Abstract
In this report we present a method to identify functional artificial lantipeptides. In vitro translation coupled with an enzyme-free protocol for posttranslational modification allows preparation of more than 1011 different lanthionine containing peptides. This diversity can be searched for functional molecules using mRNA-lantipeptide display. We validated this approach by isolating binders toward Sortase A, a transamidase which is required for virulence of Staphylococcus aureus. The interaction of selected lantipeptides with Sortase A is highly dependent on the presence of a (2S,6R)-lanthionine in the peptide and an active conformation of the protein.
Lantipeptides are produced by a range of Gram-positive bacteria and are often characterized by potent antibacterial activities.1−3 The peptide scaffold is first assembled by ribosomal translation and later acquires a series of post-translational modifications which support the bioactive conformation of the molecule. The most notable modifications are the dehydration of serine and threonine residues and the formation of lanthionine moieties (Lan) by intramolecular 1,4-addition of Cys side chains onto dehydroamino acids. These modifications are introduced by lantipeptide dehydratases and cyclases, which are characterized by surprisingly broad substrate scopes.4−8 Since the primary sequences of lantipeptides are genetically encoded, this class of natural products is amenable to directed molecular evolution and thus appears to be an excellent platform for the development of therapeutic molecules. In addition, heterologous production of both native and engineered lantipeptides9,10 has shown that such compounds should be accessible on useful scales despite their size and complex structures.
In the quest for therapeutic peptides over 50 natural lantipeptides have been described,1−3 and many more artificial Lan-containing peptides have been constructed,5,7,11−16 but a systematic method for identifying lantipeptides with tailor-made functions is missing. We therefore developed an in vitro selection method for the production and parallel screening of up to 1011 different lantipeptides. The key to this method is a protocol which allows in vitro translation of cross-linked peptides,11,17 which are linked to their coding mRNA (Figure 1).18,19 This phenotype-genotype linkage allows enrichment of rare sequences based on the functionality of their encoded and posttranslationally modified peptide. We then used solid phase peptide synthesis (SPPS) and in vitro characterization of selected lantipeptides to demonstrate that this method produces highly Lan-specific molecular interactions.
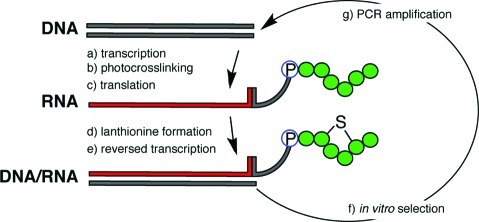
Figure 1.
Formation of mRNA-displayed lantipeptides. (a) Transcription from a synthetic DNA library coding for peptides with a conserved Lys-X2-Cys motif; (b) attachment of a puromycin (P) labeled oligonucleotide by photo-cross-linking; (c) ribosomal peptide synthesis in a reconstituted translation system, in which lysine was substituted with 4-selenalysine; (d) mRNA-peptide fusions were immobilized on oligo-dT sepharose and chemically treated to introduce Lan cross-links; (e) reverse transcription and NTA-agarose purification of the final mRNA-peptide fusions; (f) in vitro selection: the pool of 20 pmol of fusions was incubated with immobilized Sortase A SrtA; (g) retained mRNA messages were PCR amplified to serve as the DNA template for the next selection round.
In order to prepare Lan-containing peptides we used a chemically defined translation mixture in which we substituted lysine with 4-selenalysine.11,17,20 This unnatural amino acid is amenable to H2O2 induced post-translational elimination to dehydroalanine (Dha). As a template for peptide synthesis we used in vitro transcribed mRNA which was photo-cross-linked to a short puromycin-displaying oligonucleotide at the 3′-end of the open reading frame (Figure 1). This tag causes the ribosome to form a covalent bond between the C-terminus of the nascent peptide and the templating mRNA strand (Figure 1).19 The resulting peptide-mRNA fusions were immobilized on oligo-dT Sepharose under high salt conditions through base complementarity to a stretch of adenine bases on the mRNA. The immobilized fusions were then treated with a sequence of buffers to induce the formation of Lan moieties (Figure 2). During this procedure, care is required so as not to lose most of the very small quantity of fragile molecules such as picomoles of peptide-mRNA fusions.
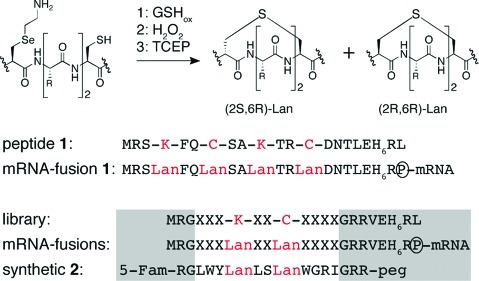
Figure 2.
(Top) Enzyme-free Lan formation.11 (1) Translated peptides were incubated with oxidized glutathione (GSHox) to protect Cys residues from irreversible oxidative damage; (2) H2O2 induced oxidative elimination of 4-selenalysine to Dha; (3) disulfide reduction initiated intramolecular 1,4-addition of Cys side chains onto Dha to produce Lan-containing peptides (TCEP: tris(2-carboxyethyl)phosphine). (Bottom) In gray: constant regions. (P): puromycin links the peptide with its coding mRNA. peg: the C-terminus of synthetic peptides was appended with a polyethylenglycol tail to increase solubility.21 5-FAM: 5-carboxyfluorescein. Lan: the two thioether-bridged residues are labeled as Lan.
To evaluate the efficiency of this step we constructed an mRNA template coding for a peptide containing two Lys-X2-LCys motifs. The corresponding 35S-methionine labeled peptide-mRNA fusions were immobilized on oligo dT sepharose and modified as outlined in Figure 2. 35S-Met scintillation counting suggested that 70% of the fusions are retained throughout the modification procedure (Figure S1), and MALDI-TOF spectrometry of the peptide portion showed that the two Lys-X2-LCys motifs have indeed been converted to Lan moieties (Figure S1).
Next, we constructed a double stranded DNA library coding for one conserved Lys-X2-Cys motif along with seven additional random residues (X) and a C-terminal histidine tag (Figure 2). In addition we included three conserved Arg residues flanking the random region to improve peptide solubility even in the absence of the mRNA tail. The codons for the conserved Val-Glu dipeptide provide two stop codons which terminate the translation of sequences with a frameshift. From this library we transcribed 1 nmol of puromycin tagged mRNA and subjected this pool to a 2.5 mL translation reaction.17,18,20 After introduction of the Lan moieties, the fusions were eluted from the sepharose support. Reverse transcription and purification on NTA agarose yielded an initial pool of 20 pmol of mRNA peptide fusions as inferred by 35S-Met scintillation counting. This corresponds to 1.2 × 1013 molecules, providing 24-fold coverage of the 5 × 1011 different sequences.
With this pool of mRNA-displayed artificial lantipeptides available, we set out to identify specific binders to Sortase A (SrtA) from Staphylococcus aureus (S. aureus).22 This Cys-dependent transamidase anchors extracellular virulence factors to the growing cell wall. The enzyme recognizes a Leu-Pro-X-Thr-Gly motif on precursor proteins, cleaves the amide bond between Thr and Gly, and links Thr with the penta-Gly tail of the cell wall precursor lipid II.22 Similar enzymes are ubiquitous among gram positive bacteria and may therefore present a target for broad-band antibacterials.23 However, development of effective low molecular weight inhibitors has proven to be difficult.23−28 This challenge coupled with the well-behaved nature of the catalytic domain of SrtA makes this protein an ideal test case for a first in vitro selection of artificial lantipeptides (Figure S2).
We immobilized 1 nmol of biotinylated SrtA on streptavidin-coated beads and coincubated with the 20 pmol pool of mRNA-displayed lantipeptides for 1 h. The beads were washed to remove approximately 98% of the specific 35S-Met activity. The retained nucleic acids were eluted in 100 mM NaOH and amplified by PCR. The product DNA was used as a template to generate a new mRNA pool for the next selection round. To suppress coselection of peptides which bind to the streptavidin-coated beads rather than to SrtA, we used a slightly different protocol for the next four rounds: we eluted specific SrtA binders by incubating the beads in a 0.2 mM solution of soluble SrtA for 2 h. Enrichment of SrtA binders was monitored in each round by quantifying the fraction of the pool retained by the SrtA-coated beads after a standardized washing protocol (Figure S3). In round five, 25% of the fusions bound to immobilized SrtA suggesting that a significant portion of this pool codes for SrtA-binding mRNA-peptide fusions. Since this fraction did not increase after an additional selection round, we cloned and sequenced individual members of the pool after round five.
From 42 clones we retrieved 36 sequences which coded for a central Lys-X2-LCys motif as designed in the initial library. These peptides are characterized by mainly hydrophobic residues with a particular overrepresentation of Trp. While the N-terminus of the randomized segment features a conserved Leu-Trp dipeptide, the C-terminus is more variable and splits the sequences into two families represented by the sequences 2 to 6 and 7 to 10 (Table 1, Figure S4).
Table 1. Synthesized Artificial Lantipeptides.
| peptidea | variable regionb | conditionsc | Kd [μM]d |
|---|---|---|---|
| 2(2S,6R) | LWYLanLSLanWGRI | 3.0 ± 0.2 | |
| ″ | ″ | no Ca2+ | >1000 |
| ″ | ″ | SrtBe | >300 |
| ″ | ″ | no pegf | 1.3 ± 0.1 |
| 2(2R,6R) | ″ | >300 | |
| 2(2R,6S) | ″ | >300 | |
| 2(2S,6S) | ″ | >300 | |
| 2linearg | LWYdA LSC(me)WGRI | >400 | |
| 3(2S,6R) | LWYLanLSLanWGFI | 4.0 ± 0.5 | |
| 4(2S,6R) | LWYLanFSLanWGRI | 9.9 ± 0.8 | |
| 5(2S,6R) | LWYLanLSLanWGRV | 11 ± 1 | |
| 6(2S,6R) | LWYLanLTLanWGRI | 32 ± 3 | |
| 7(2S,6R) | LWTLanLRLanVFPW | >100 | |
| 8(2S,6R) | LWDLanWRLanNFLW | >200 | |
| 9(2S,6R) | LWGLanWSLanVFPW | >1000 | |
| 10(2S,6R) | LWDLanWRLanVFRW | >1000 |
The (2R,6R)-Lan containing isomers of peptides 3 to 10 showed no detectable affinity for SrtA (Kd > 200 μM).
The displayed sequence corresponds to the variable region which is flanked by N- and C-terminal constant regions as depicted in Figure 2 for peptide 2.
Standard conditions are: 50 mM Tris HCl, 50 mM NaCl, 5 mM CaCl2, 1.2 mM TCEP.
Binding constants were determined by fluorescence polarization. Each data point represents an average of at least three independent measurements.
SrtA was replaced by Sortase B (SrtB).
2(2S,6R) without C-terminal peg-tail.
dA: d-Ala. C(Me): S-methyl l-Cys. The stereochemistry of 2linear therefore corresponds to (2S,6R).
Since in vitro translation does not provide enough material to verify peptide structure and function we picked nine selected sequences and prepared the corresponding peptides with a C-terminal polyethylenglycol (peg) tail21 by standard Fmoc-SPPS (Table 1). In a biomimetic approach,29−33 we first synthesized linear peptides containing a Dha-X2-LCys motif (Figures S5, S6 and Table S2) which cyclized after purification upon raising the pH above 8.0. Cyclization produced the (2S,6R)-Lan and (2R,6R)-Lan containing peptides with a significant excess of one isomer (60–80% de). Dha-X2-LCys motifs are known to preferentially form the (2S,6R)-Lan irrespective of the remaining peptide context.30−33 To confirm the predicted stereochemistry we resynthesized peptide 2 replacing the Dha-X2-LCys motif with a DCys-X2-Dha or LCys-X2-Dha motif. The latter two motifs are predicted to cyclize stereorandomly to mixtures of (2S,6S)-Lan and (2S,6R)-Lan or (2R,6S)-Lan and (2R,6R)-Lan containing molecules (2(2S,6S), 2(2S,6R), 2(2R,6S), and 2(2R,6R)).34 Despite the changes in their initial sequence, the four cyclic isomers differ only by one or two stereocenters. By HPLC comparison of the three cyclization reactions each containing a different pair of isomers, we were able to assign the stereochemistry of all four possible isomers and confirmed the major cyclization product arising from the Dha-X2-LCys motif as the (2S,6R)-Lan isomer (Figure S7).
Binding affinities were measured by fluorescence polarization using fluorescein-labeled versions of all four isomers of 2 (Figure S8). Only 2(2S,6R) binds to SrtA, with a Kd of 3 ± 0.2 μM, whereas all other isomers fail to exhibit detectable binding affinity (Table 1). Also, the linear version of 2(2S,6R) in which the (2S,6R)-Lan was replaced by d-Ala and S-methyl l-Cys (2linear) does not bind to SrtA either (Table 1). In addition, 2(2S,6R) has no detectable affinity to SrtB, a staphylococcal homologue of SrtA with similar biological functions but only 22% sequence similarity.35 We also prepared 2(2S,6R) without peg-tail. This peptide binds SrtA with a similar affinity as 2(2S,6R), suggesting that this tail does not contribute to the observed binding (no peg, Table 1).
SrtA depends on millimolar concentrations of Ca2+ for efficient substrate recognition.36 This cation does not contact the substrate directly but rather immobilizes two active site loops by coordinating several of their carboxylate side chains. Binding of 2(2S,6R) is similarly Ca2+ dependent, suggesting that its binding site overlaps with the SrtA active site. Regardless, 2(2S,6R) at a concentration of up to 1 mM is unable to inhibit SrtA catalyzed cleavage of a peptide substrate, even though this reaction is characterized by a relatively high Km of 80 μM (data not shown). However, if kcat is faster than dissociation of the substrate/enzyme complex (koff), the substrate affinity could be significantly higher than indicated by Km (koff/kon ≪ kcat/kon), making it impossible for 2(2S,6R) to compete for binding.
Finally, we determined the affinities of peptides 3(2S,6R) to 10(2S,6R) (Table 1). Most members of family 1 (2(2S,6R) to 6(2S,6R)) bind SrtA with a Kd ranging from 3 to 11 μM (Figure S4). As an outlier, 6(2S,6R) is a 10-fold weaker binder than 2(2S,6R) as the result of a Ser to Thr mutation in the lanthionine macrocycle. None of the corresponding (2R,6R)-Lan containing peptides showed detectable affinity for SrtA (Kd > 200 μM). Again, the Lan induced macrocycle appears crucial for the observed binding activity and represents a novel scaffold amenable to optimization by chemical derivatization. Neither isomer of the second family (7 to 10, Table 1) recognizes SrtA, suggesting that these sequences emerged by way of an alternative selective pressure.
The method outlined in this communication allows the identification of functional molecules from a random pool of artificial lantipeptides. In a first demonstration we found molecules with highly specific affinities toward a potential drug target from S. aureus. Unlike other peptide cyclization protocols37−40 Lan formation offers alterations in the backbone stereochemistry in addition to conformational rigidity. Natural lantipeptides generally contain (2S,6R)-Lan due to enzyme imposed stereocontrol. In contrast, enzyme-free introduction of Lan can access (2S,6R)-Lan, (2R,6R)-Lan, and (2R,6S)-Lan containing molecules which may compete for function during in vitro selection. As shown here, deconvolution of the stereochemical information after in vitro selection is relatively straightforward, using a biomimetic approach to SPPS of lantipeptides. Therefore we conclude that the herein presented method should allow a search for functional lantipeptides in both sequence and geometry space.
Acknowledgments
We thank M. C. T. Hartman, B. Seelig for insightful advice, and S. Gentz for technical assistance. F.T.H. was partly supported by the International Max Planck Research School in Chemical Biology; J.W.S. is an Investigator of the Howard Hughes Medical Institute; F.P.S. is supported by the “Professur für Molecular Bionic”. F.T.H. and F.P.S. thank R. S. Goody for financial support.
Supporting Information Available
Figures S1–S8, Tables S1 and S2, details on in vitro translation/selection, SrtA production, SPPS, isomer analysis, and fluorescence spectroscopy are provided in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Willey A. M.; van der Donk W. A. Annu. Rev. Microbiol. 2007, 61, 477–501. [DOI] [PubMed] [Google Scholar]
- Lubelski J.; Rink R.; Khusainov R.; Moll G. N.; Kuipers O. P. Cell. Mol. Life Sci. 2008, 65, 455–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A.; Hindre T.; Haras D.; Le Pennec J. P. FEMS. Microbiol. Rev. 2007, 31, 134–167. [DOI] [PubMed] [Google Scholar]
- Chatterjee C.; Patton G. C.; Cooper L.; Paul M.; van der Donk W. A. Chem. Biol. 2006, 13, 1109–1117. [DOI] [PubMed] [Google Scholar]
- You Y. O.; Levengood M. R.; Ihnken L. A. F.; Knowlton A. K.; van der Donk W. A. ACS Chem. Biol. 2009, 4, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D.; Hill C.; Cotter P. D.; Ross R. P. Mol. Microbiol. 2010, 78, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Oldach F.; Al Toma R.; Kuthning A.; Caetano T.; Mendo S.; Budisa N.; Süssmuth R. D. Angew. Chem., Int. Ed. 2011, 50, 1–5. [DOI] [PubMed] [Google Scholar]
- Zhang X. G.; van der Donk W. A. J. Am. Chem. Soc. 2007, 129, 2212–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Yang X.; Garg N.; van der Donk W. A. J. Am. Chem. Soc. 2011, 133, 2338–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano T.; Krawczyk J. M.; Mösker E.; Süssmuth R. D.; Mendo S. Chem. Biol. 2011, 18, 90–100. [DOI] [PubMed] [Google Scholar]
- Seebeck F. P.; Ricardo A.; Szostak J. W. Chem. Commun. 2011, 47, 6141–6143. [DOI] [PubMed] [Google Scholar]
- Goto Y.; Iwasaki K.; Torikai K.; Murakami H.; Suga H. Chem. Commun. 2009, 3419–3421. [DOI] [PubMed] [Google Scholar]
- Moll G. N.; Kuipers A.; Rink R. Antonie van Leeuwenhoek 2010, 97, 319–333. [DOI] [PubMed] [Google Scholar]
- Rink R.; Kluskens L. D.; Kuipers A.; Driessen A. J. M.; Kuipers O. P.; Moll G. N. Biochemistry 2007, 46, 13179–13189. [DOI] [PubMed] [Google Scholar]
- Rink R.; Kuipers A.; de Boef E.; Leenhouts K. J.; Driessen A. J. M.; Moll G. N.; Kuipers O. P. Biochemistry 2005, 44, 8873–8882. [DOI] [PubMed] [Google Scholar]
- Rink R.; Wierenga J.; Kuipers A.; Muskens L. D.; Driessen A. J. M.; Kuipers O. P.; Moll G. N. Appl. Environ. Microbiol. 2007, 73, 1792–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck F. P.; Szostak J. W. J. Am. Chem. Soc. 2006, 128, 7150–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K.; Hartman M. C.; Szostak J. W. J. Am. Chem. Soc. 2005, 127, 11727–11735. [DOI] [PubMed] [Google Scholar]
- Roberts R. W.; Szostak J. W. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y.; Inoue A.; Tomari Y.; Suzuki T.; Yokogawa T.; Nishikawa K.; Ueda T. Nat. Biotechnol. 2001, 19, 751–755. [DOI] [PubMed] [Google Scholar]
- Becker C. F. W.; Oblatt-Montal M.; Kochendoerfer G. G.; Montal M. J. Biol. Chem. 2004, 279, 17483–17489. [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K.; Liu G.; Hung T. T.; Schneewind O. Science 1999, 285, 760–763. [DOI] [PubMed] [Google Scholar]
- Maresso A. W.; Schneewind O. Pharmacol. Rev. 2008, 60, 128–141. [DOI] [PubMed] [Google Scholar]
- Clancy K. W.; Melvin J. A.; McCafferty D. G. Biopolymers 2010, 94, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel B. A.; Bentley M.; Kruger R. G.; McCafferty D. G. J. Am. Chem. Soc. 2004, 126, 3404–3405. [DOI] [PubMed] [Google Scholar]
- Oh K. B.; Kim S. H.; Lee J.; Cho W. J.; Lee T.; Kim S. J. Med. Chem. 2004, 47, 2418–2421. [DOI] [PubMed] [Google Scholar]
- Scott C. J.; McDowell A.; Martin S. L.; Lynas J. F.; Vandenbroeck K.; Walker B. Biochem. J. 2002, 366, 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suree N.; Yi S. W.; Thieu W.; Marohn M.; Damoiseaux R.; Chan A.; Jung M. E.; Clubb R. T. Bioorg. Med. Chem. 2009, 17, 7174–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor A. B. Org. Biomol. Chem. 2011, 26, 7606–7628. [DOI] [PubMed] [Google Scholar]
- Okeley N. M.; Zhu Y. T.; van der Donk W. A. Org. Lett. 2000, 2, 3603–3606. [DOI] [PubMed] [Google Scholar]
- Burrage S.; Raynham T.; Williams G.; Essex J. W.; Allen C.; Cardno M.; Swali V.; Bradley M. Chemistry 2000, 6, 1455–1466. [DOI] [PubMed] [Google Scholar]
- Toogood P. L. Tetrahedron Lett 1993, 34, 7833–7836. [Google Scholar]
- Polinsky A.; Cooney M. G.; Toypalmer A.; Osapay G.; Goodman M. J. Med. Chem. 1992, 35, 4185–4194. [DOI] [PubMed] [Google Scholar]
- Zhou H.; van der Donk W. A. Org. Lett. 2002, 4, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K.; Ton-That H.; Su K.; Schneewind O. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik M. T.; Suree N.; Ilangovan U.; Liew C. K.; Thieu W.; Campbell D. O.; Clemens J. J.; Jung M. E.; Clubb R. T. J. Biol. Chem. 2006, 281, 1817–1826. [DOI] [PubMed] [Google Scholar]
- Timmerman P.; Beld J.; WPuijk W. C.; Meloen R. H. ChemBioChem 2005, 6, 821–824. [DOI] [PubMed] [Google Scholar]
- Verdine G. L.; Hilinski G. L. Methods Enzymol. 2012, 503, 3–33. [DOI] [PubMed] [Google Scholar]
- Hayashi Y.; Morimoto J.; Suga H. ACS Chem. Biol. 2012, 3, 607–613. [DOI] [PubMed] [Google Scholar]
- Touati J.; Angelini A.; Hinner M. J.; Heinis C. ChemBioChem 2011, 12, 38–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.