Abstract
The molecular events leading to human embryonic stem cell (hESC) differentiation are the subject of considerable scrutiny. Here, we characterize an in vitro model that permits analysis of the earliest steps in the transition of hESC colonies to squamous epithelium on basic fibroblast growth factor withdrawal. A set of markers (GSC, CK18, Gata4, Eomes, and Sox17) point to a mesendodermal nature of the epithelial cells with subsequent commitment to definitive endoderm (Sox17, Cdx2, nestin, and Islet1). We assayed alterations in the transcriptome in parallel with the distribution of immunohistochemical markers. Our results indicate that the alterations of tight junctions in pluripotent culture precede the beginning of differentiation. We defined this cell population as “specified,” as it is committed toward differentiation. The transitional zone between “specified” pluripotent and differentiated cells displays significant up-regulation of keratin-18 (CK18) along with a decrease in the functional activity of gap junctions and the down-regulation of 2 gap junction proteins, connexin 43 (Cx43) and connexin 45 (Cx45), which is coincidental with substantial elevation of intracellular Ca2+ levels. These findings reveal a set of cellular changes that may represent the earliest markers of in vitro hESC transition to an epithelial phenotype, before the induction of gene expression networks that guide hESC differentiation. Moreover, we hypothesize that these events may be common during the primary steps of hESC commitment to functionally varied epithelial tissue derivatives of different embryological origins.
Introduction
Dissecting the process of human embryonic stem cell (hESC) differentiation is of considerable interest due to its biological significance as well as its therapeutic potential, such as in cell replacement therapy [1]. The onset of differentiation in hESCs, however, remains poorly understood. Most of the large-scale gene expression studies of hESC differentiation, for example, were performed on embryoid bodies (EBs) [2,3], the stage when the presence of all embryonic and extra-embryonic layers is already manifested. Evidently, inhibition of the basic fibroblast growth factor (FGF2 or bFGF) signaling pathway [4] as well as FGF2 withdrawal cause a mild and gradual differentiation [5], thus leading to detectable changes in gene expression patterns before visible morphological changes [2]. Therefore, in order to detect signals or processes responsible for the earliest steps in hESC differentiation, we induced specification by omitting FGF2 from the experimental cultures (FGF2−). After the first detection of differentiation, before the manifestation of embryonic and extra-embryonic layers, cell colonies were determined as having a “specified” fate.
We established that in our experimental conditions all hESC lines were committed to differentiate into a simple (squamous) epithelium with the appearance of thin, flat, elliptical cells, with subsequent development of definitive endoderm (DE) and trophectodermal markers.
The epithelium is a tissue that lines the cavities and surfaces throughout the body. In general, it is accepted that the epithelium arises from the cells which originate from the inner cell mass (ectoderm, endoderm, mesoderm, and extra-embryonic endoderm) and those from the trophectoderm (TE) (extra-embryonic ectoderm). The primitive endoderm forms a layer on the top of the inner cell mass (ICM) in blastocysts and diverges to visceral and parietal extraembryonic endoderm, thus contributing to respective components of the yolk sac. The DE is one of the 3 principal germ layers formed in epiblasts that gives rise to the epithelial lining of the respiratory and digestive tracts contributing to the lungs, liver, thyroid, thymus, and pancreas. TE cells function as progenitor cells of a trophoblast lineage. Although the capacity of hESCs to differentiate into the trophoblast has been known for some time, it has mainly been characterized by human chorionic gonadotropin (hCG) secretion and reverse transcription–polymerase chain reaction (RT-PCR) data [6].
Here, we show that the alterations of tight junctions (TJs) precede the beginning of differentiation. In addition, we observe that significant up-regulation of keratin-18 and decrease in the functional activity of gap junctions, along with down-regulation of 2 gap junction proteins, connexin 43 (Cx43) and connexin 45 (Cx45), are coincident with substantial elevation of intracellular Ca2+ levels during the onset of differentiation. Our results reveal a “chain of events” highlighting development of characteristic immunohistochemical (IHC) markers in the context of cellular morphology. Based on our findings, we hypothesize that inter- and intracellular changes, including cell junctions, adhesion, and remodeling of cytoskeleton, may precede the activation of germ layer specific transcription factors on differentiation to squamous epithelium. Additionally, we suggest that primary differentiation events highlighted in our study may be a general phenomenon for hESC commitment to the functionally varied epithelial tissues from different embryological origins. These new insights of the earliest onset of hESC differentiation will facilitate the maintenance of hESCs in a pluripotent state and enhance the directed differentiation along epithelial lineages and DE, in particular.
Materials and Methods
Cells cultures
hESC lines H1, H9, H7, H14 (WiCell), CM3, CM7, CM14 (established at CMRC), and 2 iPS lines SR2 and DSV2 were used for this study. SR2 and DSV2 induced plouripotent stem cell (iPSC) lines were derived from MRC-5 fibroblasts (ATCC) and AG06872 fibroblasts (Coriell), by over-expressing Oct4, Sox2, Nanog, and cMyc using retroviral vector plasmids (manuscript submitted). The cells were grown in StemPro medium (Invitrogen) on a matrigel substrate (BD Bioscience). The confluent cultures of hESCs growing on 10 cm dishes were split to 6 experimental 10 cm dishes mechanically by using the StemPro EZ Passage tool (Invitrogen). Control cell samples were taken at the time of cell splitting (DC). FGF was withdrawn (FGF–)the day after plating (D0), whereas other dishes were grown with 20 ng/mL of FGF (FGF+). The culture medium was collected from the dishes regularly and stored at–80°C. For RNA analysis, confluent cultures were lifted by using 0.05% trypsin (Cellgro) at different days of proliferation (D2, D4, D6 etc.), washed in ice-cold phosphate-buffered saline (PBS), and pellet stored at −80°C. RNA was collected at the time point when (FGF−) cultures exhibited the first sights of differentiation distinguished at about 5% of plating area. TPP (Techno Plastic Products, Trasadingen, Switzerland) tube counters were used to count cells in clusters at the time of re-plating, and a single-cell suspension was counted with the help of an automatic cell counter, Nexcelom (SelectScience), during cell lifting for the analysis. Cell confluence and morphology was observed daily.
Immunocytochemistry
The cells were washed with PBS, fixed with 4% paraformaldehyde (PFA) for 5 min. In order to establish the expression of Oct-4, hESC were additionally permeabilized with cold (−20°C) methanol for 5 min. After 30 min incubation at 37°C with blocking solution (Protein Block), cells were incubated with primary antibodies (1:100) for 1 h at 37°C. Immunostaining was done with secondary antibodies conjugated with fluorescein isothiocyanate or tetramethyl-rhodamine iso-thiocyanate (TRITC) (1:100) and incubated for 1 h at 37°C. Similarly, rabbit polyclonal anti-human Oct-4 (Santa Cruz), connexin 43 (Chemicon), and connexin 45 (Chemicon) antibodies were used to establish expression of these marker proteins (see Supplementary Data for the list of antibodies; Supplementary Data are available online at www.liebertonline.com/scd). Mounting medium containing 4', 6-diamidino-2-phenylindole (Vector Laboratories) was used for counterstaining nuclei, and the specimens were visualized with a Nikon fluorescent microscope. Alkaline phosphatase substrate kit (Vector Laboratories) was used for detection of alkaline phosphatase activity according to the manufacturer's instructions.
Scrape loading/dye transfer assay
Briefly, the cells were washed in a PBS buffer, which was replaced with a solution of Lucifer yellow (1 mg/mL, Sigma) and rhodamine–dextran (1 mg/mL; Molecular Probes) in PBS. The colonies were dissected with a scalpel blade and incubated for 5 min with a dye. Due to its low molecular weight (522 Da), Lucifer yellow diffuses from cell to cell through functional gap junctions. On the other hand, rhodamine–dextran (RD, 10,000 Da) is too large to diffuse through gap junctions, and, thus, served as a negative control (Supplementary Fig. S2[1, 2C]). The dye diffusion was documented on epifluorescence images, which were acquired on a Leica DM IRB inverted microscope system (Wetzlar) by using a Hamamatsu ORCA-ER digital camera controlled with Improvision Openlab software version 5.0.2. Scale bars were calibrated to each objective magnification and added after acquisition. Light microscopic images were acquired with a Nikon D100 digital SLR camera on an inverted Leica DM IRB microscope.
Manipulation and measurement of internal Ca2+ concentration
To image changes in intracellular Ca2+, hESCs were loaded with Fluo4 NW by using Calcium Assay Kit F 36206 (Molecular Probes) in solution supplemented with 2.5 mM Probenecid (Molecular Probes) for 40 min. at room temperature. Confocal images were acquired by using a Zeiss LSM 510 META Laser Scanning Microscope system with a 488 nm argon lazer. By varying the width of the pinhole of the detectors, the observed fluorescence was localized to a known thickness of observed tissue, the depth of the field was transmitted, and the differential interference contrast (DIC) images were adjusted. Scale bars were integrated into the image during acquisition. Epifluorescence images were acquired on a Leica DM IRB inverted microscope system with excitation 494 nm and emission at 516 nm, by using a Hamamatsu ORCA-ER digital camera controlled with Improvision Openlab software version 5.0.2. Scale bars were calibrated to each objective magnification and added after acquisition. The detected increase of Ca+ concentration was within the molar range of 100–200 nmole. All images were imported into Adobe Photoshop (Adobe System) for final image composition and contrast adjustment.
Total mRNA isolation
Total mRNA isolation from the cell lines was performed with the PureZOL RNA isolation reagent (Bio-Rad), according to the manufacturer's instructions. Purity and integrity of the isolated RNA was assessed on the ND-1000 Spectrophotometer (Thermo Fisher Scientific).
Real-time PCR
Real-time quantitative RT-PCR was performed by using an IQ5 Cycler (Bio-Rad Laboratories) according to the manufacturer's instructions. Primers were designed by using the Primer Express program version 1.5 (Applied Biosystems), and obtained from Integrated DNA Technologies (Coralville). The specificity of the primers was documented by RT-PCR and resulted in a single product with the predicted length. cDNAs were constructed by using IScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer's instruction. Reactions were performed by using IQ SYBR Green Supermix kit (Bio-Rad Laboratories) according to the manufacturer's instruction. Each reaction was performed in triplicate, using 250 nM primers, cDNA sample corresponding to 0.25 ng of total RNA, in a total volume of 25 μL. 100 nM primers for GUSB RNA (RealTimePrimers.com) were used as a reference for each of the cDNA samples. The PCR conditions were as follows: 1 cycle at 95°C for 3 min, 40 cycles at 95°C for 30 s, 60°C for 30 s, 72°C for 30 s, followed by a melting curve from 55°C to 95°C. We used the Livak method to calculate the relative gene expression levels (Bio-Rad Laboratories, Inc. Real-Time PCR Applications Guide p-41). Primer sequences are provided next. Keratin 18 (K18) L:TGAGACGTACAGTCCAGTCCTT; R:GCTCCATCTGTAGGGCGTAG. Chorionic gonadotropin alpha polypeptide (CGA L:CCCACTCCACTAAGGTCCAA; R:TCCATTCCAGAAAATCAGCA. Glial cells missing homolog 1 (GCM1) L:CTGAAGGGGAGCACAGAGAC; R:TCTGTGATTCCTCCCAGACC.
Microarray data preparation
Platform: NimbleGen array, 47634 tags (human). Images were processed in NimbleGen software, resulting in quantile-normalized signal intensities for each probe. Therefore, no additional preprocessing and normalization procedures were applied to the data.
Similarities between samples
Similarities between expression profiles of both FGF+ and FGF- cell lines were examined by using Cohen's kappa statistic at both “gene level” and “biological process level” [7,8]. For this purpose, we discretized the expression values as follows: signal intensity values less than global signal mean across 10 samples were considered non-expressed (value 0), and values exceeding global signal mean were considered expressed (value 1). For the gene level, we calculated pair-wise kappa values for all possible pairs of samples. Since kappa values have values less than 1 and the closer it is to 1, the stronger is the agreement between examined profiles, we used values 1-kij as distances for hierarchical clustering (average linkage was applied). In order to use “biological process level,” we made enrichment of “expressed” gene lists for all 10 samples by gene ontogeny (GO) processes and GeneGo processes. Processes with enrichment P values less than 0.05 were considered significant (value=1), others were assigned the zero value. Then, we calculated kappa values and conducted hierarchical clustering as just described.
Fold change analysis
The fold change (FC) value of every tag was calculated as signed ratio of its expression value in the FGF sample to the expression value in the corresponding FGF+ sample:
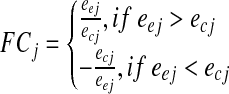 |
for each tag j, where e denotes expression value, e and c in subscripts represent corresponding case (FGF−) and control (FGF+) expression values. So, we worked with 5 samples of FC values, each corresponding to expression changes in a single cell line. We used a minimal absolute FC value of 1.5 as a threshold for significant expression change.
Enrichment analysis
In order to reveal common underlying functional changes during the onset of differentiation, we conducted pair-wise ontology enrichment analysis (EA) of the 5 gene lists (each gene list contain differentially expressed genes, which passed the 1.5-FC threshold for a correspondent pair of FGF2+\FGF2− cell lines) followed by comparison between distributions by Cohen Kappas. We tested several functional ontologies, including canonical pathway maps, GO processes, and disease biomarkers by using MetaCore data mining suite [9]. Significance of enrichment was assessed by using hypergeometric distribution (P value <0.05).
Hidden nodes analysis
We defined a gene as important for regulation of the gene list that we were interested in if the number of shortest paths between genes of interest containing the examined node was higher than expected by chance [10]. Significance of the node's importance was estimated by using hypergeometric distribution with parameters r, number of shortest paths between any 2 nodes of interest containing currently examined genes; R, total number of shortest paths between all possible pairs of genes of interest; n, total number of shortest paths between any 2 objects in the network containing examined gene; and N, total number of shortest paths between all possible pairs of objects in the database:
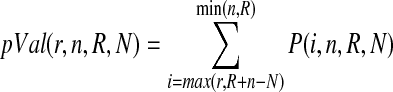 |
Differentiating pathways
We used a collection of more than 100,000 canonical pathways obtained from GeneGo's curated canonical pathway map collection. For each pathway, we calculated Euclidean distances between expression profiles of its participants in all possible pairs of the 5 FGF+ and 5 FGF- samples. Then, we addressed concordance in changes of the pathway's behavior by using t-test between sets of intra-group distances versus inter-group distances. Due to the very large number of pathways examined, we decided not to use multiple testing corrections to avoid significant loss of power. Pathways with t-test P value less than 0.001 were considered discriminative.
Results
Description and characterization of a model for early hESC differentiation
We have adapted FGF2 withdrawal as a model of the earliest changes in hESC differentiation through the simultaneous imaging of both undifferentiated and differentiating parts of the colonies, thus allowing the detection of a transitional zone (Materials and Methods). This modification allowed us to observe a gradient of differentiation, or consecutive steps, with undistorted morphology of the cells to establish sequential patterns of protein markers through immunohistochemistry (IHC). Additionally, undifferentiated cells imaged simultaneously with the cells undergoing differentiation served as an internal control for immunoassaying and other experimental techniques.
Even though stem cell lines vary significantly in their retention of pluripotency and display a tendency to spontaneously differentiate (data not shown), within 4 to 7 days, all 7 hESC lines and 2 iPSC lines growing in FGF2 conditions displayed similar morphological changes correlated with epithelial commitment (Supplementary Fig. S1, Supplementary Table S1). Five hESC lines of different ethnic backgrounds (H9, H7, H14; and CM7 and CM14, established at CMRC [11–13]) were subjected to transcriptome profiling analysis. Analysis of each corresponding pair of FGF2+/− cultures were performed at the time points when (FGF2−) cultures exhibited the first detectable morphological changes distinguished at approximately 5% of the culture area, which comprised less than 1% of the total cell number (considering lower cell density of flattened epithelial cells). The experiments were terminated at this point to ensure that the differentiated cells would not contribute significantly to transcriptome profiling analysis, which was designed to uncover the earliest events at the onset of differentiation.
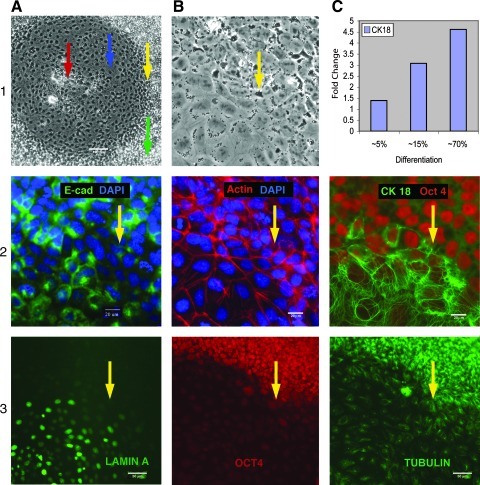
We assayed the process of transition of pluripotent cells to squamous epithelial morphology. Immunostaining with antibodies against Oct4, AP, SSEA4, TRA1–60, TRA1–81, and Sox2 reveals a gradual down-regulation of these pluripotency markers in the differentiating areas of the FGF2− colonies [Oct4 is shown (Figs. 1[3B]; 2C and Fig. 2[2B])] along with up-regulation of an early differentiation marker, lamin A/C (Fig. 1[3A]). Adherens junctions (AJs), a distinct feature of epithelial cells, extend into the cytoplasm of the adhering cells, through bonds between the cadherin-catenin machinery and the actin cytoskeleton [14]. Staining for E-cadherin showed that AJs were present, to some extent, in hESCs (as described in Ref. [15]), and became pronounced on differentiation (Fig. 1[2A]). At the same time, F-actin displayed prominent peripheral bands typical of epithelial cells on differentiation. Cytoplasmic stress fibers became apparent when cells acquired epithelial morphology (Fig. 1[1, 2B]). A circumferential actin belt, which extends around the inner surface of the cell membrane at the level of TJs, is critical for their formation and maintenance. B-Catenin was localized to cellular membranes and nuclei in differentiating cells, but in undifferentiated cells, it was predominantly present in the nuclei (Supplementary Fig. S2 [1, 2A]).
FIG. 1.
A general outline of differentiation is shown (1A). Blue arrow indicates a typical area of flat cells of squamous epithelium. Red arrow—the central clusters of advanced differentiation. Green arrow—undifferentiated cells, yellow arrow—the cells of transitional zone. E-Cadherin (CM14) staining showing that adherens junction was present in hESCs and compromised in transitional zone, but became pronounced on differentiation (2A). F-actin cytoskeleton (H14) extended around the inner surface of the cell membrane typical of the epithelial cells “circumferential actin belt.” Cytoplasmic stress fibers became apparent when cells acquired epithelial morphology (1, 2B). CK18 (H14) is drastically up-regulated on epithelial differentiation; overlay with Oct4 (2C). RT-PCR analysis (H9) of CK18 expression showing gradual increase of the proportion of epithelial cells on differentiation progression (1C). Expression of Lamin A/C emerging in differentiating areas (3A) concurrent with Oct 4 down-regulation (3B). A developing network of tubulin in the differentiated cells (H14) is shown (3C). hESC, human embryonic stem cell; RT-PCR, reverse transcription–polymerase chain reaction.
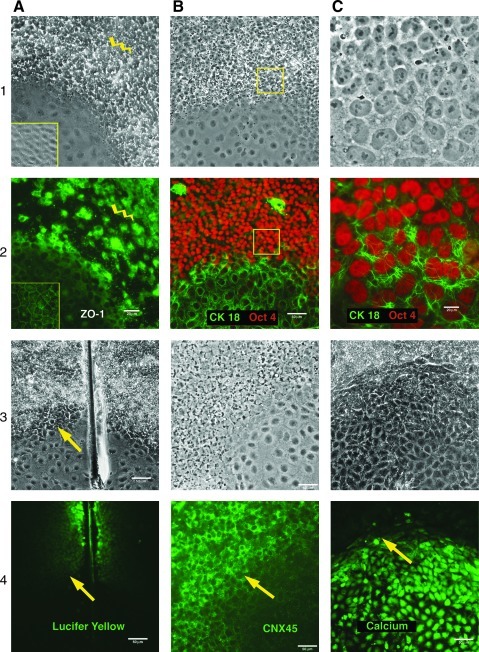
FIG. 2.
There was considerable expression of ZO-1 (1, 2A) in undifferentiated cultures (CM14); however, expression was lost near the border of differentiated areas (yellow lightening). ZO-1 was associated with tight junction as differentiation progressed to a defined epithelial phenotype (yellow inset). Oct4 (1, 2B) was down-regulated in differentiated area (H14); however, it remained intense within the zone of surrounding cells (pretransitional zone) where ZO-1 expression was altered [compare with (1, 2A)]. Higher magnification (1, 2C) of the cells in a transitional zone (yellow inset in 2B) with up-regulation of CK18 on the background of pluripotent morphology of hESCs with high levels of Oct4 expression (2C), although some evidence of cell “flattening” and nuclear elongation became apparent (1C). Scrape loading/dye transfer assay (CM14) (3, 4A). LY actively diffused through the functional gap junctions of the pluripotent cells; however, the LY diffusion was undetectable between the differentiated cells. The colonies (CM14) underwent a progressive loss of Connexin 45 expression in the differentiated cells (3, 4B). The green fluorescence (Fluo4) revealed significant elevation of intracellular Ca2+ level in the differentiating parts of the hESC colonies (H14) (3, 4C). The arrows indicate the border between the transitional zone and the differentiated cells. LY, lucifer yellow.
Membrane polarization, associated with specialized intercellular junctions, is a hallmark of epithelial cells. We immunostained ZO-1 protein to visualize TJs. There is considerable expression of ZO-1 in undifferentiated cultures as previously described [16]; however, it is lost near the border of differentiated areas before visible differentiation actually begins (Fig. 2[1, 2A]). Oct4 staining remains stable and intense within the same zone of surrounding cells (Fig. 2[1, 2B]). ZO-1 was down regulated in a transitional zone and became narrowly associated with TJs as differentiation progressed to a defined epithelial phenotype. The tubulin network is remodeled in differentiated areas (Fig. 1[3C]), whereas vimentin is absent in differentiated cells with some random traces in undifferentiated cultures (Supplementary Fig. S2[3,4C]). Nestin, routinely used for detection of neural stem cells but also characteristic for progenitor epithelial cells, is strongly expressed in differentiated regions (Fig. 3[1A]). We found a dramatic up-regulation of the simple epithelial marker CK18 in the differentiating cells (Fig. 1[2C]; Fig. 2[1, 2B]). Remarkably, CK18 expression is profound and emerges very early, before the down–regulation of Oct4 expression (Fig. 2[1, 2C]). Therefore, we used it as a quantitative measure of the percentage of cells that are undergoing differentiation by RT-PCR analysis. CK18 expression directly correlates with differentiation progression, thus showing a gradual increase of the proportion of epithelial cells after removal of bFGF (Fig. 1[1C]). Overall, the epithelial markers just described confirm our assessment of differentiation as a transition of pluripotent cells to the squamous epithelium phenotype.
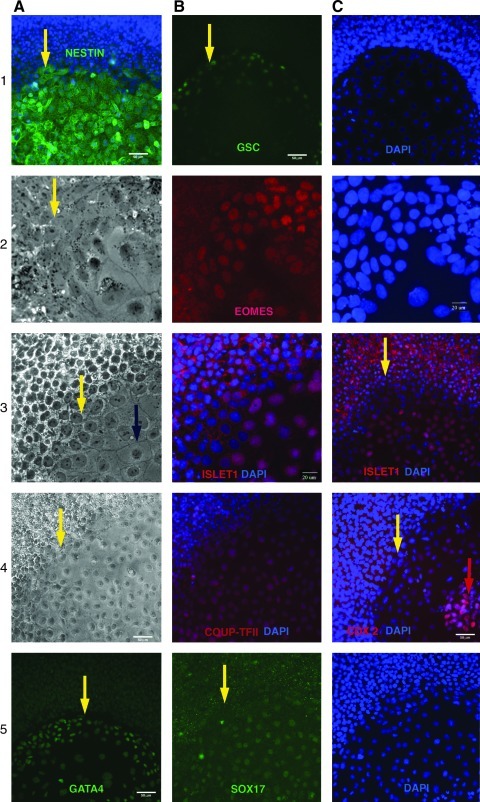
FIG. 3.
Lineage marker analysis. Marker of progenitor cells Nestin is up-regulated in epithelial cells (DSV2) overlay with DAPI is shown (1A). Mesendodermal markers Goosecoid protein (H14) (1B,C) and Eomes (DSV2) (2A–C) are expressed in the border with undifferentiated cells and are downregulated when the differentiation progresses, and cells acquire epithelial morphology. Markers of primitive and definitive endoderm: Gata4 (H14) is more pronounced in the regions adjusted to pluripotent cells (4A), COUP-TFII (H9) is expressed throughout the differentiated colony (4A, B) in a similar manner as Sox 17 (DSV2) (5B, C). Markers of definitive endoderm Islet 1 (3A–C) have a similar pattern and are expressed in the nuclei of a vast majority of differentiated epithelia cells (DSV2). Islet 1, however, also accumulated in the perimembrane space but not in the nuclei of undifferentiated cells. Cdx2 is primarily detected in the central (older) parts of differentiated colonies (H14) overlaid with DAPI (4C). Blue arrows indicate a typical area of flat squamous epithelial cells. Red arrows indicate central clusters of advanced differentiation. Yellow arrows indicate cells of transitional zones.
We pursued further investigation of gap junctions, whose involvement in early ESC differentiation has been recently suggested [17], and tested gap junction intercellular communication (GJIC) by the scrape loading/dye transfer assay as described [18] (Fig. 2[3, 4A]). As expected, GJIC appears to be active in undifferentiated cells. In stark contrast, however, the GJIC activity was undetectable in the differentiating cells in our model. Two gap junction proteins, Cx43 and Cx45, predominantly comprise gap junctions in hESCs [19]. In order to test whether Cx43 and Cx45 may be responsible for inactivation of GJIC at the onset of differentiation, we performed IHC staining of these proteins. Indeed, both connexins were down-regulated in the differentiating parts of the FGF2− colonies, along with the pluripotency markers (Fig. 2[3, 4B]; Supplementary Fig. S2[1, 2C]). Since decreases in overall cell coupling could trigger gap junction hemichannel-mediated Ca2+ waves, [20], we tested Ca2+ concentration in undifferentiated and differentiating parts of the FGF2− colonies. Ca2+ is one of the most ubiquitous second messengers in a wide variety of cell types [21], and there is an emerging body of evidence for an active role of Ca2+ signaling in stem cell development [22]. Importantly, we documented a significant increase in Ca2+ concentration within the molar range of approximately 100–200 nmole in the differentiating parts of the hESC colonies and in the cells of the transitional zone (Fig. 2[3, 4C]). These results suggest that the remodeling of TJs, decreases of GJIC, and a concurrent increase in intracellular Ca2+ concentration are molecular events that take place at the very beginning of hESC differentiation into a squamous epithelium.
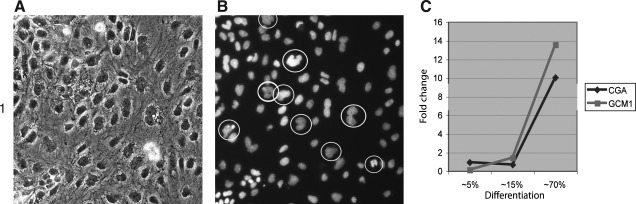
We further assayed the markers to describe the lineage commitment of epithelial differentiation in FGF free conditions and detected a set of markers that can be attributed to both TE and mesoendoderm specifications, such as Nestin (Fig. 3[1A]), gradual up-regulation of Cdx2 (Fig. 3[4C]), Eomes (Fig. 3[2A–C]), integrin alpha 6 (Supplementary Fig. S2[4B]), and a slight upregulaion of Gata3 (not shown). Expression of other markers of the trophoblast lineage, such as hCG (Supplementary Fig. S2[3A]) and glial cell missing protein (GCM1) (Supplementary Fig. S2[3B]), is relatively weak in IHC assay and was largely detected in the cells that gradually piled up in the center of flattened areas. However, RT-PCR detection showed notable up-regulation of CG alpha (10-fold) and GCM1 (14-fold) (Fig. 4C) when differentiation spread to 70% of the culture area. Additionally, multinucleated cells typical for syncytiotrophoblast were seen in the culture after several days of differentiation (Fig. 4A, B). However, TE marker SSEA1 was negative. Studying additional markers of mesendodermal specification, we detected Goosecoid protein GSC) expression (Fig. 3[1B, C]) and Eomes (Fig. 3[2A–C]) on the border with undifferentiated cells. Moreover, Eomes expression begins as soon as ES cells start to flatten and disappeared when they acquired epithelial morphology. Epithelial cell expressed endoderm markers COUP-TFII (Fig. 3[4A, B]), Gata4 (Fig. 3[5A]), Sox17 (Fig. 3[5B, C]), and DE marker Islet1 in the nucleus; whereas undifferentiated cells had the protein accumulated in perimembrane space (Fig. 3[3A–C]). Notably, (perimembrane) expression of Islet1 disappears in transitional zone, and then becomes nuclear in epithelial cells. Expression of Cdx2 became pronounced in central, more differentiated parts of the colonies (Fig. 3[4C]). Immunostaining for integrins reveals a uniform staining for alpha 5 (fibronectin receptor) and alpha 6 (receptor for laminin) at the basal cell membrane interface in undifferentiated cells, whereas alpha 5 showed a discontinuous staining (Supplementary Fig. S2[4A]) and alpha 6 distribution became polarized on differentiation. (Supplementary Fig. S2[4B]). Markers of extraembryonic endoderm Laminin β1, AFP, and Lefty were negative (not shown). We did not detect development of mesoderm markers such as ACTC1 and vimentin (Supplementary Fig. S2[3, 4C]). Neuronal ectoderm specification was ruled out by using antibodies against tubulin III, Map2; non-neuronal ectoderm: p63, Gata2, and Pankeratin (not shown). Several markers, such as Gata2, Gata4, Pankeratin, and Brachyury, had similar low background fluorescence in the differentiating areas and the rest of the colonies and were not informative. We did not detect any differences in these markers when comparing the pluripotent cells in FGF with FGF free conditions.
FIG. 4.
Multinucleated cells (H14) typical for syncytiotrophoblasts are seen in the epithelial culture (A, B). RT-PCR analysis (H9) showed notable up-regulation of CG alpha and GCM1 when differentiation spreads to 70% of culture aria (C). GCM1, glial cells missing homolog 1.
Functional analysis of expression profiles in the 5 cell line pairs
To continue our elucidation of the earliest events in hESC specification, we performed transcriptome profiling analysis. Since the number of statistically significant differentially expressed genes is relatively small even between undifferentiated hESCs and differentiating EBs [2,3], we focused on commonly altered signaling and metabolic pathways rather than on individual genes. We hypothesized that during the onset of differentiation, hESC lines of various origins may follow stepwise progression in common with each other, which are not necessarily reflected at the level of individual genes but rather at the level of pathways and pathway groups. The pathways are represented in our analysis by linear sequences of protein interactions that originate and terminate at biologically meaningful points (from membrane receptors or their ligands to transcriptional factors or their immediate targets). This approach is in line with a quickly evolving paradigm which states that, indeed, biologically related protein groups (pathways and networks), but not gene content per se, define molecular underpinnings for complex phenotypes. The “pathway paradigm” has become mainstream in several OMICs (genomics, proteomics, etc.) fields such as disease genome resequencing [23–25], genome wide association studies [26,27] and gene expression [28,29]. It was also shown that pathways and other functional groups represent a more suitable metric for comparison between OMICs datasets than gene content [30]. Further, applications of pathway descriptors have recently been positively received [31].
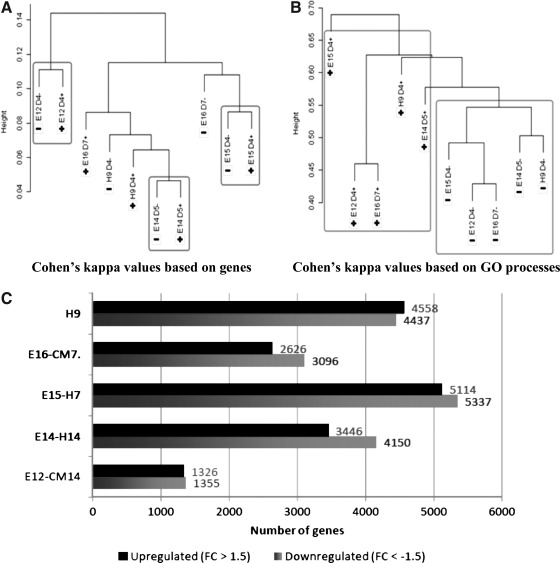
We first applied unsupervised clustering of the gene expression profiles from each of the FGF2+ and FGF2− pairs in order to evaluate similarities between them. Expression profiles in different cell lines appeared to be very heterogeneous, and clustering failed to separate the samples based on the FGF2 status. There were no significant differences in expression between FGF+ and FGF− groups by any statistical test we tried, including both paired and unpaired Welch t-test with false discovery rate (FDR) correction. Instead, most of the FGF2+ and FGF2− samples from each individual pair clustered together, thus suggesting that the individual differences among the hESC lines masked the common difference between the FGF2+ and FGF2− samples (Fig. 4A). In parallel, we applied a recently described method of unsupervised clustering of expression datasets based on ontology enrichment rather than on gene content [30] (see Material and Methods for details). Interestingly, the P value distribution pattern of pathways and processes did reflect the differentiation-related changes, separating the FGF2+ from FGF2− samples in all 5 pairs (Fig. 4B). A similar approach of functional descriptors (transcription regulation networks) was recently applied to distinguish expression profiles between human iPSCs and their parent somatic cell lines [32].
Next, to evaluate the hypothesis that during the onset of differentiation, hESC lines of various origins follow shared steps at the level of pathways rather than genes, we applied conventional statistical procedures to detect the genes with significantly altered expression patterns between the FGF2+ and FGF2− samples in all the pairs. In agreement with the results of unsupervised clustering, no such genes were detected by either Welch t-test or moderated t-test with empirical Bayesian shrinkage of variance [33] followed by FDR-controlling adjustment [34] with a conventional significance cutoff of 0.05. Omitting FDR adjustment, only 14 up-regulated and 7 down-regulated genes passed the threshold of 1.5- FC (Supplementary Table S2). Interestingly, TRPC4, 1 of the 14 up-regulated genes, is involved in transport of Ca2+ from extracellular space into cytosol [35].
In order to reveal common underlying functional changes during the onset of differentiation, we conducted pair-wise ontology enrichment analysis (EA) of the 5 gene lists (each gene list contains differentially expressed genes, which passed 1.5-FC threshold for a correspondent pair of FGF2+\FGF2− cell lines) (Fig. 4C) (see the details in the Material and Methods). Twenty-three pathway maps (MetaCore suite, GeneGo, Inc.) were significantly changed in all 5 pairs. Most pathways belonged to cytoskeleton and cell adhesion processes, followed by development- and immune-related pathways (Supplementary Fig. S3). We exported genes differentially expressed in at least 1 pair of FGF2+\FGF2− samples from pathway maps, which belong to cytoskeleton/cell adhesion (7 maps) and development-related processes (7 maps) (Table 1). The cytoskeleton/cell adhesion list had 217 genes, and the list of “development-related” genes contained 186 genes, with 56 genes shared between these 2 lists (Supplementary Table S3). We applied a recently developed algorithm of “hidden nodes” for deducing the most relevant upstream regulators for these genes [10]. We identified 771 and 1039 “hidden nodes” with P value <0.05 for the “cytoskeleton/cell adhesion” and the “development-related” gene lists, respectively. These 2 processes overlapped in 444 “hidden nodes” genes, which were further subjected to enrichment analysis (EA) in the ontology of canonical pathways (Supplementary Table S4). FGFR1 signaling was among the highest ranked pathways in the EA distribution (P value=6.17·10−28), with almost all genes involved in signaling pathways from FGF2 and FGFR1, being on the list of the important regulatory “hidden nodes” (Supplementary Fig. S4). This EA result was in accord with the model design, where withdrawal of FGF2 from the media triggers the onset of epithelial differentiation.
Table 1.
Groups of Commonly Affected Pathway Maps in All 5 Differentiated Cell Lines (Based on Enrichment Analysis)
| |
Significance score [−log10(P value)] |
||||
|---|---|---|---|---|---|
| Pathway map title | E12 | E14 | E15 | E16 | H9 |
| Development | |||||
| Development_transcription regulation of granulocyte development | 4.14 | 1.39 | 3.53 | 1.46 | 3.48 |
| Development_FGF-family signaling | 3.87 | 2.23 | 4.39 | 1.86 | 6.78 |
| Development_WNT signaling pathway. Part 2 | 2.74 | 4.25 | 7.64 | 4.46 | 5.93 |
| Development_A2B receptor: action via G-protein alpha s | 2.20 | 3.30 | 1.36 | 2.14 | 2.24 |
| Development_delta- and kappa-type opioid receptors signaling via beta-arrestin | 2.18 | 1.47 | 3.64 | 1.40 | 2.70 |
| Development_PDGF signaling via STATs and NF-kB | 1.67 | 4.61 | 2.35 | 1.46 | 3.48 |
| Development_PACAP signaling in neural cells | 1.39 | 4.43 | 3.79 | 3.54 | 1.69 |
| Cytoskeleton remodeling and cell adhesion | |||||
| Cytoskeleton remodeling_Neurofilaments | 3.85 | 2.53 | 2.04 | 5.24 | 1.34 |
| Cytoskeleton remodeling_Keratin filaments | 2.95 | 2.73 | 4.92 | 3.09 | 2.42 |
| Cell adhesion_endothelial cell contacts by junctional mechanisms | 2.82 | 2.43 | 5.41 | 1.83 | 2.33 |
| Cell adhesion_gap junctions | 2.53 | 4.92 | 5.33 | 4.51 | 2.48 |
| Cytoskeleton remodeling_TGF, WNT and cytoskeletal remodeling | 2.39 | 5.17 | 7.48 | 3.73 | 6.41 |
| Cell adhesion_ECM remodeling | 2.12 | 4.35 | 1.60 | 3.85 | 3.57 |
| Cell adhesion_tight junctions | 1.50 | 3.38 | 5.66 | 2.41 | 3.61 |
Fourteen out of 23 pathway maps, statistically significantly perturbed in all 5 cell lines, belong to 3 general biological processes: development, cell adhesion, and cytoskeleton rearrangement. These maps were used as a source for composition of process-specific lists of genes with expression changes.
FGF, fibroblast growth factor; PDGF, platelet-derived growth factor; NF-kB, nuclear factor kappa B; TGF, transforming growth factor; ECM, extracellular matrix; WNT, wingless signaling; STAT, signal transducers and activators of transcription; PACAP, pituitary adenylate cyclase-activating polypeptide.
In parallel, we applied a correlation algorithm (based on Euclidean distance between the expression profiles) [30] (See the details in the Materials and Methods). We identified 48 differentially expressed pathways (high redundancy of gene content) with 128 genes at significance level 0.0001 (Supplementary Table S5). This gene list significantly overlapped with the content of 23 “common” pathway maps (Supplementary Table S6). Overall, 16 and 21 genes belonged to the cytoskeleton/cell adhesion and development-related processes, respectively (the gene intersections with Fisher's test P value 7.59·10−4 and 1.13·10−7). Thus, despite high heterogeneity between the FGF2+/− pairs masking the onset of differentiation, we have shown that the same pathways and processes of cytoskeleton remodeling and cell adhesion were commonly altered in the earliest steps of hESC epithelial differentiation.
Discussion
In vitro model of the onset of hESC TE differentiation
Signals and mechanisms responsible for the onset of differentiation in hESCs remain largely unknown. In an attempt to address this challenge, we developed a model of the onset of hESC squamous epithelium differentiation. Our approach, based on the imaging of a transitional zone of differentiation, allowed us to establish a sequential pattern of protein and mechanistic markers of hESC specification along with DE lineage. Spontaneous differentiation of hESCs in the presence of FGF2 in a feeder-free culture system was previously described in terms of epithelial to mesenchymal transition [36]. Similarly, we rarely observed epithelial areas of spontaneous differentiation when using StemPro supplemented with FGF. Of note, epithelial areas of spontaneous differentiation become more frequent in iPSCs [which are believed to be more FGF dependent (16–100 ng/mL)] if cultured in the “normal” (for hESC) FGF range of 4–6 ng/mL. In our FGF (−) experiments, all embryonic cell lines and 2 iPSC lines exhibited the onset of commitment into a squamous epithelium, despite a high level of heterogeneity in gene expression profiles between the lines (Fig. 5A), which indicates that epithelial commitment is favored at initial stages of spontaneous differentiation in the absence of FGF. In fact, FGF is one of the key factors in specification of early cell fate decisions of hESCs, and its role, along with Activin A and bone morphogenetic protein 4 (BMP4) factors, was shown by a number of investigators (eg, [37]); whereas FGF signaling works in an antagonistic relationship with BMP [38].
FIG. 5.
(A) Unsupervised clustering based on the gene content; (B) Unsupervised clustering based on the distribution of top ranked gene ontogeny (GO) processes in enrichment analysis. (C) Differentially expressed genes, which passed 1.5-FC thresholds for a corresponding pair of FGF2+\FGF2− cell lines. (Up-, down-regulated in the FGF2− cell lines). FGF2, basic fibroblast growth factor.
There are a lot of similarities between the markers of extraembryonic and DE. Based on the fact that markers of extraembryonic endoderm, such as Laminin β1, AFP, and Lefty, were negative, whereas sets of defined markers were positive, we conclude that epithelial cells transit from mesoendoderm (GSC, Eomes, CK18, and Sox17) to DE (Gata4, COUP-TFII, Cdx2, Sox17, Nestin, Integrin alpha 6, and Islet1). Endoderm commitment of epithelial cells can be further explained with the presence of 10 ng/mL Activin A in StemPro culture media formulation that was recently disclosed. Activin A mimics a Nodal signaling essential for endoderm specification during gastrulation [39] and is used for induction of endoderm differentiation in hESCs [40,41]. However, we also detected a set of markers of TE specification such as CG, GCM1, and multinucleated cells. Several other studies also reported the presence of endoderm/mesendoderm markers on induction to trophectodem with BMP4 [37,42,43], whereas withdrawal of FGF facilitated differentiation to trophectodem [37]). Similarly, squamous epithelium in our experiments may be composed of a mixture of TE and endoderm cell types.
It is worth mentioning that differentiation to a trophoblast lineage is unusual from a developmental biology standpoint, as the trophoblast is expected to develop from TE [44] rather than the ICM, from which hESCs are derived. It would be reasonable to expect the capacity to differentiate into trophoblast to be retained in ESCs established from blastomeres of early embryos [45] as opposed to hESCs (established from ICM), as their characteristics resemble mESCs derived from epiblast and represent a more advanced postimplantation (gastrulation) developmental stage [37]. Further, mouse ESC normally do not differentiate into trophoblasts without conditional gene expression (reviewed in Ref. [46]). The conditions for isolation and culture of trophoblast stem cells from human blastocysts are not yet established, and methods developed for mouse and rat embryos did not work for humans [46–48].
The conditions for hESC differentiation are not necessarily permissive for progenitor cell expansion and specification, which can cause the appearance of multinucleated cells; meanwhile, cell fusion is typical for syncytiotrophoblast, and the presence of multinucleated cells is frequently used to detect TE differentiation. For instance, DE precursors can develop during hESC differentiation in FGF-free conditions; however, particular precursor specification is gradient dependent on FGF [49]. The media developed for human trophoblast progenitor cells, recently isolated from chorion, also contains FGF [48] and Activin A inhibitor, which promote trophoblast differentiation [50]. In our experimental conditions (FGF free StemPro media formulated with Activin), differentiated cells had limited proliferative capacity, and multinucleated cells could develop as a result of distorted cytokinesis providing the culture conditions are suboptimal.
The set of markers describing hESC transition to TE is not yet fully established. In some instances, TE differentiation was described by being solely based on immunostaining with TROMA-1 (CK7) (eg, [51]), although there is a contradicting report that CK7 is not elevated in the trophoblast compared with hESCs [6]. Additionally, CK7 is not a specific marker of the TE (reviewed in Ref. [52]). The same is true for the majority of other markers used to describe TE, such as Cdx2, Gata2, Gata3, and Eomes, including hCG, which can be expressed by many types of non-placental squamous tumors [53]. In previous reports, trophoblast differentiation was achieved with EBs techniques [54] or they were not focused on a transitional zone, which precludes following marker evolution in the process of differentiation. Expression of trophoblast lineage markers (Cdx2, CG, and GCM1) and multinucleated cells are detected in our experiments. However, the epithelial cells do not express the TE marker SSEA1, and they uniformly express other markers, for examole, Coup-TF2 and Sox17, which is inconsistent with TE. This raises a question about the origin of TE markers and TE cells in our cultures. There is a possibility that trophoblast develops as a result of transdifferentiation, omitting an epithelial TE stage of development, with non-canonical marker segregation. Alternatively, TE marker expression is a component of hESC differentiation toward a mesendodermal direction and does not represent TE per se.
Focusing on early differentiation events, we notice that CK18 became dramatically up-regulated at the beginning of the differentiation process. In fact, up-regulation of CK18 (along with an increase of Ca2+ in the cytosol) began in the transitional zone, which still retained a strong Oct4 signal and undistorted morphology (Fig. 3[1, 2C]). Interestingly, the coordinated expression of CK18 and E-Cadherin described here has been documented during epithelial differentiation of different embryonic layers, thus serving as a general characteristic of epithelial differentiation. These 2 markers were used to characterize TE development [55,56], hESC differentiation to non-neural epidermal epithelial cells (ectodermal) [56], and endodermal germ layers [57]. CK8, which was co-expressed with CK18 and also up-regulated in our experiments, was the first marker detectable during differentiation of basal cells to respiratory epithelium (endodermal germ layer) [59]. Additionally, CK18/8 and Cdx2 have been used to characterize the mesenchymal-to-epithelial transition. Since these markers were shown in epithelial cells of different embryonic origins, it is conceivable that hESCs undertake similar initial differentiation steps on commitment to epithelial fate regardless of the nascent embryonic lineage and subsequent specification.
Preexistent cellular polarization of hESCs could provide the predilection for hESCs to spontaneously differentiate to epithelium. The ICM in blastocysts is not polarized, and polarization of hESCs is presumably an in vitro artifact [14], which promoted transition to an epithelial phenotype as a default route of spontaneous differentiation. The possibility that TJs can direct differentiation has been suggested [14] based on the observation that depolarization of hESCs enhanced differentiation into hematoendothelial cells (mesoderm). The fact that ZO-1 is lost near the border with the region of differentiation likely suggests that remodeling of TJs preceded the differentiation event. There is no other tool available at this point to confirm the commitment of “specified” cells to differentiation, because conventional markers of differentiation are not yet expressed, and the hESCs retained pluripotent morphology and high levels of Oct4 expression. Alteration of ZO-1 distribution could, therefore, be a sign of commitment of regions to differentiate, which would add TJ to the cohort of the earliest markers of differentiation.
It is currently accepted that during early postimplantation development, GJIC become restricted and maintained only within cells of the same lineage [15]. For example, after implantation, GJIC remain active, thus supporting dye transfer within the ICM; however, the dye spread to the surrounding trophoblast is not transferred to other cells [60]. Remarkably, the results of our study documented a significant decrease of GJIC between pluripotent and differentiated cells as among the earliest events in the onset of hESC differentiation, thus suggesting a key role in the onset of compartmentalization. Such GJIC dependant compartmentalization was also recently shown in spontaneously differentiated hESC cultures [61]. The demonstrated decrease of gap junction activity along with the IHC results suggests that down-regulation of Cx43/Cx45 may, indeed, be responsible for the functional inactivation of gap-junctions. We believe that this is the first time that such changes in gap junction activity have been documented at the onset of hESC differentiation to squamous epithelium. Of note, Cx43 knock-down mouse ESCs exhibited down-regulation of several stem cell markers along with up-regulation of differentiation markers [15], with Cx43 down-regulation observed during hESC differentiation described as epithelial to mesenchymal transition [36].
Gap junctions are the only intercellular junctions that support cytoplasmic continuity between cells allowing direct transfer of signaling molecules and metabolites to neighboring cells, including small ions, second messengers, and amino acids, among others [15,20]. During each cell cycle, gap junctions are actively regulated, which leads to coupling, uncoupling, and hemichannel opening [20]. Since hESCs have a very short overall cell cycle length, predominantly due to dramatically shortened G1 cell cycle phase [62], it is conceivable that the onset-related perturbations in cell cycle could lead to disruption of the coupling and uncoupling of gap junctions between different cell populations. Consequently, it could have an impact on the intercellular communication and lead to the distinct communication compartments in the hESC colony followed by the commitment to differentiation.
Since it was shown that a decrease in overall cell coupling could trigger gap junction hemichannel-mediated Ca2+ waves [20], we hypothesized that these 2 events may be interconnected in our model, as both GJIC and Ca2+ signaling exhibited the same borders of the squamous epithelium compartment inside the hESC colonies. To the best of our knowledge, this is the first time that such a significant increase in Ca2+ concentration was documented at the onset of hESC differentiation. An increase in Ca2+ concentration is regulated by ion channels and results in a set of local or global signaling events triggered by Ca2+-sensitive mediators [22]. One of the first Ca2+ oscillations in early development occurs at the site of sperm-egg interaction. Presumably, these oscillations trigger the cell cycle via CaM/CaMKII pathways [21]. Moreover, calcium signals play a key role throughout development, including left–right axis determination [22]. Of relevance, an increase in Ca2+ signaling is necessary and sufficient to switch the fate of ectoderm cells in the embryo from epidermis to neural tissue [63]. However, it is not yet clear which mechanisms generate such an increase in Ca2+ signaling and/or trigger the inactivation of gap junctions.
Our results indicate that a dramatic decrease of GJIC, along with down-regulation of Cx43/Cx45, up-regulation of CK18, and an increase in cellular calcium concentration precedes up-regulation of the TE -specific transcription factor -Cdx2. These findings raise an important question regarding the “stream of events” leading to the up-regulation of specific transcription factors on hESC commitment to the squamous epithelium and, possibly, during specification of epithelial cells in general, which would trigger activation of appropriate tissue-specific transcription factors.
Functional analysis of hESC expression profiles
Using a variety of statistical methods to analyze our microarray data, we did not detect any known markers of pluripotency as being affected by the onset of differentiation, which suggests that all the changes highlighted in this study may indeed reflect the earliest steps of differentiation. High heterogeneity between the cell lines is likely responsible for the small number of commonly altered genes, as several thousands of differentially expressed genes passed the 1.5-FC thresholds for individual pairs of FGF2+\FGF2− cell lines (Fig. 4C). Low representation (∼1%) of differentiating cells in the FGF2− colonies among morphologically identical FGF2+/FGF2− pairs also limits the number of commonly altered genes.
However, despite high heterogeneity between the pairs masking the onset of differentiation, we have shown that the same pathways and processes of cytoskeleton remodeling and cell adhesion, in particular, Gap junctions, TJs, and Keratin filaments (Table 1), were commonly altered during the earliest steps of hESC differentiation. These findings were consistent with the results of immunohistochemistry for ZO-1, Cx43/45, F-actin, CK18, nestin, and tubulin. Our data also suggested that these processes are tightly interlinked. The results of independent methods of functional analysis confirmed that the pathways in these processes were, indeed, key differentiators between the FGF2+ and FGF2− cell lines, where 4 out of 7 cytoskeleton/cell adhesion pathway maps and 6 out of 7 development-related pathway maps (with 3–5 canonical pathways on each map) were involved in differentiation (Supplementary Table S7). Interestingly, CK18 belongs to one of the most affected processes involved in the cytoskeleton remodeling (Table 1- Cytoskeleton remodeling_Keratin filaments). These results were also in line with the studies on preimplantation embryo development where gene families encoding for cell polarity, cell junctions, cytoskeletal formation, and ion transport are involved in cell lineage specification in the blastocyst [64]. As mentioned earlier, we established membrane association and nuclear localization of b-catenin, (Supplementary Fig. S2[3, 4A]) in both pluripotent and differentiated cells, which suggested a role for an active Wnt/b-catenin pathway. The nuclear localization of b-catenin has been described during embryo development in some progenitors and cancer cells [65]. Interestingly, we found that among 56 genes shared between the cytoskeleton/cell adhesion and the “development-related” gene lists, 11 belong to the Wnt pathway (Supplementary Table S3).
In summary, analysis of the earliest steps in the transition of hESC colonies to squamous epithelium revealed an alteration of TJs, significant decrease in the functional activity of gap junctions along with down-regulation of 2 gap junction proteins, Cx43 and Cx45, which was coincident with substantial elevation of intracellular Ca2+ levels.In addition, we observed significant up-regulation of keratin-18 during the onset of differentiation. We hypothesize that these events may be common during primary steps of hESC commitment to the functionally varied epithelial tissue derivatives of different embryological origins. Further investigation of the onset of epithelial differentiation in hESCs, including functional implications of TJs, gap junctions, and Ca2+ signaling, along with the associated processes, will lead to the development of much needed methods to control the differentiation process, and will ultimately benefit the fast-growing field of regenerative medicine.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Philip Hockberger and Dr. Gregory Filatov for their help in establishing calcium assays and discussion of the experimental data; Dr. Louise Laurent for critical reading of the article; and Shauna Houlihan, Eugenia Krotova, and Mariana Perepitchka for technical help. Research was supported in part by NHLBI, RC1HL100168 (M.J.C.H.,V.G.); NIH, CA121205 (M.J.C.H.); NIH EY020946, George M. Eisenberg Foundation for Charities (PMI).
Author Disclosure Statement
The authors indicate no potential conflict of interest.
References
- 1.Murry C. Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Cai J. Chen J. Liu Y. Miura T. Luo Y. Loring JF. Freed WJ. Rao MS. Zeng X. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem cells. 2006;24:516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya B. Cai J. Luo Y. Miura T. Mejido J. Brimble SN. Zeng X. Schulz TC. Rao MS. Puri RK. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and differentiating embryoid bodies. BMC Dev Biol. 2005;5:22–38. doi: 10.1186/1471-213X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallier L. Alexander M. Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 5.Eiselleova L. Matulka K. Kriz V. Kunova M. Schmidtova Z. Neradil J. Tichy B. Dvorakova D. Pospisilova S. Hampl A. Dvorak P. A complex role for FGF-2 in self-renewal, survival, and adhesion of human embryonic stem cells. Stem Cells. 2009;27:1847–1857. doi: 10.1002/stem.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu RH. Chen X. Li DS. Li R. Addicks GC. Glennon C. Zwaka TP. Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 7.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 8.Siegel S, editor; Castellan NJ, editor. Nonparametric Statistics for the Behavioral Sciences. McGraw-Hill; Boston, MA: 1988. [Google Scholar]
- 9.Nikolsky Y. Kirillov E. Zuev R. Rakhmatulin E. Nikolskaya T. Functional analysis of OMICs data and small molecule compounds in an integrated “knowledge-based”. platform. In: Nikolsky Y, editor; Bryant J, editor. Protein Networks and Pathway Analysis. Vol. 563. Springer Protocols; New York, N.Y: 2009. pp. 177–196. [DOI] [PubMed] [Google Scholar]
- 10.Dezso Z. Nikolsky Y. Nikolskaya T. Miller J. Cherba D. Webb C. Bugrim A. Identifying disease-specific genes based on their topological significance in protein networks. BMC Syst Biol. 2009;3:36. doi: 10.1186/1752-0509-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurent LC. Nievergelt CM. Lynch C. Fakunle E. Harness JV. Schmidt U. Galat V. Laslett AL. Otonkoski T, et al. Restricted ethnic diversity in human embryonic stem cell lines. Nat Methods. 2010;7:6–7. doi: 10.1038/nmeth0110-06. [DOI] [PubMed] [Google Scholar]
- 12.Malchenko S. Galat V. Seftor EA. Vanin EF. Costa FF. Seftor RE. Soares MB. Hendrix MJ. Cancer hallmarks in induced pluripotent cells: new insights. J Cell Physiol. 2010;225:390–393. doi: 10.1002/jcp.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurent LC. Ulitsky I. Slavin I. Tran H. Schork A. Morey R. Lynch C. Harness JV. Lee S, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada S. Pokutta S. Drees F. Weis WI. Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiprilov EN. Awan A. Desprat R. Velho M. Clement CA. Byskov AG. Andersen CY. Satir P. Bouhassira EE. Christensen ST. Hirsch RE. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol. 2008;180:897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krtolica A. Genbacev O. Escobedo C. Zdravkovic T. Nordstrom A. Vabuena D. Nath A. Simon C. Mostov K. Fisher SJ. Disruption of apical-basal polarity of human embryonic stem cells enhances hematoendothelial differentiation. Stem Cells. 2007;25:2215–2223. doi: 10.1634/stemcells.2007-0230. [DOI] [PubMed] [Google Scholar]
- 17.Wong RCB. Pera MF. Pébay A. Role of Gap Junctions in Embryonic and Somatic Stem Cells. Stem Cell Rev. 2008;4:283–292. doi: 10.1007/s12015-008-9038-9. [DOI] [PubMed] [Google Scholar]
- 18.Wong RC. Pebay A. Nguyen LT. Koh KL. Pera MF. Presence of functional gap junctions in human embryonic stem cells. Stem Cells. 2004;22:883–889. doi: 10.1634/stemcells.22-6-883. [DOI] [PubMed] [Google Scholar]
- 19.Carpenter MK. Rosler ES. Fisk GJ. Brandenberger R. Ares X. Miura T. Lucero M. Rao MS. Properties of four human embryonic stem cell lines maintained in a feeder-free culture system. Dev Dyn. 2004;229:243–258. doi: 10.1002/dvdy.10431. [DOI] [PubMed] [Google Scholar]
- 20.Elias LAB. Kriegstein AR. Gap junctions: multifaceted regulators of embryonic cortical development. Trends Neurosci. 2008;31:243–250. doi: 10.1016/j.tins.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitaker M. Calcium at Fertilization and in Early Development. Physiol Rev. 2006;86:25–88. doi: 10.1152/physrev.00023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slusarski DC. Pelegri F. Calcium signaling in vertebrate embryonic patterning and morphogenesis. Dev Biol. 2007;307:1–13. doi: 10.1016/j.ydbio.2007.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leary RJ. Lin JC. Cummins J. Boca S. Wood LD. Parsons DW. Jones S. Sjöblom T. Park BH, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16230. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons DW. Jones S. Zhang X. Cheng-Ho Lin J. Leary RJ. Angenendt P. Mankoo P. Carter H. Mei Siu I, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones S. Zhang X. Parsons DS. Lin JC. Leary RJ. Angenendt P. Mankoo P. Carter H. Kamiyama H, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo L. Peng G. Zhu Y. Dong H. Amos CI. Xiong M. Genome-wide gene and pathway analysis. Eur J Hum Genet. 2010;18:1045–1053. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong M. Yang X. Kaplan LM. Molony C. Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ideker T. Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi W. Bugrim A. Nikolsky Y. Nikolskya T. Brennan RJ. Characteristics of genomic signatures derived using univariate methods and mechanistically-anchored functional descriptors for predicting drug and xenobiotic-induced nephrotoxicity. Tox Mech Methods. 2008;18:267–276. doi: 10.1080/15376510701857072. [DOI] [PubMed] [Google Scholar]
- 30.Shi W. Bessarabova M. Dosymbekov D. Dezso Z. Nikolskaya T. Dudoladova M. Serebryiskaya T. Bugrim A. Guryanov A, et al. Functional analysis of multiple genomic signatures demonstrates that classification algorithms choose phenotype-related genes. Pharmacogenomics. 2010;10:310–323. doi: 10.1038/tpj.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolsky Y. Bessarabova M. Kirillov E. Dezso Z. Shi W. Nikolskaya T. Pathways and networks as functional descriptors for human disease and drug response endpoints. In: Dehmer M, editor; Emmert-Streib F, editor; Graber A, editor; Salvador A, editor. Applied Statistics for Biological Networks; Methods in System Biology. Willey & Sons; Chicester, West Sussex, England: 2011. pp. 415–442. [Google Scholar]
- 32.Saiyo S. Onuma Y. Ito Y. Toyoda THM. Akutsu H. Nishino K. Chikazawa E. Fukawatase Y. Miyagawa Y, et al. Potential linkages between the inner and outer cellular states of human induced pluripotent ste cells. Proceedings of the The Fourth International Conference on Comp. Systems Biology (ISB2010); Suzhou, China. Sep 9–11;; 2010. pp. 381–388. [Google Scholar]
- 33.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Benjamini Y. Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 35.Plant TD. Schaefer M. TRPC-4 and TRPC-5: receptor-operated Ca2+- permeable nonselective cation channels. Cell Calcium. 2003;33:441–450. doi: 10.1016/s0143-4160(03)00055-1. [DOI] [PubMed] [Google Scholar]
- 36.Ullmann U. In't Veld P. Gilles C. Sermon K. De Rycke M. Van de Velde H. Van Steirteghem A. Liebaers I. Epithelial-mesenchymal transition process in human embryonic stem cells cultured in feeder-free conditions. Mol Hum Reprod. 2007;13:21–32. doi: 10.1093/molehr/gal091. [DOI] [PubMed] [Google Scholar]
- 37.Vallier L. Touboul T. Chng Z. Brimpari M. Hannan N. Millan E. Smithers LE. Trotter M. Rugg-Gunn PP. Weber A. Pedersen RA. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greber B. Lehrach H. Adjaye J. Fibroblast growth factor 2 modulates transforming growth factor beta signaling in mouse embryonic fibroblasts and human ESCs (hESCs) to support hESC self-renewal. Stem Cells. 2007;25:455–464. doi: 10.1634/stemcells.2006-0476. [DOI] [PubMed] [Google Scholar]
- 39.Lowe LA. Yamada S. Kuehn MR. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- 40.Osafune K. Chen AE. Melton DA. Directed differentiation of human embryonic stem cells into early endoderm cells. In: Sullivan S, editor; Cowan C, editor; Eggab K, editor. Human Embryonic Stem Cells -The Practical Handbook. Wiley; Chicester, West Sussex, England: 2007. pp. 179–187. [Google Scholar]
- 41.D'Amour K. Agulnick A. Eliazer S. Kelly O. Kroon E. Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 42.Erb TM. Schneider C. Mucko SE. Sanfilippo JS. Lowry NC. Desai MN. Mangoubi RS. Leuba SH. Sammak PJ. Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. 2011;20:1601–1614. doi: 10.1089/scd.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He S. Pant D. Schiffmacher A. Meece A. Keefer CL. Lymphoid enhancer factor 1-mediated Wnt signaling promotes the initiation of trophoblast lineage differentiation in mouse embryonic stem cells. Stem Cells. 2008;26:842–849. doi: 10.1634/stemcells.2007-0356. [DOI] [PubMed] [Google Scholar]
- 44.Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 45.Ilic D. Giritharan G. Zdravkovic T. Caceres E. Genbacev O. Fisher SJ. Krtolica A. Derivation of human embryonic stem cell lines from biopsied blastomeres on human feeders with minimal exposure to xenomaterials. Stem Cells Dev. 2009;18:1343–1350. doi: 10.1089/scd.2008.0416. [DOI] [PubMed] [Google Scholar]
- 46.Rossant J. Stem cells from the mammalian blastocyst. Stem Cells. 2001;19:477–482. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- 47.Galat V. Binas B. Iannaccone S. Postovit LM. Debeb BG. Iannaccone P. Developmental potential of rat extraembryonic stem cells. Stem Cells Dev. 2009;18:1309–1318. doi: 10.1089/scd.2009.0115. [DOI] [PubMed] [Google Scholar]
- 48.Genbacev O. Donne M. Kapidzic M. Gormley M. Lamb J. Gilmore J. Larocque N. Goldfien G. Zdravkovic T. McMaster MT. Fisher SJ. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 2011;29:1427–1436. doi: 10.1002/stem.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ameri J. Ståhlberg A. Pedersen J. Johansson JK. Johannesson MM. Artner I. Semb H. FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells. 2010;28:45–56. doi: 10.1002/stem.249. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z. Zhang W. Chen G. Cheng L. Liao J. Jia N. Gao Y. Dai H. Yuan J. Cheng L. Xiao L. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem. 2008;283:24991–25002. doi: 10.1074/jbc.M803893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mali P. Ye Z. Hommond HH. Yu X. Lin J. Chen G. Zou J. Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- 52.Golos TG. Giakoumopoulos M. Garthwaite MA. Embryonic stem cells as models of trophoblast differentiation: progress, opportunities, and limitations. Reproduction. 2010;140:3–9. doi: 10.1530/REP-09-0544. [DOI] [PubMed] [Google Scholar]
- 53.Iles RK. Purkis PE. Whitehead PC. Oliver RT. Leigh I. Chard T. Expression of beta human chorionic gonadotrophin by non-trophoblastic non-endocrine 'normal' and malignant epithelial cells. Br J Cancer. 1990;61:663–666. doi: 10.1038/bjc.1990.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harun R. Ruban L. Matin M. Draper J. Jenkins NM. Liew GC. Andrews PW. Li TC. Laird SM. Moore HD. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum Reprod. 2006;21:1349–1358. doi: 10.1093/humrep/del017. [DOI] [PubMed] [Google Scholar]
- 55.Goossens K. Van Soom A. Van Poucke M. Vandaele L. Vandesompele J. Van Zeveren A. Peelman LJ. Identification and expression analysis of genes associated with bovine blastocyst formation. BMC Dev Biol. 2007;7:64. doi: 10.1186/1471-213X-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kan NG. Stemmler MP. Junghans D. Kanzler B. de Vries WN. Dominis M. Kemler R. Gene replacement reveals a specific role for E-cadherin in the formation of a functional trophectoderm. Development. 2007;134:31–41. doi: 10.1242/dev.02722. [DOI] [PubMed] [Google Scholar]
- 57.Aberdam E. Barak E. Rouleau M. de LaForest S. Berrih-Aknin S. Suter DM. Krause KH. Amit M. Itskovitz-Eldor J. Aberdam D. A pure population of ectodermal cells derived from human embryonic stem cells. Stem Cells. 2008;26:440–444. doi: 10.1634/stemcells.2007-0588. [DOI] [PubMed] [Google Scholar]
- 58.Tada S. Era T. Furusawa C. Sakurai H. Nishikawa S. Kinoshita M. Nakao K. Chiba T. Nishikawa S. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 59.Rock JR. Randell SH. Hogan BLM. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Models Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lo CW. Gilula NB. Gap junctional communication in the preimplantation mouse embryo. Cell. 1979;18:399–409. doi: 10.1016/0092-8674(79)90059-x. [DOI] [PubMed] [Google Scholar]
- 61.Sharovskaya YY. Lagarkova MA. Kiselev SL. Chailakhyan LM. Gap junctional intercellular communication in human embryonic stem cells during spontaneous differentiation. Dokl Biol Sci. 2009;427:387–390. doi: 10.1134/s0012496609040243. [DOI] [PubMed] [Google Scholar]
- 62.Becker KA. Ghule PN. Therrien JA. Lian JB. Stein JL. van Wijnen AJ. Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 63.Moreau M. Leclerc C. The choice between epidermal and neural fate: a matter of calcium. Int J Dev Biol. 2004;48:75–84. doi: 10.1387/ijdb.15272372. [DOI] [PubMed] [Google Scholar]
- 64.Watson AJ. Natale DR. Barcroft LC. Molecular regulation of blastocyst formation. Anim Reprod Sci. 2004;82–83:583–592. doi: 10.1016/j.anireprosci.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Willert K. Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.