Abstract
Background
Irritable bowel syndrome (IBS) is a chronic, episodic gastrointestinal disorder that is prevalent in a significant fraction of western human populations; and changes in the microbiota of the large bowel have been implicated in the pathology of the disease.
Methods
Using a novel comprehensive, high-density DNA microarray (PhyloChip) we performed a phylogenetic analysis of the microbial community of the large bowel in a rat model in which intracolonic acetic acid in neonates was used to induce long lasting colonic hypersensitivity and decreased stool water content and frequency, representing the equivalent of human constipation-predominant IBS.
Key Results
Our results revealed a significantly increased compositional difference in the microbial communities in rats with neonatal irritation as compared with controls. Even more striking was the dramatic change in the ratio of Firmicutes relative to Bacteroidetes, where neonatally irritated rats were enriched more with Bacteroidetes and also contained a different composition of species within this phylum. Our study also revealed differences at the level of bacterial families and species.
Conclusions & Inferences
The PhyloChip is a useful and convenient method to study enteric microflora. Further, this rat model system may be a useful experimental platform to study the causes and consequences of changes in microbial community composition associated with IBS.
Keywords: enteric microflora, irritable bowel syndrome, PhyloChip, visceral hypersensitivity
INTRODUCTION
Irritable bowel syndrome (IBS) is a chronic, episodic gastrointestinal disorder that is characterized by abdominal pain and altered bowel habits.1 Irritable bowel syndrome prevalence is estimated to be 10–15% in Western countries2–6 comprising 25–50% of all referrals to gastroenterologists.7 The gastrointestinal tract harbors a complex and diverse microbial community, which plays important roles in host nutrition, immune function, health and disease, and it is hypothesized the IBS disease phenotype is associated with a change in colonic microbiota and/or host factors such as mucosal function and immunity.8–17 This is strengthened by reports that luminal antibiotics or probiotic treatment may be effective in alleviating symptoms in patients with IBS.18 Indirect evidence for alterations in the microflora of humans with IBS comes from changes in colonic fermentation patterns have been described in patients with IBS.19,20 Traditional culture methods, even though limited, were the first to show significant changes in enteric microflora.21–24 Most recently, several investigators have used culture-independent methods including real-time PCR and high throughput 16S ribosomal RNA gene sequencing to demonstrate significant differences in the microbiome of IBS patients.25–30
Despite these recent advances, research in this area remains limited both by the sensitivity of the microbiological techniques and the lack of suitable experimental model systems to explore various disease states. Studying the enteric microflora in humans with IBS for instance, is subject to many confounding factors including the inherent heterogeneity of the syndrome, variations in diet, antibiotic use and host genetic factors. Therefore, elucidating a causative role for microflora changes in IBS presents a serious challenge. The task is somewhat simpler in rodent models that mimic the human condition. While several models exist, we focused on one of the most well accepted animal models of ‘irritable bowel syndrome’, induced by neonatal colonic irritation.31 This paradigm results in these animals being sensitized to colorectal distention as adults, without any evidence of overt colonic injury, thus mimicking IBS in humans. We have shown several specific molecular attributes to the colonic hypersensitivity observed in these animals including a possible role for hydrogen sulfide, a metabolic product of sulfate reducing bacteria in the colon.32
We therefore hypothesized that the enteric microflora of adult rats with colonic hypersensitivity would differ from that of controls. Further, we proposed that a comprehensive and relatively simple way of studying the microflora using a 16S rRNA gene DNA microarray called the PhyloChip.33,34 This microarray consists of 500 000 oligonucleotide probes capable of identifying 8743 of Bacteria and Archaea, and provides a comprehensive census for presence and relative abundance of most known prokaryotes in a massive parallel assay.
MATERIALS AND METHODS
Irritable bowel syndrome rats
Male Sprague–Dawley rats were used in all the experiments (Harlan, Indianapolis, IN, USA). The animal protocol was approved by the Institutional Animal Care and Use Committee of the Stanford University Medical Center. Thirteen neonatal rats underwent colonic exposure to dilute acetic acid (IBS) or saline (control) as previously described.31
Colonic sensitivity and stool water content
Visceral hypersensitivity was measured at around 8 weeks by grading the response of rats to colorectal distention (CRD) as previously described by us and others.31,32 Under mild sedation, CRD was performed by rapidly inflating an intrarectal balloon to various pressures: 10, 20, 30, 40, 50, 60, 70 & 80 mmHg, for a 20-s stimulation period followed by a 2-min rest. Behavioral responses to CRD were measured by visual observation of the abdominal withdrawal reflex (AWR) by blinded observer and the assignment of an AWR score as follows:
= Normal behavior without response
= Contraction of abdominal muscles
= Lifting of abdominal wall
= Body arching and lifting of pelvic structures
In a separate set of experiments, stools were collected during a 4-h period. After weighing (wet weight), pellets were dried under vacuum overnight and dry weight was measured.
Luminal and mucosal sampling
At 11 weeks, a separate set of 13 rats were euthanized over the course of 2 days for microbial community profiling of the luminal matter and the corresponding mucosal layer across the cecum, proximal, middle and distal colon of each rat. The 13 rats were dissected over the course of 2 days. On day 1, three control and four IBS rats (seven in total) were euthanized and then aseptically dissected to remove luminal matter and mucosal tissue. On day 2, three control rats and three IBS rats (six in total) were euthanized and aseptically dissected in the same matter. To observe the potential for microbial community intra-bowel variation within each rat model and inter-bowel variation between the IBS and control, colonic luminal matter, approximately 250–450 mg of luminal matter was collected separately from the cecum, proximal colon, middle colon and distal colon of each rat. Upon dissection, each luminal sample was immediately placed in a sterile cryogenic tube, without preservatives and snap-frozen in liquid nitrogen. After collecting each luminal sample, a mucosal sample was immediately dissected from the same location. Once the luminal matter was removed from the respective bowel segment the mucosal tissue was gently rinsed of obvious fecal matter using sterile water. After gentle rinsing, the mucosal tissue sample was removed and immediately placed in a sterile cryogenic tube and then snap-frozen in liquid nitrogen without preservatives. All samples were stored at −80 °C until further processing.
Luminal sample processing
Genomic 16s rDNA was extracted from 250 mg frozen luminal matter using the UltraClean® Fecal DNA Isolation Kit (MoBio, Inc., Carlsbad, CA, USA) as per the manufacturer’s instructions. After extracting the DNA from each luminal sample, DNA concentration and purity was determined using a NanoDrop® ND-1000 UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Fifty nanogram of extracted DNA samples were then pooled in equimolar ratios into an 11 μL volume by dissection day (day 1 or day 2), IBS or control rat and bowel section (cecum, proximal, middle & distal). This process generated four IBS pooled samples and four pooled control extractions for each dissection day totaling 16 pooled extractions for analysis. The pooled extractions from each dissection day were kept independent of each other to observe the potential for sampling day bias in later analysis. Additionally, to compare whether or not the pooled DNA extractions are truly representative of our populations, one IBS rat and one control rat was selected for non-pooled/independent PhyloChip characterization of each part of the large bowel. These samples were extracted and processed in the same manner as described above but without pooling of the extracted genomic DNA. This non-pooling of samples generated four control independent DNA extractions and four IBS independent DNA extractions. The final numbers for genomic DNA extractions include 16 luminal matter Genomic DNA pooled extractions from day 1 and day 2 and eight independent/non-pooled samples resulting in 24 luminal matter extractions be to be characterized on the Phylo-Chip microarray.
Mucosal sample processing
DNA was extracted from 25 mg of frozen mucosal tissue layer using the Qiagen® DNeasy Blood & Tissue Kit (Qiagen, Inc., Carlsbad, CA, USA) as per the manufacturer’s instructions. After extracting the DNA from each mucosal sample, DNA concentration and purity was determined using a NanoDrop® ND-1000 UV-Vis Spectrophotometer (Thermo Scientific). Fifty nanogram of DNA samples were then pooled in equimolar ratios by dissection day (day 1 or day 2), IBS or control rat and bowel section (cecum, proximal, middle & distal) totaling four IBS pooled DNA extraction and four pooled control DNA extractions, each with a 200 ng final DNA concentration in an 11 μL volume. The extracted mucosal tissue samples resulted in four pooled control extractions and four pooled IBS extractions, each from the respective bowel areas, totaling eight genomic DNA extractions to be used for microarray analysis.
PCR amplification and sample preparation for 16S rRNA gene PhyloChip analysis
All 32 individually processed luminal matter and mucosal gDNA extractions, described above, were then PCR amplified. Primers 27f (5′-AGAGTTTGATCMTGGCTCAG) and 1492r (5′-GGYTACCTTGTTACGACTT) were used to amplify the 16S rRNA gene regions of bacteria.34 Archeal primers were not used for this study and thus not characterized further with the PhyloChip. Polymerase chain reaction (PCR) was carried out using the TaKaRa Ex Taq system (Takara Bio Inc., Otsu, Japan). The amplification protocol was previously described.34 Three temperatures (48, 51.9 and 58 °C) and 25 cycles were used for the annealing step. The 32 luminal and mucosal DNA 16S rRNA amplicons were each hybridized on an Affymetrix PhyloChip, and visualized.
Microarray processing
Microarray analysis was performed using the PhyloChip, a high-density phylogenetic Affymetrix microarray developed by Lawrence Berkeley National Lab. The protocols were previously reported.35 Briefly, amplicons were purified using SpinPrep™ PCR Clean-Up Kit (EMD Chemicals Inc., Gibbstown, NJ, USA) and eluted to a volume of 40 μL. The DNA amplicons were fragmented with DNAse, biotin labeled, denatured, and hybridized to the DNA microarray at 48 °C overnight (>16 h). The arrays were subsequently washed and stained. Reagents, conditions, and equipment are detailed elsewhere.36 Arrays were scanned using a GeneArray Scanner (Affymetrix, Santa Clara, CA, USA). The CEL files obtained from the Affymetrix software that produces information about the fluorescence intensity of each perfect match (PM) probe, mismatch probe (MM), and control probes.
PhyloChip image data analysis
Thirty-two ‘.CEL’ files obtained from the Affymetrix GeneChip Operating Software were analyzed using a custom-tailored package using R,37 containing the annotation of all the probes on the PhyloChip. We used Bioconductor to do the data manipulation and normalizations.38 We used variance stabilization39 and a between chip smoothing to standardize the chips.
Statistical analysis
Examination of the microbial communities associated with the various bowel sections (cecum, proximal, middle and distal colon) revealed no statistically significant differences between the community composition and geographic location within both the IBS and control rat model. Additionally, when comparing the mucosal layer to the luminal matter communities within the IBS group and within the control group no statistically significant microbial community variation was observed. Examination of the potential for sampling day and pooling bias demonstrated that neither pooling of extracted DNA nor day of sampling influenced the statistics (data not shown). As there appeared to be fewer differences between the mucosa and luminal matter and little variation between the various bowel locations, we chose to concentrate the subsequent studies on a comparison between the two animal models rather than biogeography within the large bowel. We therefore paired the samples of the same type and location between sensitized and control rats. Thus, the actual analysis includes all the data using this pairing. The PhyloChip uses approximately 24 probe-pairs (25-mer oligonucleotides) for each probe set to classify organisms into 8364 bacterial groups. A probe set is used to identify organisms at the 97% or higher 16S rRNA gene sequence identify and is hereafter given the operational definition of species. To estimate species relative abundances we first ranked the number of species abundances within each array. Species present on the array were ranked from 1 (low abundance) to 8364 (maximum abundance). A threshold was then established at the ranking of 6000. All species with abundance rankings at or below the threshold were considered noise and set to a value of 6000. These were also given an attribute of 0 in the present/absent column. In order to identify the most significant changes in the microbial community composition between control and IBS microarrays we used a paired difference rank analysis. Paired rank analysis examines pairs of data (IBS vs control chips) in association with their corresponding bowel sections. If species were present (i.e. gave a hybridization signal) in at least 11 out of the 32 arrays, those species were then retained and considered as species of interest. This paired ranked difference analysis allows us to examine and focus on the phylotypes that are significantly differentially expressed between IBS and control groups. Consequently, this procedure restricted the number of different species that were considered to be of interest to 2700. We performed the hypergeometric test40 in order to rank families according to significance of the observed species presence–absence patterns in both control and IBS groups independently. Details of our statistical methodology can be found in the supplemental data file (Data S1).
RESULTS
Colonic hypersensitivity
A significant increase in AWR grade in response to increasing CRD pressure was observed in the IBS rats indicating colonic hypersensitivity (Fig. 1A) compatible with previously published results from our laboratory.31,32
Figure 1.
(A) Effect of neonatal acetic acid treatment on sensitivity to colorectal distention (CRD) in 8-week-old rats. AWR (abdominal withdrawal reflex) scores (y-axis) as a function of distention pressure, (x-axis). AA = acetic acid treated; saline = saline treated; n = 6 each (P < 0.001; Welch 2-sample t-test). (B) Effect of neonatal acetic acid treatment on stool water content, pellet number and bodyweight. These results are normalized to the average values in control (P10 saline) group (*P < 0.05).
Stool water content
We measured pellet water content and total pellet output over a period of 4 h (Fig. 1B). Pellet water content in IBS rats was reduced by 32% (0.86 ± 0.5 vs 1.3 ± 0.7 g, P = 0.009, n = 20) compared to controls. Irritable bowel syndrome rats produced fewer pellets compared to controls, but this did reach statistical significance (5.6 ± 0.5 vs 7.0 ± 0.7, P = 0.103, n = 20). There was no difference in the bodyweights of rats in the two groups.
Changes in colonic microflora
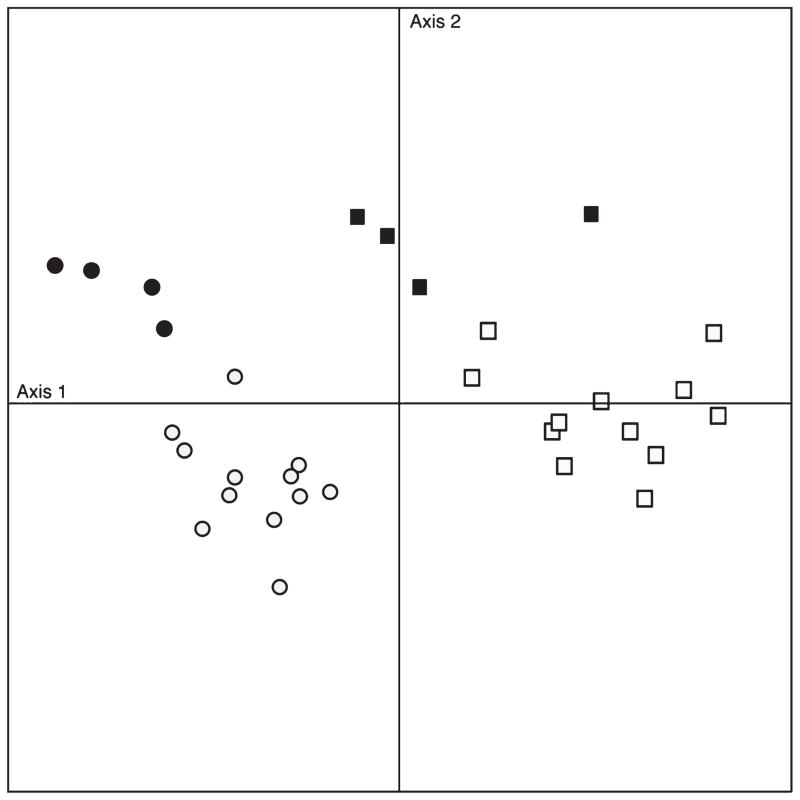
From the 2700 species of interest, we examined 40 (the top 20 different up and 20 down) for which the adjusted P-values were significant (<0.05). We then analyzed the relationship of relative abundance to the two types of samples (Fig. 2). 65.6% of the variance was captured by the first two components, which are abundance of species (axis-1) and composition of IBS and control (axis-2). The graph shows that significant differences in the composition of the microbial community exist between IBS-induced rats and control group. Interestingly, mucosa populations are intermediary. Because of this intermediate phenotype and because there appears to be fewer differences between the mucosal samples of the two groups, in the subsequent analysis we concentrated on the comparison between the two animal groups rather than the biogeography within the large bowel.
Figure 2.
Principal component analysis (PCA) of the top 40 extreme microbial species of irritable bowel syndrome (IBS)-induced and control rats. IBS luminal matter samples are indicated in open squares and IBS mucosal samples are closed squares. Control luminal matter samples are indicated in open circles and IBS mucosal samples are closed circles. X-axis represents the abundance of species, axis Y represents the community composition.
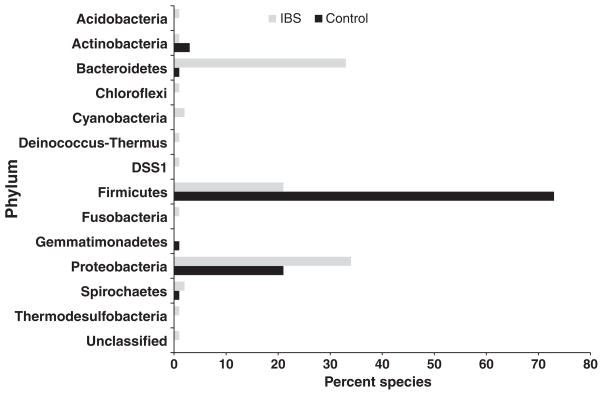
We then assigned the 200 species with the highest paired ranked difference to their respective families according to the phylogenetic groupings within Greengenes and PhyloTrac.41 This analysis revealed significant differences in the overall composition of the community. As shown in Fig. 3, a disparity in representation between the species that were more abundant in the control relative to the IBS-induced rats was observed. Microbial species in the IBS-induced rats fall into 12 distinct phyla while those in the control rats fall into only five. For example, over 70% of the top 200 ranked species in the control rats were from the Firmictues phylum. Interestingly, no Acetobacteria, Acidobacteria, Chloroflexi, Spirochaeates, Planctomycetes, or Cyanobacteria-related bacteria were detected, which were found at varying abundance in the IBS-induced rats. At this point the significance of the presence of Chloroflexi-like or Cyanobacteria-like phylotypes in the large bowel is unclear, and might merely indicate the presence of closely related species with unrelated physiologies. The data, however, illustrate a general increase in abundance of diverse bacterial groups in the large bowel of IBS-induced rats.
Figure 3.
Percentage of the top 150 ranked species associated with their representative phylum in irritable bowel syndrome (IBS)-induced and control rats.
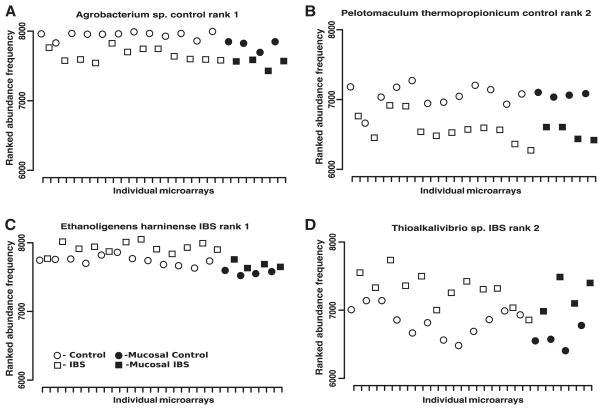
Out of the 200 most significant species we then identified those that showed the most dramatic differences in relative abundance. Table 1 displays the top 20 most significant phylotypes in IBS-induced and control rats, respectively. Most interestingly, the species most similar to Ruminococcus and the family Lachnospiraceae appear to dominate the top-ranked species list in the control rats. Moreover, the phylum Firmicutes was found to be more abundantly represented in the control rats relative to IBS-induced rats. Interestingly, the composition of the Firmicutes phylotypes in IBS-induced rats was markedly different than that of control rats. In the former, we found that species of Clostridium and Prevotella dominated the top-ranked community within the families of Prevotellaceae, and Clostridiaceae. The IBS rats once again demonstrate a higher degree of compositional differences on the phylum and at the family and species levels, respectively. A paired ranked difference plot was generated for each of the two top-ranked species from Table 1. Fig. 4 shows the comparison of the ranked abundances for each of the different samples paired. An abundance of around 8000 (out of 8300), for example, indicates that this species is highly abundant within the sample. Fig. 3 shows the top-ranked Agrobacterium and Pelotomaculum within the control group and the top-ranked Ethanoligenens and Thioalkalivibrio within the IBS groups.
Table 1.
Top 20 significant differentially present phylotypes in IBS-induced and control rats. Each species has been assigned to its representative phylum and family identifier. This table was generated using paired differences above the 6000 threshold
| Control rats
|
IBS-induced rats
|
||||||
|---|---|---|---|---|---|---|---|
| Rank | Representative species | Phylum | Family | Rank | Representative species | Phylum | Family |
| 1 | Agrobacterium | Proteobacteria | Rhizobiaceae | 1 | Ethanoligenens harbinense | Firmicutes | Clostridiaceae |
| 2 | Pelotomaculum thermopropionicum | Firmicutes | Peptococcaceae/Acidaminococcus | 2 | Thioalkalivibrio sp. | Proteobacteria | Ectothiorhodospiraceae |
| 3 | Clostridium sp. | Firmicutes | Clostridiaceae | 3 | Streptococcus bovis | Firmicutes | Streptococcaceae |
| 4 | Ruminococcus lactaris | Firmicutes | Lachnospiraceae | 4 | Helicobacter canadensis | Proteobacteria | Helicobacteraceae |
| 5 | Clostridium methylpentosum | Firmicutes | Clostridiaceae | 5 | Simplicispira | Proteobacteria | Comamonadaceae |
| 6 | Ruminoccoccus hydrogenotrophica | Firmicutes | Lachnospiraceae | 6 | Ethanoligenens sp. | Firmicutes | Clostridiaceae |
| 7 | Eubacterium rectale | Firmicutes | Lachnospiraceae | 7 | Enterococcus faecalis | Firmicutes | Enterococcaceae |
| 8 | Tetrasphaera | Actinobacteria | Intrasporangiaceae | 8 | Prevotella buccae | Bacteroidetes | Prevotellaceae |
| 9 | Anaerobiospirillum | Proteobacteria | Succinivibrionaceae | 9 | Clostridium sp. | Firmicutes | Clostridiaceae |
| 10 | Ruminococcus gnavus | Firmicutes | Lachnospiraceae | 10 | Propionispira | Firmicutes | Acidaminococcaceae |
| 11 | Ruminococcus hydrogenotrophica | Firmicutes | Lachnospiraceae | 11 | Clostridium sp. | Firmicutes | Clostridiaceae |
| 12 | Colwellia rossensis | Proteobacteria | Alteromonadaceae | 12 | Rickettsia bellii | Proteobacteria | Caedibacteraceae |
| 13 | Ruminococcus gnavus | Firmicutes | Lachnospiraceae | 13 | arabacterioides | Bacteroidetes | Bacteroidaceae |
| 14 | Paenebacillus | Firmicutes | Paenibacillaceae | 14 | Prevotella | Bacteroidetes | Prevotellaceae |
| 15 | Clostridium elementii | Firmicutes | Clostridiaceae | 15 | Blastococcus | Actinobacteria | Geodermatophilaceae |
| 16 | Ruminococcus sp. | Firmicutes | Lachnospiraceae | 16 | Halanaerobacter chitinivorans | Firmicutes | Halobacteroidaceae |
| 17 | Chryseobacterium sp. | Bacteroidetes | Flavobacteriaceae | 17 | Prevotella sp. | Bacteroidetes | Prevotellaceae |
| 18 | Roseburia inulinivorans | Firmicutes | Lachnospiraceae | 18 | Wolbachia sp. | Proteobacteria | Anaplasmataceae |
| 19 | Clostridium carvoxi | Firmicutes | Clostridiaceae | 19 | Clostridium sp. | Firmicutes | Clostridiaceae |
| 20 | Oribacterium | Firmicutes | Lachnospiraceae | 20 | Clostridium sp. | Firmicutes | Clostridiaceae |
Figure 4.
Paired ranked difference plots of the top two control and irritable bowel syndrome (IBS) species. The x-axis indicates the individual samples. The y-axis shows the paired ranked difference in species abundance, highest rank being 8300 and lowest 6000, of the corresponding control and IBS samples. Plots (A) & (B) show the first and second ranked control species. Plots (C) & (D) show the first and second ranked IBS species.
Among the significant observed species within our IBS and control groups, we wanted to next characterize whether or not these tended to belong to certain families. To do this, we needed to take into consideration that all families are not equally represented on the PhyloChip and account for that bias when estimating the significance of the families present in the 2700 species of interest. We used the hypergeometric test, which focuses only on the number of significantly present probes and ignores the magnitude of changes of the fluorescence intensity, allowing us to observe whether or not the families appear as significant in proportion to their presence. The top 200 as most significantly different ranked species were analyzed to test whether the 200 most present species within each of the IBS and control groups are of a significant family and phyla. Tables 2 and 3 show the results of the hypergeometric family analysis. Notably, the 200 significant phylotypes in both IBS and control each do not appear to be a random representation of the 2700 member universe. The results showed that the families of Oxalobacteraceae, Prevotellaceae, Burkholderiaceae, Sphingobacteriaceae were significantly overrepresented in IBS rat. Conversely, the most significantly enriched family in control rats were Lachnospiraceae, including Ruminococcus sp., followed by Erysipelotrichaeceae and Clostridiaceae. There was also clearly a high increase in many Bacteroidetes in the IBS model. Applying the hypergeometric test on the phyla level (Tables 2 and 3) supported the qualitative observation of Fig. 3 showing the higher representation of Bacteroidetes in the IBS model, and the higher representation of Firmicutes in the healthy controls.
Table 2.
Hypergeometric test data for the IBS-induced rats
| Family | Expected A | Observed A | Expected B | Observed B | Observed sum |
|---|---|---|---|---|---|
| Acidobactriaceae | 18 | 18 | 1 | 1 | 19 |
| Bacillaceae | 67 | 72 | 5 | 0 | 72 |
| Bacteroidaceae | 22 | 22 | 2 | 2 | 24 |
| Burkholderiaceae | 13 | 11 | 1 | 3 | 14 |
| Clostridiaceae | 186 | 190 | 13 | 9 | 199 |
| Enterobacteraceae | 37 | 40 | 3 | 0 | 40 |
| Flavobacteriaceae | 38 | 33 | 3 | 8 | 41 |
| Lachnospiraceae | 289 | 299 | 20 | 10 | 309 |
| Oxalobacteracae | 7 | 4 | 1 | 4 | 8 |
| Paenibacillaceae | 13 | 14 | 1 | 0 | 14 |
| Peptococcuaceae | 37 | 38 | 3 | 2 | 40 |
| Peptostreptococcaceae | 49 | 52 | 3 | 0 | 52 |
| Prevotellaceae | 54 | 36 | 4 | 22 | 58 |
| Rhodobacteraceae | 39 | 40 | 3 | 2 | 42 |
| Sphingomondaceae | 22 | 22 | 1 | 1 | 23 |
| Spirochaetaceae | 32 | 30 | 2 | 4 | 34 |
The family that is most significantly over-represented is indicated in bold. The family that is most significantly under-represented is indicated in bold and italics.
Table 3.
Hypergeometric test data for the control rats
| Family | Expected A | Observed A | Expected B | Observed B | Observed sum |
|---|---|---|---|---|---|
| Acidobacteriaceae | 18 | 19 | 1 | 0 | 19 |
| Aerococcaceae | 18 | 19 | 1 | 0 | 19 |
| Altermonadaceae | 27 | 28 | 2 | 1 | 29 |
| Bacillaceae | 66 | 69 | 4 | 1 | 70 |
| Bacteroidaceae | 23 | 24 | 1 | 0 | 24 |
| Bifidobacteriaceae | 12 | 13 | 1 | 0 | 13 |
| Bradyrhizobiaceae | 11 | 12 | 1 | 0 | 12 |
| Burkholderiaceae | 13 | 14 | 1 | 0 | 14 |
| Clostridiaceae | 183 | 182 | 12 | 13 | 195 |
| Comamonadaceae | 21 | 22 | 1 | 0 | 22 |
| Desulfovibrionaceae | 18 | 18 | 1 | 1 | 19 |
| Enterobacteraceae | 35 | 36 | 2 | 1 | 37 |
| Erysipelotrichaceae | 9 | 9 | 1 | 1 | 10 |
| Flavobacteriaceae | 35 | 36 | 2 | 1 | 37 |
| Flexibacteraceae | 23 | 24 | 1 | 0 | 24 |
| Helicobacteraceae | 10 | 11 | 1 | 0 | 11 |
| Lachnospiraceae | 244 | 222 | 16 | 38 | 260 |
| Lactobacillaceae | 37 | 38 | 2 | 1 | 39 |
| Peptostreptococcaceae | 46 | 47 | 3 | 2 | 49 |
The family that is most significantly over-represented is indicated in bold. The family that is most significantly under-represented is indicated in bold and italics.
DISCUSSION
As we have shown previously,31,32 rats sensitized with acetic acid developed colonic hypersensitivity as adults. In addition, we now show that these rats have decreased water content of their stool and a trend towards fewer stools. Thus, these rats have a colonic phenotype that models human IBS, perhaps of the constipation-predominant type. The most important findings of our study relate to the use of the 16S rRNA PhyloChip, a high-density microarray that represents a relatively new, culture-independent approach with several advantages over customary alternatives to studying bacterial communities – it is rapid, replicable, highly sensitive (capable of detecting taxa in parallel to as little as 0.01% of the total abundance and can identify approximately 8500 bacterial taxa (defined as groups of microorganisms that share at least 97% 16S rRNA sequence identity) in a single assay. The Phylo-Chip therefore represents a powerful technique to study alterations in disease states or in response to interventions, as has recently been described. We used this approach to profile the bacterial community in a well-established rodent model of irritable bowel syndrome with the intent of gaining insight into possible clues to the pathogenesis of this disorder. Our results suggest that there are several significant changes in the colonic microflora of diseased rats that are generally consistent with what has been described in the literature of human IBS.
Our initial principal component analysis suggested marked differences between the two experimental groups, with most of the variation explained by the relative abundance of species and specific differences in microbial phylogeny. We collected samples from several regions along the longitudinal axis of the colon and the radial axis (i.e. mucosal vs luminal) as regionally distinct selective conditions may influence bacterial ecology at a given point in the longitudinal or radial axis.42 Further, distinct ‘functional’ roles have been suggested for ‘planktonic’ (luminal, free-living) bacteria versus those associated with the mucosa, with a clearer role for the former in fermentation and digestion of nutrients. On the other hand, mucosaadherent bacteria may play a greater role in epithelial health and modulation of mucosal immune responses.43 Distinct microbial populations have been previously observed in healthy individuals.44–46 Our results showed that neither longitudinal nor radial axial sampling made a major contribution to the observed differences between the IBS and control groups, suggesting global changes in the enteric microflora with the induction of IBS, compatible with what has recently been reported in humans with IBS.30 However, our sample size may have been too small to detect a statistically significant difference, and subtle changes may have been missed.
Amongst the most significant changes observed are those related to the composition of the microbial community. The microflora of IBS-induced rats is significantly more diverse relative to that of control rats. The literature on humans with IBS is conflicted in this regard with some studies reporting increased diversity, 27,47 while others have suggested a decrease.28,30 Even more striking was the dramatic change in the ratio of Firmicutes to Bacteroidetes, two of the major phyla found in the colon. Again the human literature, although sparse, is contradictory with some authors suggesting that a decrease in Firmicutes is characteristic of IBS47 although a recent report suggests the opposite.28 The significance of a change in the ratio of these two major phyla is not well understood although it has also been described in the context of obesity, with abundance of Firmicutes being associated with an increased body mass index (BMI)48 and aging.49 The relevance of these changes to the development of the IBS phenotype is not known, but theoretically, differences in the metabolic profile of these two phyla may be significant with Bacteroidetes genomes containing a larger number of genes encoding for glycan-degrading enzymes relative to Firmicutes.50
Our study also revealed differences at the level of bacterial families and species in a direction that is both similar and different than what has been described in humans with various subtypes of IBS. These include various species of Prevotella, Clostridium, Streptococcus, Roseburia and Ruminoccocus and the Lachnospiraceae family to name a few.27–29,51 However, as with the human studies, it is difficult to infer a specific role for these changes in the pathogenesis of the IBS phenotype without performing additional studies. Further, it can be argued that changes at the taxonomic level are less relevant than the global metabolic profile represented by the gene diversity in the bacterial community, which may not correlate with traditional microbiological classifications.
A fundamental question is the relationship of these phylogenetic differences to the behavioral phenotype of hypersensitivity to colonic distention, which we have previously shown in several reports.31,32 A recent report suggested in a very non-specific manner that fecal flora was altered in neonatal rats subject to maternal separation and this was accompanied by colonic hypersensitivity.52 In our study, control and sensitized pups are handled in the same way so stress by itself is unlikely to be a cause. It is possible that the mild inflammation (or its chronic consequences in terms of remodeling) caused by acetic acid may permit an aberrant colonization pattern in the colon. It is also possible that sensitization and abnormal activity of spinal afferents in the colon alters the local microenvironment facilitating the growth of certain bacterial types at the expense of others. Further experiments will be required to determine the pathogenesis of these changes in the enteric microflora and whether they in turn play a causative role in colonic hypersensitivity.
In summary, the most important contribution of this study is the finding that the use of a novel technique (PhyloChip) provides a relatively simple way to track changes in enteric microflora. Secondly, significant changes in the enteric microflora can be found in a rat model associated with the equivalent of a human IBS phenotype (colonic hypersensitivity). Many of these changes we observed are compatible with what has been described in humans. Further, it is hoped that the description of this model and technique will facilitate the stage for rigorous scientific studies to elucidate the cause and effect relationship of changes in enteric microflora to alterations in colonic physiology in this condition as these studies may be difficult or impossible to do in humans.
Acknowledgments
Supported by a grant from the Bio-X Interdisciplinary Initiative Program IIP4-32 (PI: Pasricha; co-investigator: Spormann) and NIH-5-R01GM086884-2 (PI: Holmes).
Work performed at Lawrence Berkeley National Laboratory was supported by the U.S. Department of Energy contract number DE-AC02-05CH11231.
Footnotes
AUTHOR CONTRIBUTIONS
TAN designed, planned and performed the experiments, authored and wrote the manuscript; SH planned, designed and performed the statistical analysis; participated in writing and edited the manuscript; AVA performed the statistical analysis; MS performed experiments; TDS supervised the design and use of the PhyloChip micorarray; CW supervised the design and use of the PhyloChip microarray; GLA supervised the design and use of the PhyloChip microarrayl; JW designed and performed the experiments on stool output and water content; JS aided in the design of the experiments; PJP conceived and designed the study, supervised experiments, authored and edited the manuscript; AS conceived and designed the study, supervised experiments, authored and edited the manuscript.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Data S1. Statistical methods used to analyze PhyloChip data.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Hahn BA, Yan S, Strassels S. Impact of irritable bowel syndrome on quality of life and resource use in the United States and United Kingdom. Digestion. 1999;60:77–81. doi: 10.1159/000007593. [DOI] [PubMed] [Google Scholar]
- 2.Hungin APS, Chang L, Locke GR, Dennis EH, Barghout V. Irritable bowel syndrome in the United States: prevalence, symptom patterns and impact. Aliment Pharmacol Ther. 2005;21:1365–75. doi: 10.1111/j.1365-2036.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- 3.American College of Gastroenterology Task Force of Irritable Bowel Syndrome. An evidence-based position statement on the management of irritable bowel syndrome. Am J Gastroenterol. 2009;104(Suppl 1):S1–35. doi: 10.1038/ajg.2008.122. [DOI] [PubMed] [Google Scholar]
- 4.Saito YA, Locke GR, Talley NJ, et al. A comparison of the Rome and Manning criteria for case identification in epidemiological investigations of irritable bowel syndrome. Am J Gastroenterol. 2000;95:2816–24. doi: 10.1111/j.1572-0241.2000.03192.x. [DOI] [PubMed] [Google Scholar]
- 5.Thompson WG, Irvine EJ, Pare P, et al. Functional gastrointestinal disorders in Canada: first populationbased survey using Rome II criteria with suggestions for improving the questionnaire. Dig Dis Sci. 2002;47:225–35. doi: 10.1023/a:1013208713670. [DOI] [PubMed] [Google Scholar]
- 6.Drossman DA, Zhiming L, Andruzzi E, et al. US householders survey of functional gastrointestinal disorders: prevalence, sociodemography, and health impact. Dig Dis Sci. 1993;38:1569–80. doi: 10.1007/BF01303162. [DOI] [PubMed] [Google Scholar]
- 7.Hungin AP, Whorwell PJ, Tack J, Mearin F. The prevalence, patterns and impact of irritable bowel syndrome: an international survey of 40 000 subjects. Aliment Pharmacol Ther. 2003;17:643–50. doi: 10.1046/j.1365-2036.2003.01456.x. [DOI] [PubMed] [Google Scholar]
- 8.Shanahan F. Irritable bowel syndrome: shifting the focus toward the gut microbiota. Gastroenterology. 2007;133:340–2. doi: 10.1053/j.gastro.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 9.Dunlop SP, Hebden J, Campbell E, et al. Abnormal intestinal permeability in subgroups of diarrhea predominant irritable bowel syndromes. Am J Gastroenterol. 2006;101:1288–94. doi: 10.1111/j.1572-0241.2006.00672.x. [DOI] [PubMed] [Google Scholar]
- 10.Barbara G. Mucosal barrier defects in irritable bowel syndrome. Who left the door open? Am J Gastroenterol. 2006;101:1295–8. doi: 10.1111/j.1572-0241.2006.00667.x. [DOI] [PubMed] [Google Scholar]
- 11.Aerssens J, Camilleri M, Talloen W, et al. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2008;6:194–205. doi: 10.1016/j.cgh.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbara G, Stanghellini V, De Giorgio R, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 13.Barbara G, Stranghellini V, Cremon C, De Giorgio R, Corinaldesi R, et al. Almost all irritable bowel syndromes are post-infectious and respond to probiotics: controversial issues. Dig Dis. 2007;25:245–8. doi: 10.1159/000103894. [DOI] [PubMed] [Google Scholar]
- 14.Ohman L, Isaksson S, Lundgren A, Simren M, Sjovall HA. A controlled study of colonic immune activity and beta7+ blood T lymphocytes in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:980–6. doi: 10.1016/s1542-3565(05)00410-6. [DOI] [PubMed] [Google Scholar]
- 15.Chadwick VS, Chen WX, Shu DR, et al. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778–83. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- 16.Gwee KA, Collins SM, Read NW, et al. Increased rectal mucosal expression of interleukin 1beta in recently acquired post-infectious irritable bowel syndrome. Gut. 2003;52:523–6. doi: 10.1136/gut.52.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinan TG, Quigley EM, Ahmed SM, et al. Hypothalamic-pituitary-gut axis dysregulation in irritable bowel syndrome: plasma cytokines as a potential biomarker? Gastroenterology. 2006;130:304–11. doi: 10.1053/j.gastro.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Guslandi M. Probiotic agents in the treatment of irritable bowel syndrome. J Int Med Res. 2007;35:583–9. doi: 10.1177/147323000703500501. [DOI] [PubMed] [Google Scholar]
- 19.King TS, Elia M, Hunter JO. Abnormal colonic fermentation in irritable bowel syndrome. Lancet. 1998;352:1187–9. doi: 10.1016/s0140-6736(98)02146-1. [DOI] [PubMed] [Google Scholar]
- 20.Treem WR, Ahsan N, Kastoff G, Hyams JS. Fecal short-chain fatty acids in patients with diarrhea-predominant irritable bowel syndrome: in vitro studies of carbohydrate fermentation. J Pediatr Gastroenterol Nutr. 1996;23:280–6. doi: 10.1097/00005176-199610000-00013. [DOI] [PubMed] [Google Scholar]
- 21.Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G. The fecal microbial population in the irritable bowel syndrome. Microbiologica. 1982;5:185–94. [PubMed] [Google Scholar]
- 22.Bayliss CE, Bradley HK, Jones VA, Hunter JO. Some aspects of colonic microbial activity in irritable bowel syndrome associated with food intolerance. Ann Ist Super Sanita. 1986;22:959–63. [PubMed] [Google Scholar]
- 23.Bradley HK, Wyatt GM, Bayliss CE, Hunter JO. Instability in the faecal flora of a patient suffering from food-related irritable bowel syndrome. J Med Microbiol. 1987;23:29–32. doi: 10.1099/00222615-23-1-29. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt GM, Bayliss CE, Lakey AF, Bradley HK, Hunter JO, Jones VA. The faecal flora of two patients with food-related irritable bowel syndrome during challenge with symptom-provoking foods. J Med Microbiol. 1988;26:295–9. doi: 10.1099/00222615-26-4-295. [DOI] [PubMed] [Google Scholar]
- 25.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–9. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 26.Malinen E, Rinttila T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy with real-time PCR. Am J Gastroenterol. 2005;100:373–82. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 27.Kassinen A, Krogiuskurikka L, Makivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95–105. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyra A, Rinttila T, Nikkila J, et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol. 2009;15:5936–45. doi: 10.3748/wjg.15.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Codling C, O’Mahony L, Shanahan F, Quigley E, Marchesi J. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 2010;55:392–7. doi: 10.1007/s10620-009-0934-x. [DOI] [PubMed] [Google Scholar]
- 31.Winston J, Shenoy M, Medley D, Naniwadekar A, Pasricha PJ. The vanilloid receptor initiates and maintains colonic hypersensitivity induced by neonatal colon irritation in rats. Gastroenterology. 2007;132:615–27. doi: 10.1053/j.gastro.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Xu GY, Winston JH, Shenoy M, Zhou S, Chen JD, Pasricha PJ. The endogenous hydrogen sulfide producing enzyme cystathionine-beta synthase contributes to visceral hypersensitivity in a rat model of irritable bowel syndrome. Mol Pain. 2009;5:44. doi: 10.1186/1744-8069-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson KH, Wilson WJ, Radosevich JL, et al. High-density microarray of small-subunit ribosomal DNA probes. Appl Environ Microbiol. 2002;68:2535–41. doi: 10.1128/AEM.68.5.2535-2541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane DJ. 16S/23S rRNA Sequencing. Chichester, United Kingdom: Wiley; 1991. [Google Scholar]
- 35.Brodie EL, DeSantis TZ, Dominique JC, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–98. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda N, Church GM. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J Bacteriol. 2002;184:6225–34. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 38.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 40.Rivals I, Personnaz L, Taing L, Potier MC. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2007;23:401–7. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 41.Schatz MC, Phillippy AM, Gajer P, DeSantis TZ, Andersen GL, Ravel J. Integrated microbial survey analysis of prokaryotic communities for the PhyloChip microarray. Appl Environ Microbiol. 2010;76:5636–8. doi: 10.1128/AEM.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camp J, Kanther M, Semova I, Rawls J. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langlands SJ, Hopkins MJ, Coleman N, Cummings JH. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut. 2004;53:1610–6. doi: 10.1136/gut.2003.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macfarlane S, Macfarlane G. Bacterial diversity in the human gut. Adv Appl Microbiol. 2004;54:261–89. doi: 10.1016/S0065-2164(04)54010-8. [DOI] [PubMed] [Google Scholar]
- 45.Macfarlane S, Hopkins M. Bacterial growth and metabolism on surfaces in the large intestine. Microb Ecol Health Dis. 2000;12:64–72. [Google Scholar]
- 46.Eckburg P, Bik E, Bernstein C, Purdom E. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoetendal EG, Rajilic-Stojanovic M, De Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–15. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 48.Ley RE, Turnbaugh PJ, Klein S, Gordon J. Microbial ecology – human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 49.Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123–8. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahowald MA, Rey FE, Seedorf H, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A. 2009;106:5859–64. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajilic’-Stojanovic’ M. Laboratory of microbiology; PhD thesis. Wageningen, The Netherlands: Wageningen University; 2007. Diversity of the human gastrointestinal microbiota: novel perspectives from high throughput analyses; pp. 1–214. [Google Scholar]
- 52.O’Mahony SM, Marchesi JR, Scully P, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263–7. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]