variants. Thus, the faithful sorting of CENP-A away from bulk H3 is predicted to involve strict recognition by Scm3/HJURP.
Recognition by HJURP was initially proposed to be conferred by a specific region of CENP-A (Foltz et al., 2009), termed the CENP-A targeting domain (CATD) for its ability to confer centromere targeting when substituted into conventional H3 (Black et al., 2004). An H3 chimera containing the CATD (H3CATD) co-purifies HJURP from mammalian cells, binds directly to recombinant HJURP, and requires HJURP expression for its targeting to centromeres (Foltz et al., 2009). The view that the CATD harbors the specificity determinants has been sharply challenged, however, by recent structural analysis of the CENP-A/H4/HJURP ternary complex, where it was not apparent how any of the 22 amino acid changes within the CATD relative to H3 imparts binding specificity by HJURP (Hu et al., 2011). Indeed, in this recent study, it was concluded that the artificial nature of the H3CATD chimera contributed to its access to the HJURP pathway, and that the primary specificity determinant, Ser68, resides outside of the CATD (Hu et al., 2011). This proposal was supported by GST pull-down experiments showing that CENP-A with the S68Q mutation (glutamine is at the corresponding position in H3) fails to bind to HJURP while H3 with the Q68S mutation gains recognition by HJURP (Hu et al., 2011). In addition to this recent structural work, two orthologous fungal complexes (Cse4/H4/Scm3 from S. cerevisiae (Zhou et al., 2011) and K. lactis (Cho and Harrison, 2011)), were solved at high resolution. While the proposed recognition residues on the yeast Cse4 proteins were within the CATD region (Cho and Harrison, 2011; Zhou et al., 2011), they are not conserved with the corresponding residues in mammalian CENP-A proteins. Thus, despite major insight through crystal and NMR structures, there are contradicting data on the means by which mammalian HJURP recognizes CENP-A/H4.
We now use cell-based, biochemical, and biophysical approaches to identify the role of the CATD in binding to HJURP and define the major structural and dynamic requirements for CENP-A incorporation into centromeric chromatin. During the course of these studies we also made the surprising finding that HJURP binding transmits stability to a large portion of the histone fold domains of both CENP-A and H4, strongly suggesting that HJURP function extends beyond the classical definition of a ‘histone chaperone’—shielding its substrate from aggregation with nucleic acids prior to nucleosome assembly—to also include stabilizing the folded state of CENP-A/H4 for a substantial portion of the cell cycle.
Results
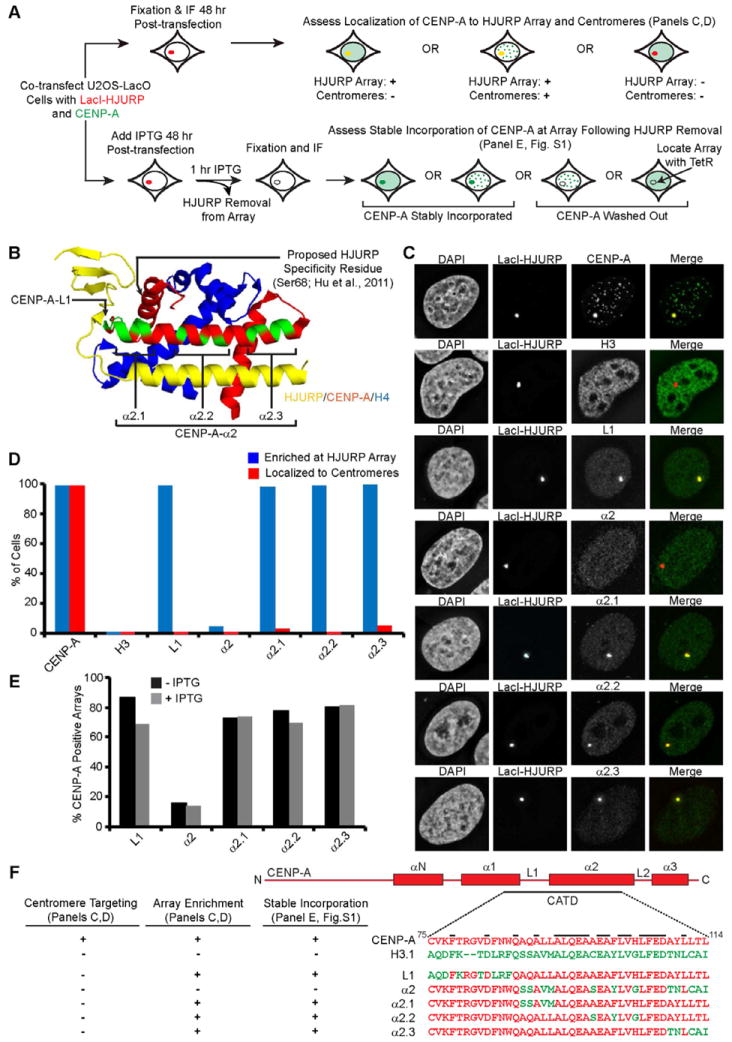
HJURP Recognition of CENP-A is Solely Dependent on Residues in the CATD
Five sets of mutations (called L1, α2, α2.1, α2.2, α2.3) where CATD residues are replaced with the corresponding residues from histone H3 each abolish efficient CENP-A targeting to centromeres (Black et al., 2004; Shelby et al., 1997), but it had not been tested whether or not any of these affect recognition by HJURP (Fig. 1A,B). While both the loop 1 (L1) and α2 helix of CENP-A are in contact with HJURP in a crystal structure (Hu et al., 2011), Xu and colleagues concluded that there are no good candidate side-chain substitutions in this entire region of CENP-A where H3 residues would preclude binding. To directly test this proposal, we employed a cell-based approach that we recently described that monitors HJURP-association at an ectopic site and HJURP-mediated stable assembly of CENP-A into chromatin (Barnhart et al., 2011). This approach utilizes the LacO/I chromosome tethering system, where LacI-fused proteins can be targeted to a chromosomally incorporated LacO array and subsequently removed by the addition of the LacI allosteric effector molecule, IPTG (Belmont, 2001; Janicki et al., 2004). Endogenous (Barnhart et al., 2011) or exogenously expressed CENP-A protein (Fig. 1C,D) is efficiently recruited to the HJURP-containing array. Importantly, the centromere targeting of CENP-A is independently measured within individual cells.
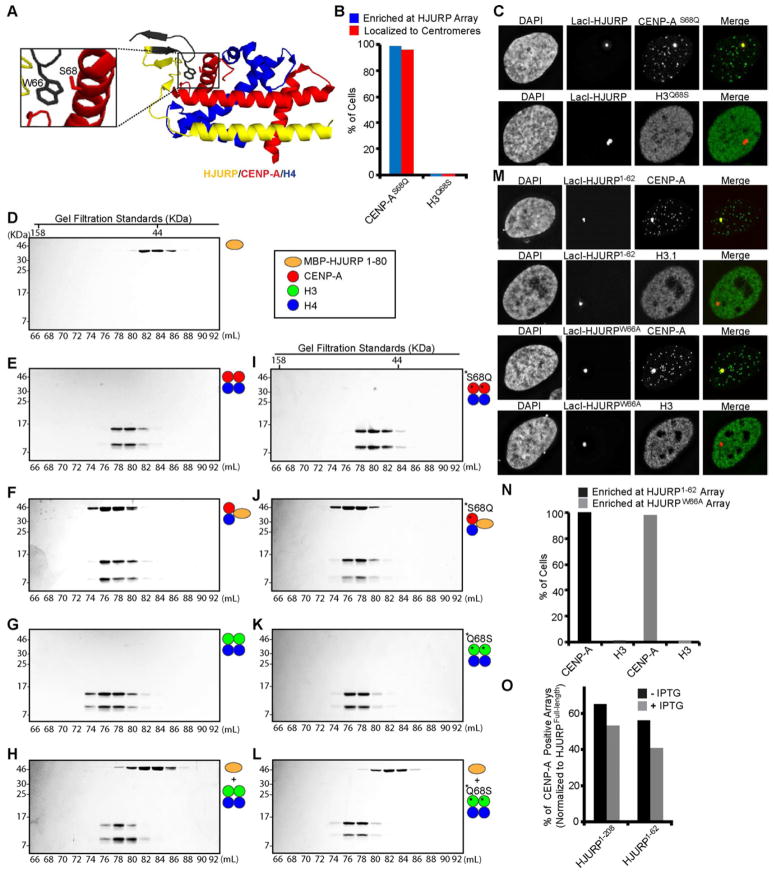
Figure 1. HJURP binding is insufficient for centromere targeting of CENP-A.
(A) Scheme for HJURP chromosome tethering assay in U2OS-LacO-TRE cells. In this system, the mCherry-LacI-HJURP fusion protein is expressed at similar levels to endogenous HJURP (Fig. S1B) (B) Structure of HJURP/CENP-A/H4 complex (PDB 3R45; Hu et al., 2011) highlighting CATD residues swapped for H3 (green) in the various mutant versions. The location of the proposed HJURP specificity determinant (Ser68; Hu et al., 2011) is also shown. (C) Representative images of CENP-A, H3, or mutant versions of CENP-A introduced along with LacI-HJURP into U2OS-LacO-TRE cells. (D) Quantification of the sub-nuclear localization of the indicated histone constructs. In each case, 100 cells were counted and the results are representative of multiple independent experiments. (E) Quantification of stable CENP-A incorporation into the HJURP-containing array. Cells were analyzed 48 h after co-transfection of mCherry-LacI-HJURP, GFP-TetR, and HA-tagged CENP-A mutant proteins and a 1 h treatment with (gray) or without (black) 15 mM IPTG. (F) Sequences of the CENP-A mutants used in these experiments and a summary of our results. Black bars indicate residues shared in both H3 and CENP-A. See also Figure S1.
We found that four (L1, α2.1, α2.2, α2.3) of the five mutants that fail to target to centromeres are robustly recruited to HJURP-containing arrays while the fifth (α2) fails to enrich at the HJURP-containing array or at centromeres (Fig. 1C,D). In addition, all four of the mutants that retain robust HJURP recognition and recruitment to the ectopic chromosomal array are stably incorporated into the ectopic chromosomal locus after LacI-HJURP removal (Figs. 1E,F and S1A), suggesting their failure to target to centromeres is not due to an inability to undergo HJURP-mediated chromatin assembly. Perhaps the simplest explanation of these results is that the CATD provides the primary HJURP recognition determinants, and that a combination of CENP-A-specific residues spanning the entire α2 helix are required for HJURP binding. However, based on the proposal of Xu and colleagues that information outside of the CATD provides the principal HJURP specificity determinant (Hu et al., 2011), our data do not exclude an alternative explanation such as that the chimeric α2 CENP-A mutant fails to recruit to the HJURP-containing array due to some type of structural incompatibility. To address this alternative explanation, we tested in our cell-based approach (Fig. 1A) the CENP-AS68Q and H3Q68S mutations that were reported in GST-pull down experiments to cause the two histone variants to switch allegiance: CENP-AS68Q was reported to eliminate HJURP binding and H3Q68S was reported to confer HJURP binding (Hu et al., 2011). In the crystal structure, there is a hydrophobic pocket formed in a β-sheet portion of HJURP that accommodates the side-chain of Ser68 in CENP-A, but where there is predicted unfavorable packing (particularly due to clashing with Trp66 of HJURP) with Gln68 in the analogous position of H3 (Fig. 2A)(Hu et al., 2011). In contrast to the earlier GST-pull down experiments (Hu et al., 2011), we found that substitutions at this position (S68Q in CENP-A and Q68S in H3) had no measurable effect on centromere targeting nor on recruitment to the HJURP-containing arrays: CENP-AS68Q behaves the same as WT CENP-A and H3Q68S behaves the same as WT H3 (Figs. 2B,C & S2M,O).
Figure 2. CENP-A Ser68 is not a recognition determinant for HJURP binding.
(A) Highlight of the contact point between CENP-A Ser68 and HJURP Trp66 (PDB 3R45; Hu et al., 2011). The region of HJURP deleted in the HJURP1–62 version is labeled in black. (B) Quantification of the sub-nuclear localization of the indicated mutant histone constructs. WT histones were tested in parallel and those data are shown in Fig. S2M,O. (C) Representative images of the indicated mutant histone constructs introduced along with LacI-HJURP into U2OS-LacI-TRE cells. (D–L) SDS-PAGE of the indicated fractions from SEC of the indicated protein mixes. (M) mCherry-LacI-HJURP1–62 or full length mCherry-LacI-HJURPW66A was co-transfected into U2OS-LacO-TRE cells with HA-CENP-A or HA-H3 and cells were analyzed at 48 hours. WT histones were tested in parallel and those data are shown in Fig. S2N,P. (N) Quantification of CENP-A and H3 recruitment to HJURP1–62 and HJURPW66A arrays. (O) Quantification of stable incorporation of endogenous CENP-A into the HJURP-containing array. Cells co-transfected with mCherry-LacI-HJURP1–62 and GFP-TetR were treated 48 h post-transfection with (gray) or without (black) 15 mM IPTG for 1 h, and assessed for recruitment of endogenous CENP-A to the array. The values shown are normalized to the level of CENP-A recruitment to mCherry-LacI-HJURPFull-length. See also Figure S2.
Our results in this cell-based test may indicate that the mutations (CENP-AS68Q and H3Q68S) behave differently in cells than in direct binding reactions as purified components. Alternatively, we considered the possibility that the bead-based pull-down approaches with histone proteins (Hu et al., 2011) were problematic and precluded definitive information on the specificity of this reaction, and that Ser68 is neither necessary nor sufficient for HJURP recognition. Indeed, background binding in pull-downs apparently required the use of differential detergent conditions depending on the histone complexes under investigation (Hu et al., 2011). To avoid such potential experimental vagaries, we monitored the formation of HJURP/CENP-A/H4 complexes using purified components and size exclusion chromatography (SEC; Fig. 2D–L). MBP-HJURP (a.a. 1–80) undergoes a nearly quantitative shift to a larger species upon binding to CENP-A/H4 (Fig. 2D,F). (CENP-A/H4)2 heterotetramers also shift to a larger HJURP/CENP-A/H4 heterotrimer species, but the magnitude of the shift is predictably less pronounced (Fig. 2E,F). In the case of MBP-HJURP and H3/H4, none of the components shift from their original chromatography behavior upon mixing, indicating that there is no detectable binding (Fig. 2G,H). For the mutant versions, CENP-AS68Q/H4 quantitatively forms a trimer with HJURP both at moderate and high salt (Fig. S2I–K [300 mM NaCl]; Fig. 2I,J [1 M NaCl]), and H3Q68S/H4 fails to bind to HJURP (Fig. 2K,L).
In addition to these findings with mutant versions of CENP-A and H3, mutation of HJURP to remove the steric clashing with Gln68 of H3 that was initially proposed to confer specificity (Hu et al., 2011) has no detectable effect on HJURP recognition in our cell-based approach (Figs. 2M,N & S2N,P) or in ternary complex formation monitored by SEC (Fig. S2A–H). These mutants include the removal of the side-chain of HJURP (Trp66) that is proposed to clash with Gln68 of H3 (HJURPW66A; Figs. 2M,N & S2F–H) or removal of two β-strands, including the one containing Trp66 (HJURP1–62; Figs. 2M,N & S2A–E). Remarkably, we found that the small N-terminal portion of HJURP (a.a. 1–62) is nearly as efficient as the much larger domain of HJURP previously identified as sufficient (a.a. 1–208; (Barnhart et al., 2011)) in mediating the stable incorporation of endogenous CENP-A into an ectopic chromosomal locus (Figs. 2O & S2L). Together, our findings in cells and using purified components show that Ser68 of CENP-A is neither necessary nor sufficient for HJURP recognition and subsequent deposition into chromatin.
HJURP Binding at the α1 Helix of CENP-A Generates Stability to Most of the Histone Fold Helices of CENP-A and H4
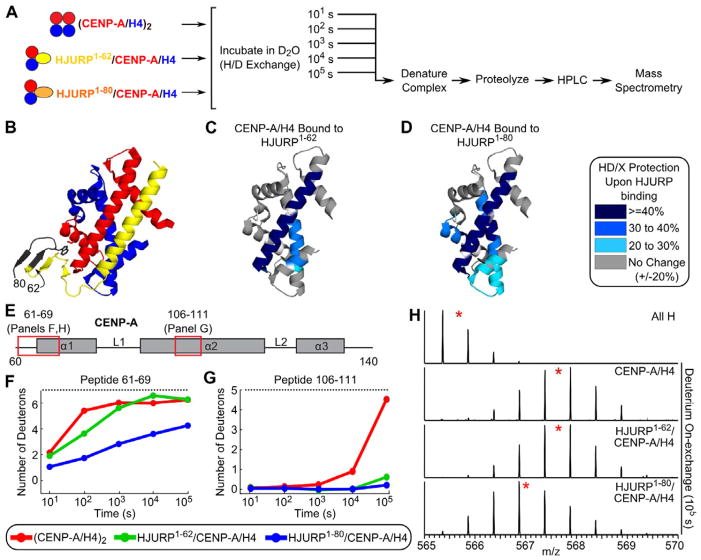
An intact hydrophobic pocket, wherein Trp66 is the key HJURP residue, is clearly dispensable for CENP-A recognition (Fig. 2), but its association with the α1 helix of CENP-A is nonetheless a striking structural feature of the ternary complex (Hu et al., 2011). To test the impact of this interaction on prenucleosomal CENP-A/H4, it is necessary to compare HJURP/CENP-A/H4 trimers harboring a version of HJURP with the entire interaction interface intact (i.e. HJURP1–80) to a version of this complex where the hydrophobic pocket, including Trp66, is removed (i.e. HJURP1–62) (Fig. 3A). Hydrogen-deuterium exchange (H/DX) is a powerful solution-based approach to access information about protein structure, dynamics, and folding (Englander, 2006). We and others have coupled H/DX to mass spectrometry (MS) in order to access dynamic information on macromolecular interactions, providing key insight to complement known static structures (Hansen et al., 2011; Mendillo et al., 2009; Lee et al., 2004; Panchenko et al., 2011). In order to determine how HJURP binding affects CENP-A/H4 dynamics, we measured levels of CENP-A/H4 protection in complex with HJURP relative to independent measurements on (CENP-A/H4)2 heterotetramers using the conditions optimized for CENP-A/H4 proteolysis and peptide recovery at the experimental steps following H/DX (Black et al., 2004). HJURP1–62 (Fig. 3B) protects CENP-A/H4 locally at the region of contact, i.e. the L1 and the α2 helix of CENP-A as well as portions of the α2 and α3 helices of H4 (Figs. 3C,E–H and S3A–D). This protection is clear even at the latest time point (105 s; Fig. 3C,G; see a representative example of this in the peptide highlighted in Fig. 3G indicating long-term stability of the interaction between HJURP1–62 and CENP-A/H4). Our analysis further indicates that HJURP1–62 does not interact with the α1 helix of CENP-A, since there is no detectable difference in H/DX rates in this version of the trimer relative to the (CENP-A/H4)2 heterotetramer.
Figure 3. HJURP interactions with the α1 helix of CENP-A stabilize the histone fold domains of both CENP-A and H4.
(A) Experimental scheme for determining H/DX of protein complexes at various time points. (B) Structure of HJURP/CENP-A/H4 complex (PDB 3R45; Hu et al., 2011). The region of HJURP absent from the HJURP1–62 version is labeled in black. (C–D) Protection from H/DX upon binding HJURP1–62 (C) or HJURP1–80 (D) is mapped onto the structure of CENP-A/H4 (PDB 3NQJ; Sekulic et al., 2010). These data correspond to the 105 s time point. Labeling is as indicated in the legend, indicating the consensus behavior of all overlapping peptides at each position (white indicates the small number of positions lacking peptide coverage). (E) Diagram of CENP-A secondary structure with red boxes corresponding to locations of example peptides shown in F–G. (F–G) Comparison of H/DX for the indicated CENP-A peptides from each of the indicated complexes. Dotted lines indicate maximum levels of deuteration determined from fully-deuterated samples. (H) Raw mass spectrometry data for the CENP-A peptide shown in (F). Red star indicates peptide centroid value. ‘All H’ is data from the non-deuterated control sample. See also Figure S3.
Strikingly, HJURP1–80 directs major H/DX protection that extends much further through the histone fold domains of both CENP-A and H4 (Fig. 3D,E–H & S3A–D) despite adding new contact points only at the α1 helix of CENP-A relative to HJURP1–62. The increased protection upon binding HJURP1–80 was observed in experiments performed in buffers spanning a large range of ionic strengths (Fig. S3E–I). Within the CENP-A α1 helix, many of the helix residues are strongly protected (Fig. 3F; taking 100–1000 times as long to reach the same level of deuteration as when bound to HJURP1–62), strongly suggesting that protection is a result of restricting transient unfolding of the entire helix that accompanies exchange of amide protons. The stability gained in CENP-A α1 when bound to HJURP1–80 transmits stability through the complex to additional protection observed in portions of all histone fold helices in H4 compared to the HJURP1–62-containing trimer (Figs. 3C,D, & S3A–D). In total, HJURP1–80 binding generates >20% H/DX protection at the 105 s timepoint in 73 residues of the CENP-A and H4 histone fold domains compared to just 42 residues when CENP-A/H4 is bound by HJURP1–62 (comparing data sets with similar [90–95%] extensive peptide coverage). So rather than serving as a molecular recognition platform, the interface gained by the inclusion of a.a. 63–80 of HJURP (in the HJURP1–80-containing complex) greatly restricts the conformational flexibility of CENP-A/H4.
Three Surface-exposed Residues Within the CATD are Sufficient for Recognition by HJURP
Since the CATD carries the information to confer HJURP binding, we next sought to identify the specific CATD residues conferring recognition by HJURP. We started by using the crystal structure of the HJURP/CENP-A/H4 complex (Hu et al., 2011) to predict which of the 22 a.a. specific to the CATD of CENP-A are critical specificity determinants. We focused on six candidate residues on the surface of the CATD, one in L1 (Asn85) and five in the α2 helix (Ala88, Gln89, Leu92, His104, and Leu112)(Fig. 4A,B). Indeed, a version of H3 carrying these six substitutions (construct H3HJURP Surf.) is sufficient for recruitment to the HJURP-containing chromosomal array (Fig. 4C,D). We noted, however, that unlike the H3CATD which is stably incorporated at both the HJURP-containing array (Fig. S4A) and endogenous centromeres, recognition of H3HJURP Surf. by HJURP (either in the presence or absence of mCherry-LacI-HJURP) was not accompanied by delivery to the centromere (Figs. 4C,D & S4B,C). To determine the minimal set of residues that confer HJURP recognition we generated a set of swap mutations of the six CENP-A positions in the H3HJURP Surf. construct. We tested first if the single substitution of His104 into histone H3, replacing a glycine, could confer access to HJURP, since this is perhaps the single most dramatic side-chain addition and highly compatible with the contacting HJURP surface (construct H3G104H)(Fig. 4A). We found, however, that this single alteration is insufficient to confer HJURP recognition (Fig. 4C,D). Furthermore, the N-terminal three substitutions (Asn85, Ala88, and Gln89; construct H3R85N/S88A/S89Q) or C-terminal three substitutions (Leu92, His104, and Leu112; construct H3M92L/G104H/C112L) are not sufficient for HJURP recognition when separated from each other (Fig. 4C,D). These findings indicate that CENP-A-specific contact points within both the N- and C-terminal portions of the CATD are required for recognition by HJURP. Indeed, combining the C-terminal two substitutions (His104 and Leu112) with either one of the two most dramatic substitutions at the N-terminal portion of the CATD (Asn85 or Gln89) is sufficient to target CENP-A to the chromosomal HJURP array (Fig. 4C,D; constructs H3R85N/G104H/C112L and H3S89Q/G104H/C112L).
Figure 4. As few as three residues within the CATD are sufficient for HJURP specificity.
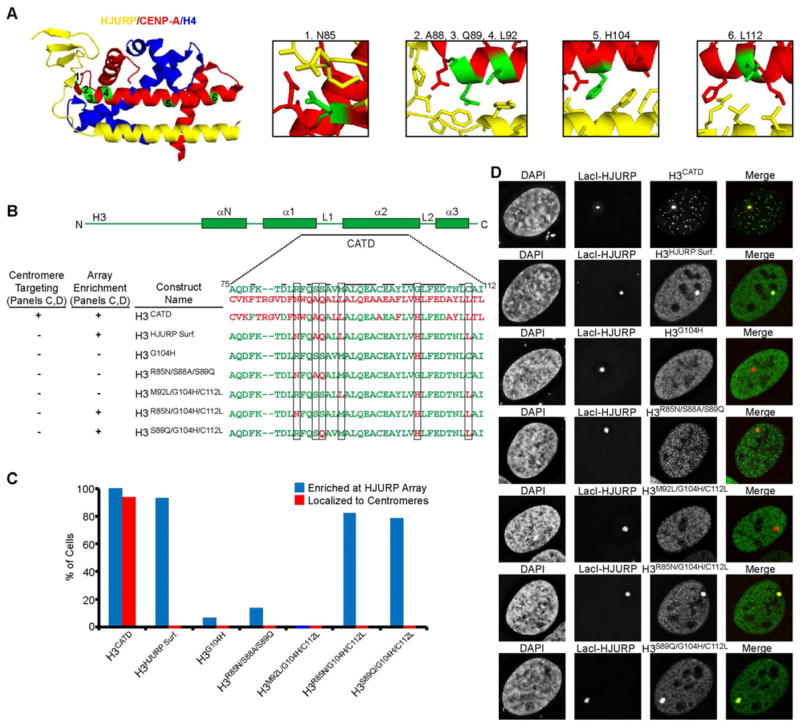
(A) Structure of HJURP/CENP-A/H4 structure (PDB 3R45; Hu et al., 2011) highlighting six candidate residues within the CATD to confer HJURP specificity. (B) Sequences of versions of H3 carrying the indicated CATD residues and summary of our results. Black bars indicate residues shared in both H3 and CENP-A, and boxes highlight the six candidate residues. (C) Quantification of the sub-nuclear localization of the indicated histone constructs. In each case, 100 cells were counted and the results are representative of multiple independent experiments. (D) Representative images of the indicated histone constructs introduced along with mCherry-LacI-HJURP into U2OS-LacO-TRE cells. See also Figure S4.
CENP-A Hydrophobic Stitch Residues are Required for Efficient Incorporation into Centromeres
Our finding that robust HJURP binding is insufficient for centromere targeting of CENP-A or the H3 gain-of-function mutants (Figs. 1 and 4) suggests that other CATD features exist that are vital for generating centromere-specifying nucleosomes. With the first available high-resolution structure of CENP-A (in the context of the (CENP-A/H4)2 heterotetramer) in hand, we initially described three distinguishing features conferred by the CATD (Sekulic et al., 2010). One unique feature is the bulged L1 that generates a surface of opposite charge (Fig. S5A) (Sekulic et al., 2010) that is exposed on the face of the nucleosome (i.e. not occluded by DNA binding; (Tachiwana et al., 2011)). Mutation of the L1 residues to reverse the surface charge, however, does not affect CENP-A centromere targeting (Fig. S5B,C), and engineered reduction of the L1 bulge was reported to reduce stability but not efficient targeting (Tachiwana et al., 2011). Another unique feature is rotation at the CENP-A/CENP-A interface of the heterotetramer, but the residues (His104 and Leu112) implicated in accommodating the rotation (Sekulic et al., 2010) are also key for recognition by HJURP (Fig. 4C,D). In addition, it is not yet clear to what extent the tendency to form a rotated CENP-A/CENP-A interface affects CENP-A-containing nucleosomes since in the available crystal structure of the nucleosome the CENP-A/CENP-A interface is rotated outwards to a conventional orientation (Tachiwana et al., 2011).
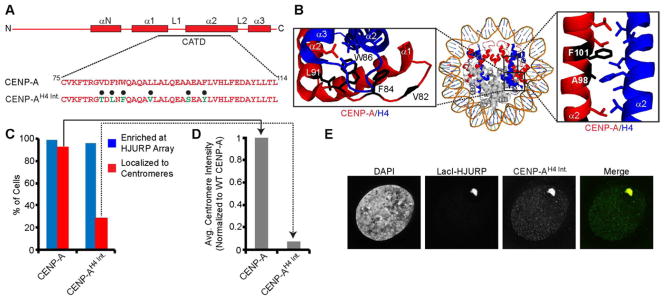
The third unique feature encoded by the CATD is conferred by six residues that contact histone H4 and generate hydrophobic stitches (Sekulic et al., 2010). The hydrophobic stitches reduce 10-fold the conformational flexibility at the CENP-A/H4 interface (Fig. 5A,B)(Black et al., 2004). In addition, the CENP-A-specific side-chain interactions with H4 are nearly identical in the sub-nucleosomal heterotetramer and nucleosome structures (Fig. S5D)(Sekulic et al., 2010; Tachiwana et al., 2011). In order to test the requirement of the hydrophobic stitches in centromeric chromatin assembly, we mutated all six CENP-A hydrophobic residues to their H3 counterparts: V82T, F84L, W86F, L91V, A98S, F101Y. The resulting mutant protein (CENP-AH4 Int.) enriches at the HJURP-containing array in 93% of cells but loses centromere targeting in the majority (71%) of cells (Figs. 5C–E & S5E,F). The remaining 29% of cells have detectable centromere accumulation, although the level of accumulation is reduced ~20-fold compared to WT CENP-A (Fig. 5D). Therefore, we conclude that the hydrophobic stitch residues fulfill a requirement for efficient CENP-A assembly that is downstream of HJURP binding.
Figure 5. CENP-A hydrophobic stitch residues are required for assembly into centromeric chromatin.
(A) Sequence of the CENP-AH4 Int. mutant containing six H3 residues, highlighted with black circles. (B) Structure of the CENP-A-containing nucleosome (PDB 3AN2; Tachiwana et al., 2011) highlighting the six CENP-A a.a. positions substituted with H3 residues in CENP-AH4 Int.. (C) Quantification of the sub-nuclear localization of CENP-AH4 Int.. (D) The average centromere intensity was measured for the population of cells in (C) with indicated histone localizing to the centromere. Intensity is normalized to wild-type CENP-A. (E) Representative images of CENP-AH4 Int. introduced along with LacI-HJURP into U2OS-LacO-TRE cells. See also Figure S5.
An Intact CENP-A/CENP-A Interface is Required for Nucleosome Formation
CENP-A/H4 exists as a heterotetramer in solution with two copies of CENP-A (Black et al., 2004) and there are two copies of CENP-A in the available nucleosome crystal structure (Tachiwana et al., 2011). Binding of HJURP/Scm3 occludes the CENP-A/CENP-A interface (Cho and Harrison, 2011; Hu et al., 2011; Zhou et al., 2011), but HJURP assembles initial products in vitro that are nonetheless thought to include two copies of human CENP-A (or its yeast counterparts; Barnhart et al., 2011; Dechassa et al., 2011; Shuaib et al., 2010; Mizuguchi et al., 2007; Xiao et al., 2011). In any eukaryote, the composition of the centromeric nucleosome in vivo is far less clear, with proposed models including an octameric nucleosome containing two copies each of CENP-A/H4/H2A/H2B (mammals, budding yeast, and insects; Camahort et al., 2007; Sekulic et al., 2010; Shelby et al., 1997; Foltz et al., 2006; Zhang et al., 2011), a tetrasome containing two copies each of CENP-A/H4 (yeast; Williams et al, 2009), nucleosome-like particles lacking H2A/H2B but retaining Scm3 after assembly into DNA (yeast; Mizuguchi et al, 2007), and, most radically, a hemisome containing one copy each of CENP-A/H4/H2A/H2B (insects; Dalal et al., 2007) that wraps DNA with the reverse handedness of conventional nucleosomes (insects and yeast; Furuyama and Henikoff, 2009). In models including two copies of CENP-A, an intact CENP-A/CENP-A interface is key, whereas it would be unused, and therefore dispensable, in a hemisome with a single copy of CENP-A (Dalal et al., 2007; Furuyama and Henikoff, 2009).
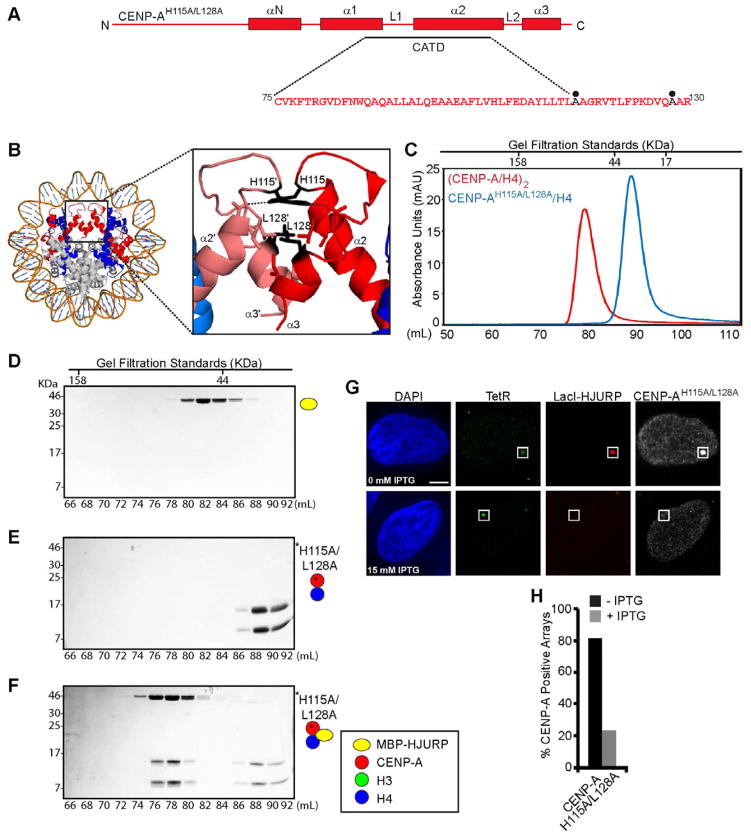
In (CENP-A/H4)2 heterotetramers that spontaneously form upon co-expression in bacteria or after stepwise assembly into nucleosomes, the two CENP-A chains are held together by hydrophobic interactions of several side-chains (including those from Leu111, Leu128, and Ile132) and an intermolecular salt bridge between His115 on one chain and Asp125 on the other (Sekulic et al., 2010; Tachiwana et al., 2011)(Fig. 6A,B). In this way, the CENP-A/CENP-A interface is held together in a nearly identical fashion as the H3/H3 interface in the conventional nucleosome (Luger et al., 1997; Sekulic et al., 2010; Tachiwana et al., 2011). We designed a mutant version of CENP-A (CENP-AH115A/L128A) in which the salt-bridge is broken and the otherwise hydrophobic interface is weakened (Fig. 6A,B). Recombinant CENP-AH115A/L128A/H4 chromatographs as a single peak consistent with a uniform dimeric species (the calculated MW of the heterodimer is 27.5 kDa with WT CENP-A/H4 chromatographing as a uniform heterotetramer of calculated MW=55 kDa; Fig. 6C). Gel filtration of CENP-AH115A/L128A/H4 mixed with MBP-HJURP shows robust formation of a HJURP/CENP-AH115A/L128A/H4 ternary complex (Fig. 6D–F). This finding clearly indicates that HJURP can trimerize with CENP-A/H4 that initially exists as either a (CENP-A/H4)2 heterotetramer (Fig. 2F) or a CENP-A/H4 heterodimer (Fig. 6F). Using our cell-based approach, we find that the CENP-A dimer mutant is clearly enriched at the HJURP array, but is nonetheless severely compromised in targeting to centromeres (Fig. S6A,B). At the HJURP array, CENP-AH115A/L128A fails to stably incorporate into chromatin at the array following IPTG treatment, with its targeting dependent on a persistent LacI-HJURP/LacO interaction (Fig. 6G,H). These data suggest a strong requirement for an intact CENP-A/CENP-A interface in HJURP-mediated chromatin assembly at an ectopic chromosome locus.
Figure 6. An intact CENP-A/CENP-A interface is a requirement for HJURP-mediated nucleosome assembly.
(A) Diagram of the CENP-AH115A/L128A mutant protein. Black circles indicate mutated residues. (B) Structure of the CENP-A nucleosome (PDB 3AN2; Tachiwana et al., 2011), with inset depicting residues H115 and L128 within the CENP-A/CENP-A interface. (C) Gel filtration chromatograph comparing elution profiles of (CENP-A/H4)2 and (CENP-AH115A/L128A/H4). (D–F) SDS-PAGE of the indicated fractions from SEC of the indicated protein mixes. (G) Representative images of cells co-transfected with mCherry-LacI-HJURP, GFP-TetR, and HA-tagged CENP-AH115A/L128A and treated with or without IPTG for 1 h prior to processing for immunofluorescence. (H) Quantification of CENP-AH115A/L128A stable incorporation into the HJURP-containing array with (gray) or without (black) 15 mM IPTG. At least 30 cells were counted over multiple experiments. See also Figure S6.
Discussion
Regarding the proposal that the CATD plays a vital role in centromere propagation, we now provide evidence that it contains the exposed recognition residues for HJURP association. While a surface on the α1 helix of CENP-A, lying outside of the CATD, is part of the HJURP binding interface (Hu et al., 2011), it is not used for molecular recognition but rather as a contact point that confers stability to many of the α-helices of the histone folds of CENP-A and H4 (Fig. 3). Furthermore, our finding here that the interior residues that rigidify the interface between CENP-A and H4 are not involved in HJURP recognition but are required for efficient centromere localization of CENP-A (Fig. 5) strongly suggests a vital role for specialized intranucleosomal dynamics in specifying centromere location. Along with previous functional analysis that implicated the CATD in centromere function (Black et al., 2007b), our findings here strongly support the notion that the CATD provides the key features for distinguishing CENP-A pre-nucleosomal complexes from their conventional counterparts and for rigid intranucleosomal dynamics specifically required for generating centromeric chromatin (Fig. 7).
Figure 7. Summary of CENP-A features required for centromeric nucleosome assembly.
Structure of CENP-A/H4 (PDB 3NQJ; Sekulic et al., 2011) highlighting CENP-A residues required for HJURP specificity (purple), a rigid interface with H4 (yellow), and formation of an intact CENP-A/CENP-A interface required for assembling into octameric nucleosomes (cyan).
Specificity of HJURP Binding Is Achieved Through Cooperative Contact Sites Spread Across the Surface of the CATD
The six CENP-A specific residues within the CATD that we found important for HJURP recognition include one on L1 (Asn85), three on the N-terminal portion of the α2 helix (Ala88, Gln89, and Leu92), and two on the C-terminal portion of the α2 helix (His104 and Leu112)(Fig. 4A). His104 is important for recognition but is insufficient by itself (Fig. 4C,D, construct H3G104H) or in combination with substitutions at the positions corresponding to Leu92 and Leu112 (Fig. 4C,D, construct H3M92L/G104H/C112L) to confer H3 with the surface information to be recognized by HJURP. In conjunction with N-terminal substitutions (i.e. with either Asn85 or Gln89), substitutions of His104 and Leu112 are minimally sufficient to confer access to HJURP (Fig. 4C,D, Constructs H3R85N/G104H/C112L and H3S89Q/G104H/C112L). These findings lead to a model in which CENP-A-specific exposed residues at both the N- and C-terminal portion of the CATD cooperate to form the recognition surface for HJURP.
HJURP is a Protein Folding Chaperone, Not Just a Histone Chaperone
While the contact point between the C-terminal β-sheet region (i.e. a.a. 63–80) of HJURP and the α1 helix of CENP-A is not involved in discriminating CENP-A from bulk H3, our findings that this region of HJURP is responsible for transmitting broad stability to CENP-A/H4 (Fig. 3) strongly suggest that this portion of the HJURP/CENP-A interface is critical for a histone stabilizing function of HJURP. The stability induced by HJURP1–62 binding is most substantial within the CATD (Fig. 3C) since it lacks any major contact with the α1 helix of CENP-A (Hu et al., 2011). Inclusion of the contact residues in the HJURP1–80 construct spreads the induced stability, as measured by a dramatic slowing in backbone amide proton exchange, throughout much of the histone folds of both CENP-A and H4 (Fig. 3D). The term ‘molecular chaperone’ was initially used in the world of protein biochemistry to describe histone chaperones (Laskey et al., 1978), which have subsequently been largely distinguished from other classes of molecular chaperones (i.e. proteins that assist folding of their substrates by either stabilizing correctly folded proteins, or destabilizing misfolded proteins to allow a subsequent refolding attempt (Hartl et al., 2011)). Our finding with HJURP provides one example of a protein that challenges this distinction since it has the hallmarks of a histone chaperone and a protein-folding chaperone. HJURP certainly does not require ATP, a property of many protein folding chaperones such as chaperonins and heat shock proteins, but HJURP qualifies as a protein folding chaperone in the sense that it stabilizes its substrate and is not a component of the final product (similar to a proposal for trigger factor function in ribosome assembly; Martinez-Hackert and Hendrickson, 2009), as HJURP is only present at centromeres for a portion of one phase (G1) of the cell cycle (Foltz et al., 2009; Dunleavy et al., 2009).
The molecular chaperone function of HJURP is likely to be a conserved one, since Scm3 has been reported to increase the solubility of Cse4/H4 in the budding yeast system (Xiao et al., 2011). It will be interesting to investigate whether or not other so-called histone chaperones confer similar stability to their bulk histone substrates as there is within the ternary HJURP/CENP-A/H4 complex (Fig. 3), especially in light of the finding that (H3/H4)2 heterotetramers are intrinsically 10-fold more flexible than (CENP-A/H4)2 heterotetramers (Black et al., 2004). HJURP binds to CENP-A/H4 upon new CENP-A protein expression in late S/G2 phases of the cell cycle (Foltz et al., 2009; Shelby et al., 2000) and must chaperone CENP-A/H4 until new CENP-A nucleosome assembly occurs in the following G1 phase (Dunleavy et al., 2009; Jansen et al., 2007; Schuh et al., 2007). Thus, for HJURP, its role in preventing inappropriate interactions between CENP-A/H4 and other macromolecules occurs over timescales that likely exceed those for the bulk histone assembly pathways where histone deposition occurs shortly after histone synthesis. Furthermore, the degree of stability that HJURP induces to much of CENP-A/H4 is substantial, similar to the extent measured within the rigid interior of nucleosomes (Black et al., 2007a).
CATD Function Extends Beyond HJURP Recognition in Mammals
Budding yeast Cse4 lacks the positive charge of its human counterpart on its bulged loop L1 that alters the nucleosome surface (Cho and Harrison, 2011; Sekulic et al., 2010; Tachiwana et al., 2011). In mammals, this positively charged loop is a strong candidate to generate a binding surface for centromere proteins, such as CENP-N, which recognizes nucleosomes assembled with the H3CATD chimera (Carroll et al., 2009; Guse et al., 2011). While the conformational flexibility of yeast centromeric histone complexes has not been directly tested, budding yeast Cse4 is reported to lack hydrophobic stitching with histone H4 (Cho and Harrison, 2011). These points of evolutionary variation led to the proposal that the budding yeast CATD has the sole task of providing recognition by Scm3 (Cho and Harrison, 2011), perhaps as an adaptation upon the loss in some yeasts of epigenetic centromeres and the gain of DNA sequence-specified centromeres. On the other hand, in the budding yeast S. cerevisiae, the CATD is recognized by Scm3 (Zhou et al., 2011) and recruits the Psh1 E3 ubiquitin ligase (Ranjitkar et al., 2010) to CENP-A nucleosomes misincorporated at non-centromeric loci (Hewawasam et al., 2010; Ranjitkar et al., 2010), suggesting important CATD contributions before and after nucleosome assembly. The swapping at six positions of CENP-A→H3 residues at the H4 contact surface, individually subtle changes that reduce polarity or increase the size of hydrophobic side-chains (Fig. 4A), leads to a loss of faithful localization at centromeres despite robust recruitment to the HJURP-containing chromosomal array (Fig. 4C,D). Our clear delineation in function of CATD residues involved in HJURP recognition from the buried hydrophobic stitches (Fig. 7) leads to the simple conclusion that mammalian CATD function includes: 1. sorting of newly expressed CENP-A protein from bulk histones, 2. assembly at centromeres, and 3. conformational rigidity unique to centromere-specifying nucleosomes. While we prefer this model as it is based primarily on structure-based mutational analysis (Figs. 4 & 5), it is possible that additional CATD-encoded features exist that are required for function. To this point, we note that while four (L1, α2, α2.1, and α2.2) of the five defective CENP-A mutants examined in Figure 1 lack HJURP association and/or involve removal of hydrophobic stitch residues, the α2.3 mutant is clearly recognized by HJURP and contains all six of the hydrophobic stitch residues.
A Requirement of Forming Octameric Nucleosomes for CENP-A Assembly at Centromeres
There are now biochemical and structural data indicating that CENP-A assembled with purified histones onto nucleosomal DNA sequences (either centromere-derived or not) forms an octameric histone core that is wrapped in the conventional left handed manner (Dechassa et al., 2011; Sekulic et al., 2010; Panchenko et al., 2011; Barnhart et al., 2011). A potential exception to this is in budding yeast, where the >80% AT-rich centromere sequence (Clarke and Carbon, 1985) precludes efficient nucleosome formation (Camahort et al., 2009; Xiao et al., 2011) but is reportedly conducive to forming non-nucleosomal particles containing Scm3 but lacking H2A/H2B dimers (Xiao et al., 2011). The hemisome model, however, proposes that the major form of CENP-A-containing nucleosomes at centromeres contains only a single copy of each histone (Dalal et al., 2007; Furuyama and Henikoff, 2009). Our precise targeting of the CENP-A/CENP-A interface with the CENP-AH115A/L128A mutant that retains full recognition by HJURP but completely fails to track to centromeres (Fig. S6A,B) suggests that this interface is essential for stable assembly at centromeres. Furthermore, the CENP-AH115A/L128A mutant cannot stably assemble into chromatin after association on the chromosome with HJURP (Figs. 6G,H), as opposed to the stable incorporation of WT CENP-A and all other mutants that we tested that maintain HJURP association (Figs. 1 & S4). Thus, our data supports the notion that CENP-A/CENP-A interactions soon upon HJURP-mediated deposition onto DNA are required for retaining CENP-A in centromeric chromatin. This is probably a conserved feature of centromeric nucleosomes, and others have very recently reported that engineered disruption of the CENP-ACID/CENP-ACID interface reduces its centromere localization in fruit fly cells (Zhang et al., 2011).
We favor a model wherein the biochemically stable octameric form of CENP-A nucleosomes (Camahort et al., 2009; Sekulic et al., 2010; Tachiwana et al., 2011; Kingston et al., 2011) is the most prominent form at mammalian centromeres, but not at the exclusion of intermediate forms that occur during a program of G1 centromere maturation or immediately following redistribution of the CENP-A proteins on the daughter strands of the S-phase replication fork (Black and Cleveland, 2011). Nor does our work exclude models proposing two pools of CENP-A at mitotic chromosomes, with one proposal involving a major pool consisting of stable octamers for epigenetic memory of centromere location and a small population of a specialized form (i.e. tetrasomes (Williams et al., 2009), hemisomes (Dalal et al., 2007; Furuyama and Henikoff, 2009), or hexasomes (Mizuguchi et al., 2007; Xiao et al., 2011)) at the foundation of the mitotic kinetochore (Mizuguchi et al., 2007; Xiao et al., 2011). On the other hand, since the small (4–6 residue) unstructured C-terminal tail of CENP-A is sufficient when transplanted onto histone H3 to generate nucleosomal arrays capable of nucleating a functional kinetochore in frog egg extracts (Guse et al., 2011), there is a precedent for at least one system where such radical nucleosome specialization is not required for an important mitotic role of centromeric chromatin.
Conclusions
There is broad interest in how histones transmit epigenetic memory through dynamic processes such as transcription and replication, and histone chaperones play a fundamental role in directing these processes. Centromeric epigenetic memory is transmitted from cell-to-cell and through generations, so the fidelity of the CENP-A nucleosome deposition is perhaps the highest of any case of histone-based epigenetic memory. In the last year, the centromere field has quickly accumulated several high-resolution snapshots of CENP-A in various contexts (Cho and Harrison, 2011; Hu et al., 2011; Sekulic et al., 2010; Tachiwana et al., 2011; Zhou et al., 2011). We used information from these structures along with a powerful cell-based functional assay and present here a sophisticated description of the elements on CENP-A and its chaperone HJURP governing their molecular recognition. The cell-based approaches were essential for us to properly assign function to particular elements in each protein, and in the course of our studies we clarified earlier confusion on the primary determinants for HJURP recognition (Foltz et al., 2009; Hu et al., 2011). Recognition is a key step in robust propagation of centromere-specifying nucleosomes, replenishing each cell cycle the foundational chromatin that ensures proper genome transmission at cell division.
Experimental Procedures
Cell-based Experiments
U2OS-LacO-TRE cells containing 200 copies of a 256×LacO/96×tetracycline-responsive element array on chromosome 1 (Janicki et al., 2004; kindly provided by S. Janicki, Wistar Institute, Philadelphia, PA) were cultured in DME supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 100 μg/mL hygromycin. Cells were plated onto coverslips in 6-well dishes 24 h prior to transfection. Cells were co-transfected using Fugene (Roche) at a 2:1 ratio (HJURP construct:H3 or CENP-A construct). CENP-A replacement mutants (L1, α2, α2.1, α2.2, α2.3) have been described previously (Shelby et al., 1997; Black et al., 2004). CENP-A and H3 mutant constructs used for transient transfections (CENP-AS68Q, H3Q68S, H3CATD, H3HJURP Surf., H3G104H, H3R85N/S88A/S89Q, H3M92L/G104H/C112L, H3R85N/G104H/C112L, and H3S89Q/G104H/L112C) were generated in pCDNA3.1 vector with a triple hemagglutinin (HA) fusion tag on the N-terminus. CENP-AH4 Int. and CENP-AH115A/L128A were generated in a pBABE vector with an N-terminal Yellow Fluorescent Protein (YFP) tag. mCherry-LacI-HJURP1–62 and mCherry-LacI-HJURPW66A mutations were generated from full-length mCherry-LacI-HJURP (Barnhart et al., 2011). Mutants were generated by one or multiple PCR site-directed mutagenesis steps, except for constructs H3HJURP Surf. and CENP-AH4 Int., which were synthesized as complete ORFs (GenScript) and subsequently shuttled into pCDNA3.1. In all cases, the final mutant constructs were verified by DNA sequencing. Cells were fixed with 4% formaldehyde and processed for indirect immunofluorescence 48 h following transfection. For the stable incorporation assay (Barnhart et al., 2011), U2OS-LacO-TRE cells were also co-transfected with TetR-GFP to mark the array, and cells were treated at 48 h with or without 15 mM IPTG for 1 h prior to fixation. HeLa cells were cultured in DME supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin, and were transfected with WT YFP-CENP-A or YFP-CENP-AH115/L128A mutant using Effectene (Qiagen). Monoclonal anti-HA.11 antibody (clone 16B12; Covance) was used at a 1:1000 dilution, and polyclonal anti-CENP-B (clone H-65; Santa Cruz Biotechnology, Inc.) was used at a 1:1000 dilution. FITC- and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used at a 1:200 dilution. Cells were stained with DAPI and mounted with Vectashield medium (Vector Laboratories).
Images were captured using LAF software (Leica) with a charge-coupled device camera (ORCA AG; Hamamatsu Photonics) mounted on an inverted DMI6000B microscope (Leica) with a 100X 1.4 NA objective lens. Images were collected in 0.2 μM z sections, deconvolved, and projected as a single two-dimensional image using PhotoShop (version 12; Adobe) and Illustrator (version 12; Adobe). For quantification of CENP-A and H3 mutant localization, 100 cells were counted and verified with multiple independent experiments. For quantification of CENP-A mutant stable incorporation into the HJURP-containing array, at least 30 cells were counted over two independent experiments. For quantification of endogenous CENP-A recruitment to the HJURP1–62- and HJURP1–208-containing arrays, the percentage of cells containing CENP-A positive arrays was divided by the percentage previously reported for HJURPFull-length (Barnhart et al., 2011).
Recombinant Protein Preparation
The expression and purification of human histone proteins prepared from bacteria have been described (Luger et al 1997a; Black et al., 2004; Sekulic et al, 2010). Plasmids for mutant histones (CENP-AS68Q, H3S68Q, and CENP-AH115A/L128A) were generated by PCR site-directed mutagenesis. All constructs for HJURP protein expression were generated in a modified pET vector (kindly provided by G. Van Duyne, University of Pennsylvania, Philadelphia, PA) with an N-terminal Maltose Binding Protein (MBP) fusion. MBP-HJURP1–62, MBP-HJURP1–80, and MBP-HJURP1–80/W66A were expressed in the BL21 Rosetta(DE3) bacterial strain. Cells were lysed by sonication in 25 mM Tris-Cl pH 7.5, 300 mM NaCl, 5 mM βME. Lysates were incubated with amylose resin (New England Biolabs) for 2 h at 4°C and eluted with 15 mM maltose, followed by preparative SEC.
Analytical SEC
Purified recombinant HJURP protein [MBP-HJURP1–62, MBP-HJURP1–80, or MBP-HJURPW66A] and human histone complexes [(CENP-A/H4)2, (H3/H4)2, (CENP-AS68Q/H4)2, (H3S68Q/H4)2, or (CENP-AH115A/L128A/H4)] were mixed equimolar (20 μM each in 2 mL volume) in 25 mM Tris pH 7.5, 1 M NaCl (or 300 mM NaCl, where indicated), 20 mM MgCl2, 5 mM βME. Protein mixtures were pre-incubated on ice for 30 min and injected onto a HiLoad 16/60 Superdex200 column (GE Healthcare) with running buffer identical to sample buffer. Fractions were collected at 2 mL each and analyzed by 15% SDS-PAGE with Coomassie Brilliant Blue staining. The column was calibrated with gel filtration protein standards (Biorad) in identical buffer conditions.
H/DX Reactions
30 μL of protein at 1 mg/mL of the indicated protein complex sample was mixed with 90 μl D2O and incubated at 4°C. Final buffer conditions in H/DX reactions are 0.25 mg/ml protein mix, 10 mM sodium phosphate (pH=7.2), 1.75 mM βME, and either 300 mM, 500 mM, or 1M NaCl. At each timepoint (10, 102, 103, 104, and 105 s), 20 μl of each H/DX reaction was withdrawn and added to 30 μl of ice cold quench buffer (1.66 M guanidinium-HCL, 0.8% formic acid, 10% glycerol) and immediately frozen in liquid N2. Samples were stored at −80°C prior to analysis.
Protein fragmentation, mass spectrometry, and data analysis
Protein fragmentation, MS, and data analysis steps were performed similarly to those described for nucleosome H/DX (Panchenko et al., 2011). In brief, H/DX samples were thawed on ice and injected onto an immobilized pepsin column at an initial flow rate of 50 μl/min for 3 min followed by 150 μl/min for another 3 min. Pepsin (Sigma) was coupled to Poros 20 AL support (Applied Biosystems) and packed into column housings of 2 mm x 2 cm dimensions (IDEX). Protease generated fragments were collected onto a C18 HPLC trap column (2.5 x 0.5 mm, LC Packings). Peptides were eluted into and through an analytical C18 HPLC column (0.3 x 75 mm, Agilent) by a linear 12–55% buffer B gradient at 6 μl/min (Buffer A: 0.1% formic acid; Buffer B: 0.1% formic acid, 99.9% acetonitrile). The effluent was electrosprayed into the mass spectrometer (LTQ Orbitrap XL, Thermo Fisher Scientific). SEQUEST (Bioworks v3.3.1) software program (Thermo Fisher Scientific) was used to identify the likely sequence of the parent peptides using non-deuterated samples via tandem MS. The ExMS program (Kan et al., 2011) was used for data analysis. The level of H/DX at each timepoint is expressed as the number of deuterons in the peptide, or in terms of the percentage of exchange within the peptide. To correct for loss of deuterium from each peptide during the H/DX-MS analysis, measurements were made of reference samples that had been deuterated under denaturing conditions.
Immunoblot
Whole cell extracts of U2OS-LacO-TRE cells were collected 48 h post-transfection, along with untransfected control cells. 5 x 104 cells were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with an anti-HJURP antibody generated against a C-terminal fragment (1 μg/ml) and anti-β-actin (Sigma; 1:1,000) as a loading control. Antibodies were detected using a horseradish peroxidase-conjugated secondary antibody at 1:10,000 (Jackson ImmunoResearch Laboratories) and enhanced chemiluminescence (Thermo Scientific).
Supplementary Material
Research Highlights.
The CENP-A Targeting Domain (CATD) houses the residues that confer HJURP binding
HJURP transmits stability to CENP-A/H4 prior to chromatin assembly
CENP-A hydrophobic stitch residues are required for incorporation at centromeres
CENP-A assembly at centromeres requires an intact CENP-A/CENP-A interface
Acknowledgments
We thank S. Janicki (Wistar), G. Van Duyne (Penn), K. F. Sullivan (NUI Galway), D. Cleveland (UCSD), and K. Luger (Colorado State University) for reagents, J. Shorter (Penn) for helpful discussion, and the anonymous reviewer who suggested to extend our analysis of the HJURP/CENP-A/H4 trimer. This work was supported by grants from the NIH (GM082989 to B.E.B.) and the ACS (to D.R.F.), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund (to B.E.B.), and a Rita Allen Foundation Scholar Award (to B.E.B.). E.A.B. and T.P. were supported the University of Pennsylvania Structural Biology Training Grant (NIH grant GM08275). E.A.B. was also supported by an AHA predoctoral fellowship. N.S. is supported by a postdoctoral fellowship from the ACS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH. Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 2001;11:250–257. doi: 10.1016/s0962-8924(01)02000-1. [DOI] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007a;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007b;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. The structure and function of yeast centromeres. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Wexler BB, Rogers DJ, Hite KC, Panchenko T, Ajith S, Black BE. DNA binding restricts the intrinsic conformational flexibility of methyl CpG binding protein 2 (MeCP2) J Biol Chem. 2011;286:18938–18948. doi: 10.1074/jbc.M111.234609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu RM. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan ZY, Mayne L, Sevugan Chetty P, Englander SW. ExMS: Data Analysis for HX-MS Experiments. J Am Soc Mass Spectrom. 2011;22:1898–1905. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell. 2009;138:923–934. doi: 10.1016/j.cell.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Hargreaves VV, Jamison JW, Mo AO, Li S, Putnam CD, Woods VL, Jr, Kolodner RD. A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc Natl Acad Sci U S A. 2009;106:22223–22228. doi: 10.1073/pnas.0912250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci U S A. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- Willard HF. Centromeres of mammalian chromosomes. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila Centromeric Nucleosomes Requires CID Dimerization. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.12.010. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.