Abstract
In living systems, the mechanisms of inheritance involving gene expression are operated by (i) the traditional model of genetics where the deoxyribonucleic acid (DNA) transcription and messenger ribonucleic acid stability are influenced by the DNA sequences and any aberrations in the primary DNA sequences and (ii) the epigenetic (above genetics) model in which the gene expression is regulated by mechanisms other than the changes in DNA sequences. The widely studied epigenetic alterations include DNA methylation, covalent modification of chromatin structure, state of histone acetylation, and involvement of microribonucleic acids. Significance: Currently, the role of cellular epigenome in health and disease is rapidly emerging. Several factors are known to modulate the epigenome-regulated gene expression that is crucial in several pathophysiological states and diseases in animals and humans. Phytochemicals have occupied prominent roles in human diet and nutrition as protective antioxidants in prevention/protection against several disorders and diseases in humans. Recent Advances: However, it is beginning to surface that the phytochemical phenolic antioxidants such as polyphenols, flavonoids, and nonflavonoid phenols function as potent modulators of the mammalian epigenome-regulated gene expression through regulation of DNA methylation, histone acetylation, and histone deacetylation in experimental models. Critical Issues and Future Directions: The antioxidant or pro-oxidant actions and their involvement in the epigenome regulation by the phytochemical phenolic antioxidants should be at least established in the cellular models under normal and pathophysiological states. The current review discusses the mechanisms of modulation of the mammalian cellular epigenome by the phytochemical phenolic antioxidants with implications in human diseases. Antioxid. Redox Signal. 17, 327–339.
Introduction: Epigenetic Regulation of Gene Expression
Recent studies with the aid of novel and more advanced molecular tools have provided deeper insights into the nuclear architecture, including critical information on the mechanisms of normal and pathophysiological states of gene expression. Furthermore, these studies have revealed that the mechanisms of inheritance involving gene expression are operated through genetic alterations influencing deoxyribonucleic acid (DNA) transcription and messenger ribonucleic acid (mRNA) stability through modifications of the primary DNA sequences and epigenetic alterations involving the covalent modification of chromatin architecture and post-translational modifications (77). DNA in its native form is inaccessible for transcription (46). Nucleosomes, the building blocks of higher order chromatin structure, consist of 147 base pairs of DNA wrapped around an octamer of histones (H2A, H2B, H3, and H4) (46). Short stretches of linker DNA join nucleosomes to form polymers that are further organized into tightly compacted native chromatin configuration as seen in the nucleus and have an appearance of beads on a string. Chromatin has regions of transcriptionally active euchromatin and inactive heterochromatin. The interconversion of these two regions for DNA accessibility to transcription factors is determined by the epigenome components, including histone chaperones, chromatin-remodeling complexes, histone- and histone variant-modifying enzymes, DNA methylating agents, noncoding RNAs like the microribonucleic acids (miRNAs), and other epigenome constituents. Thus, the term “epigenetics” is defined as the stable and perpetual but reversible and altered active states of gene expression without modifying the primary DNA sequences (77).
Epigenetic regulation is furthermore involved in tissue-specific gene expression and silencing. There are different mechanisms that mediate and regulate the duplication of chromatin state, which depends on the chromatin domain, organism, and function. The key components involved include histone chaperones, chromatin-remodeling factors, histone-modifying enzymes, RNA components, and the DNA replication machinery itself. Mechanisms that regulate chromatin remodeling, a crucial step in epigenetic alterations, include DNA methylation, post-translational modifications of histones, including acetylation, deacetylation, phosphorylation, poly-adenosine-3′,5′-diphosphate (ADP)-ribosylation, ubiquitination, histone variants, and miRNAs. DNA methylation is brought about by enzymes called DNA methyltransferases (DNMTs), which catalyze the addition of a methyl group to cytosine (C) located 5′ to guanines (G) on the phosphodiester bond between cytosine and guanine (CpG) islands. DNA methylation, an epigenetic mechanism, is known to attenuate gene expression and has been shown to play crucial roles in cellular functions. Histones represent a class of proteins that are extremely arginine rich. Almost 14% of the histone H4 amino acids are arginine residues. Histone modification involves covalent modification of the N-terminus tail of histone in determination of the level of gene expression. Histone acetylation of conserved lysine residues in histone tails, the signature of transcriptionally active regions, is brought about by the histone acetyltransferase (HATs) and hypoacetylation by histone deacetylases (HDACs) as seen in transcriptionally inactive heterochromatin regions (67).
The roles of sirtuins (SIRTs; class III HDACs) are implicated in physiological and pathophysiological phenomena, including inflammation, cellular aging and senescence, cell proliferation, apoptosis, cell differentiation, metabolism, stem cell pluripotency, and cell cycle regulation (18). Seven different types of SIRTs have been identified such as SIRT 1–7 in mammals (7, 18). SIRT1-3 has been well characterized, whereas SIRT4 and SIRT6 have been reported to have ADP-ribosylation activity, which may be due to their deacetylation activity (7). However, SIRT 1, 2, 3, and 5 catalyze deacetylation. A tight and well-regulated balance between the activities of HATs and HDACs will maintain a controlled balance between acetylation and deacetylation of histones, and the epigenome actions under normal physiological states in the cells and any alterations in this balance would lead to pathophysiological conditions.
Oxidative Stress in Epigenetic Regulation of Gene Expression and Disease
Oxidative stress has been established as an important mechanism of either onset or progression of several disease states or disorders, including myocardial ischemia, cardiovascular diseases (CVDs), cerebrovascular diseases, neurological diseases and disorders, diabetes, renal diseases, infection and sepsis, pulmonary diseases and disorders, and obesity (3, 6, 14, 16, 28, 30, 34, 43, 59). Environmental, toxic, and dietary factors are known to cause oxidative stress by different mechanisms (40). Oxidative stress is mediated by the oxygen-derived reactive oxygen species (ROS), reactive nitrogen species (RNS), and other oxidants (40). Oxidative stress is caused by either external oxidants/pro-oxidants or intracellular oxidants. The intracellular oxidants are generated by either nonenzymatic mechanisms (often involving transition metals such as iron) or enzymatic catalysis operated by a variety of oxidases that activate oxygen and convert them into the ROS or RNS. Cellular membrane lipids containing the polyunsaturated fatty acids, proteins, and nucleic acids are vulnerable to the attack by oxidative stress leading to the pathophysiological alterations (15). It has long been established that 4-hydroxy-2-nonenal (4-HNE) is a major lipid hydroperoxide-derived aldehydic bifunctional electrophile that reacts with DNA and proteins (53, 74). The lipid peroxidation-derived electrophilic carbonyl, 4-HNE, has been shown to inhibit the class II HDAC (mitochondrial SIRT3) through a thiol mechanism, and this has implications in lipid peroxidation-mediated alterations of epigenetic regulation (25). Overall, convincing experimental evidence has shown that lipid peroxidation is also an important player in the oxidative stress-mediated epigenetic regulation of gene expression. Oxidative stress also brings alterations in the redox status of the cell, wherein the thiol-redox (glutathione [GSH] and protein thiols) system crucial for the cellular thiol-antioxidant defense system and cellular metabolic machinery is jeopardized (61), leading to the oxidative deterioration of the cell. Cellular systems have evolved several enzymatic and nonenzymatic antioxidant systems to cope with the surge of constant oxidative stress. However, the overwhelming production of ROS and RNS with their empowering oxidative stress and the endogenous antioxidant defenses are compromised and lead to the cellular demise. This is one of the often-recognized mechanisms of either disease onset or progression.
The role of epigenome and epigenetic regulation of gene expression in several human diseases, including cancer and CVDs, are becoming compellingly evident (21). At present, evidence is mounting in favor of oxidative stress modulating the epigenome, leading toward the regulation of gene expression (81). Also, recent studies have highlighted the importance of epigenetic alterations in cardiovascular, neurological, immunological, and other more complex genetic disorders and diseases (21). Taken together, it is emerging that oxidative stress-modulated regulation of the epigenetic mechanism of gene expression plays an important role in certain diseases. Understanding the mechanisms of epigenetic regulation will lead to the development of novel therapies for treatment of diseases and the development of regenerative medicine, and identification of strategies for preventive intervention. In this regard, antioxidants are seriously sought after to strengthen the cellular antioxidant defense system to combat and counteract the overwhelming oxidant generation and oxidative stress, thus attenuating or normalizing the adverse oxidant-mediated epigenetic regulation of gene expression that is responsible for either the onset or progression of the disease.
Phytochemical Antioxidants: Oxidative Stress, Disease, and Epigenome
Cellular antioxidant defense machinery has been unequivocally established as an oxidative stress-counteracting entity. Cellular antioxidants comprise both the (i) nonenzymatic molecules and (ii) enzymes. The nonenzymatic antioxidants consist of the thiol antioxidants such as GSH and other thiols and small-molecule antioxidants (vitamins C, A, D, and E). Apart from GSH, most of the antioxidants make their entry into the cells through nutrition and diet. Also, the function of these nonenzymatic antioxidants is regulated by the redox state of the cell and vice versa. Antioxidant supplementation/treatment has been adopted for either prevention of or protection against several disorders and pathophysiological states wherein oxidative stress has been established as a causative mechanism (5). The immunomodulatory and anti-inflammatory effects of polyphenols have been documented (26, 39). The beneficial effects of moderate consumption of red wine on lessening the coronary heart disease are accredited to the wine polyphenols such as anthocyanosides, catechins, proanthocyanidins, stilbenes, and phenolic compounds (19). Naturally occurring phytochemical antioxidants have occupied a prominent position as effective antioxidants for the prevention and/or treatment of several disorders and diseases among humans (32, 70, 71). The premise for this has been the antioxidant actions of the phytochemicals as free-radical scavengers, oxidative stress relievers, and lipoperoxidation inhibitors (68). Phytochemical antioxidants include simple molecule antioxidants such as vitamins C, E, and K; plant pigments such as carotenoids (β-carotene), xanthophylls, lycopene, anthocyanins, and phaeophytins; and secondary plant metabolites, including simple phenolics to more complex polyphenols (13). Some of these phytochemical polyphenols, in addition to acting as antioxidants, will also function as pro-oxidants that cause oxidative stress (42, 62). The pro-oxidant action of tea polyphenols has been linked to their anticancer actions (23, 42). Polyphenols are known for their complexing abilities (chelation) with trace metals. Polyphenols have been shown to attenuate the iron-induced DNA damage by complexation with iron and keeping the Fe in the +3 state after oxidation of Fe in the +2 state in presence of molecular oxygen (63). Two major pathways of biosynthesis of the phytochemical polyphenols in plants have been identified: (i) the shikimate pathway and (ii) the polyketide or acetate pathway (57). Structurally, the plant polyphenol has a phenolic or a benzene ring with a hydroxyl group attached, and also certain substituents such as ester and glycosides present as functional groups (73). The majority of the plant polyphenols have two or three hydroxyl groups, and hence they are called dihydric or trihydric polyphenols, respectively. The classification of plant polyphenols is based on (i) the number of phenolic or benzene rings present and (ii) the number and type of substituents present on the phenolic rings. Phytochemical polyphenols are broadly classified into (i) simple phenols (ferulic and gallic acids), (ii) stilbenes (dihydrophenols such as resveratrol), (iii) chalcones, and (iv) flavonoids (Fig. 1) (57). Flavonoids, a family of the most bioactive complex phytochemical polyphenols, are further divided into seven classes such as (i) flavonols, (ii) flavanols, (iii) flavones, (iv) flavanones, (v) flavanonols, (vi) isoflavones, and (vii) anthocyanins (73). Simple phenolic and polyphenolic secondary plant metabolites are known to play crucial roles in plants as protectants against oxidative stress and ultraviolet (UV) radiation, wound healing, defense against microbes, fungi, herbivores, plant competitors, and disease resistance in plants (2, 20). However, the mechanism of such protection and their physiological roles in plants are not completely understood. Nevertheless, flavonoids have been shown to exhibit a plethora of biological effects, including antibacterial, antiviral, analgesic, antiallergic, hepatoprotective, cytostatic, apoptotic, estrogenic, and antiestrogenic functions (31).
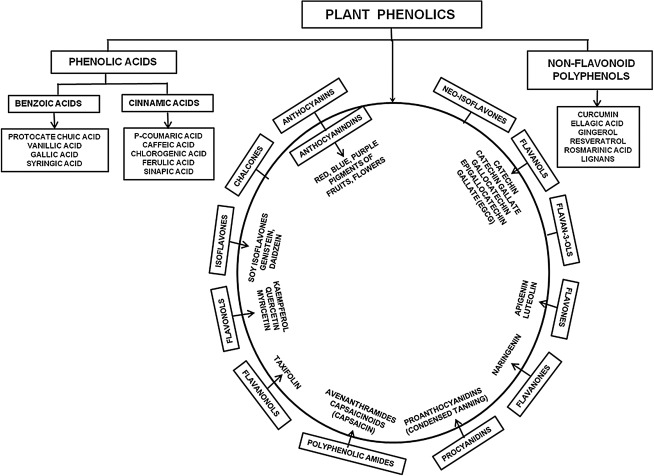
FIG. 1.
Classification of plant phenols. Here, phenols of plant origin are broadly divided into three major groups such as (i) phenolic acids, (ii) flavonoids, and (iii) nonflavonoid polyphenols. Flavonoids are further divided into several classes. Flavonoids such as kaempferol, quercetin, and myricetin fall under the class of flavonols. Genistein and daidzein are classified under the class of isoflavones and thereby are called the soy isoflavones. (-)-epigallocatechin-3-gallate (EGCG) falls under the class of flavanols. These phytochemical phenols as secondary metabolites in plants have definitive roles in plants as the ultraviolet (UV) protectants, wound-healing action, and disease and pest resistance. Primarily, these compounds have established protective actions against oxidative stress as potent, naturally occurring antioxidants.
Animals and humans obtain these phytochemical polyphenols from diet or nutritional supplementation. The majority of the phytochemical polyphenols are present in their native state in plants as polymers or as glycosylated molecules (conjugated to sugars), wherein the sugar moiety is termed as the “glycone” and the polyphenol is called the “aglycone” (76). Most of the polyphenols in the dietary plant sources occur as complex molecules linked to the sugar residues (O-glycosides), but some occur as C-glycosides. Polyphenol O-glycosides undergo hydrolysis in the lumen of the intestine, and the sugar residue is released upon the action of β-glucosidase, setting the aglycone free. On the other hand, the C-glycosides are resistant to the intestinal enzymatic hydrolysis (9). It is interesting to note that the polyphenols, after the uptake by the intestinal cells, undergo metabolic biotransformation similar to the xenobiotics by the involvement of phase-I and phase-II enzymes (9). Bacteria in the large intestine catalyze the conversion of aglycones of polyphenols causing the heterocyclic B-ring opening and cleavage after which the metabolites are absorbed and reach circulation (9). Similar to the intestine, the liver also metabolizes polyphenols, and circulating polyphenols are in the form of β-glucuronides and esters of sulfate with trace levels of aglycones. Polyphenols that are hydrophobic can easily be transported into the intestinal cells through the plasma membrane. Nevertheless, the phytochemical polyphenol metabolism is complex, and it is challenging to identify whether the parent molecule or its metabolite is biologically active. In recent times, plant polyphenols have been the attraction as effective antioxidants (from diet or supplementation) in prevention and treatment of several diseases, including CVDs, cerebrovascular diseases, Alzheimer's disease, airway disease, and cancer, with a focus to alleviate the oxidative stress as the causative mechanism in those diseases (17, 24, 38, 51, 70, 71, 80). In spite of the beneficial effects of the phytochemical polyphenolic antioxidants in humans, their mechanisms of action (physiology and pharmacology) in the mammalian systems (including humans) are just emerging. The regulation of gene expression by dietary polyphenols in cellular models such as the vascular endothelial cells is evident (56, 57). One of the noteworthy current discoveries that have emerged from the phytochemical–antioxidant interactions in mammalian model systems is their nature of modulating the mammalian epigenome (21). Totally, novel disciplines such as nutrigenetics and nutrigenomics have emerged with a focus on the phytochemical nutrient–gene interactions toward cancer prevention in which signaling pathways, networks, and epigenetic phenomena are investigated (22). Hence, this review focuses on the relevant studies on certain selected phytochemical phenolic antioxidants that have been shown to mediate the modulation of the mammalian epigenome and its relevance in the prevention of and protection against diseases.
Mechanisms of Epigenetic Regulation of Diseases: Role of Phytochemical Antioxidants
As overviewed earlier, epigenetic alterations in DNA are inheritable, which lead to the modulation of gene expression without involving any changes in the primary sequences of DNA. However, chemical alterations in the histone proteins and/or DNA by covalent modifications brought by certain enzymes are the causative mechanisms for the epigenetic alterations and subsequent modulation of gene expression. It is progressively more evident that DNA methylation, an epigenetic phenomenon catalyzed by DNMTs using the methyl donor, S-adenosylmethionine (SAM), leads to anomalous DNA methylation (hypermethylation), and this is associated with certain diseases, including cancer and CVDs (21). Methylation of CpG islands is a common occurrence in the pathological tissues such as cancer (29). Transcription of methylated genes is either arrested or suppressed. Global hypomethylation of DNA and hypermethylation at specific sites of DNA are generally encountered in the tumors (21). DNA hypomethylation is known to enhance the expression of proto-oncogenes leading to an elevated cancer risk. DNA hypermethylation is also associated with certain cancers. Diet has been known to affect the DNA methylation status, especially folate, which is the precursor for SAM, the substrate for DNMT that catalyzes DNA methylation. Dietary phytochemical polyphenols have been identified to modulate DNA methylation and subsequent epigenetic regulation of gene expression with a change in the outcome of cancer and CVDs. Wherever DNA hypermethylation catalyzed by DNMTs is encountered as a key mechanism in a disease such as cancer, DNMT inhibitors can be promising drugs in the chemotherapy of cancer (29). However, certain phytochemicals, especially the polyphenols, are emerging as modulators of DNA methylation and appear as promising drugs in the treatment of cancer and myocardial pathologies (9, 36).
Post-translation modifications of nuclear proteins, the histones, play a crucial role in the epigenetic regulation of gene expression. Two important modifications of histones are (i) acetylation and (ii) deacetylation. Histone acetylation is catalyzed by the histone acetyltransferases (HATs). Histone deacetylation is catalyzed by 11 different HDACs, which are divided into 4 classes based on their homologies (37). These HDACs are distributed in the cytoplasm, mitochondria, and nucleus. There are seven members of HDACs called SIRTs (SIRT1–SIRT7) present in the cytoplasm, mitochondria, and nucleus, which catalyze the deacetylation of histones (37). A tight balance between the acetylation and deacetylation of histones is maintained under normal states, but if that balance is altered due to the abnormal activities of either HATs or HDACs, then that will result in pathological situations as seen in cancer (37, 41). HDAC inhibitors have been focused as therapeutic molecules in cancer chemotherapy. Once again, phytochemical polyphenols have been shown to act as HDAC inhibitors with potential in prevention and therapy of certain diseases, including cancer and CVDs (10, 36).
Phytochemical Polyphenols As Modulators of Epigenome: Disease Prevention and Therapy
Polyphenols are a major group of phytochemicals with antioxidant actions, disease prevention properties, and therapeutic actions. However, polyphenols have recently captured attention as the modulators of epigenome. Although humans consume most of the polyphenols through diet, they also can be ingested or administered as isolated compounds from plant sources for prevention or treatment of certain diseases (50). Green tea contains several polyphenols, including (−)-epicatechin, (−)-epicatechin-3-gallate (ECG), (−)-epigallocatechin, and (−)-epigallocatechin-3-gallate (EGCG) (48). Green tea as a beverage is also seriously consumed worldwide as a health-promoting drink due to the presence of phytochemicals, which are considered to act as anticancer agents, retain healthy heart, and prevent cataract. EGCG is a predominant polyphenol present in green tea that has been shown to possess anticancer, antitumor, and anti-inflammatory actions (Fig. 2) (52). In a study on the human breast cancer cells (MCF-7 and MDA-MB-231 cells), EGCG and the novel prodrug of EGCG (pro-EGCG) have been shown to inhibit cell proliferation and transcription of the human telomerase reverse transcriptase (hTERT), the catalytic subunit of telomerase that is crucial in sustaining the telomere chain length and tumor formation, the latter being through the epigenetic regulation of the estrogen receptor (11, 52). In this study, hypomethylation of the hTERT promoter region and histone deacetylation through the inhibition of DNMT and HAT, respectively, have contributed to the inhibition of hTERT transcription in the breast cancer cells under the treatment of EGCG and pro-EGCG. Moreover, EGCG and pro-EGCG also have caused the chromatin remodeling (alterations) leading to hTERT-repressor binding in the regulatory sites. This study reveals that the green tea polyphenol EGCG and its novel prodrug cause inhibition of proliferation of breast cancer cells through epigenetic mechanisms involving inhibition of DNMT and HAT and resulting in DNA hypomethylation and histone deacetylation (Fig. 3). The importance of estrogen receptor-α (Erα) in clinical prognosis and in the therapy of breast cancer has been emphasized, because ERα-deficient breast cancers do not respond to the therapies aimed at the hormone targets (49). Having this as the premise, it has been revealed that in the breast cancer cells (MDA-MB-231 cells), the green tea polyphenol (EGCG) alone or in combination with the HDAC inhibitor (trichostatin A) offered reactivation of the ERα expression in the ERα-deficient breast cancer cells (49). This has also led to the estradiol action in the ERα-deficient breast cancer cells through the action of the ERα receptors in response to estradiol treatment. In this context, EGCG has been found to cause chromatin remodeling through modulation of histone acetylation and DNA methylation, leading to the reactivation of ERα (49). This study indicates that the green tea polyphenol, EGCG, can reactivate ERα toward effective treatment of hormone-resistant breast cancer. To establish the anticancer actions of EGCG against skin cancer through epigenetic regulation, a study has been conducted on the reactivation of the tumor suppressor genes (Cip1/p21 and p16INK4α) in human skin cancer cells (55). This study reveals that EGCG suppresses global DNA methylation, DNMT activity, and HDAC activity; lowers DNMT protein and mRNA; and increases histone acetylation in histones H-3 and H-4 in the skin cancer cells. Along these lines, ECGC has been observed to cause activation of expression of the tumor suppressor genes. This study presents convincing results that the green tea polyphenol, EGCG, is an epigenetic modulator that may be useful as an epigenetic drug in skin cancer therapy.
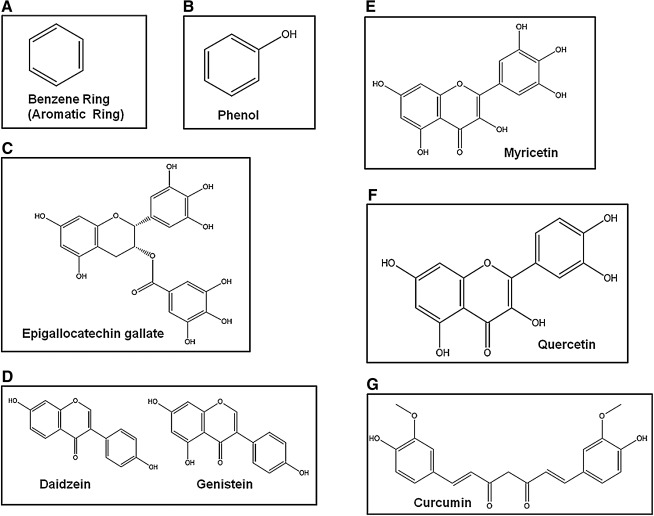
FIG. 2.
Chemical structures of different phytochemical polyphenol antioxidants (A–G). Phytochemical polyphenols have their structural origin from the simple phenolic (benzene) ring and having two or more hydroxyl (OH) groups offer their polyphenolic nature and name of their class of compounds. These are compounds that are bioactive natural products and act as free-radical quenchers, potent antioxidants, trace metal-complexing molecules, pro-oxidants, and regulators of cell proliferation. More strikingly, they are emerging as the modulators of epigenetic regulation of gene expression.
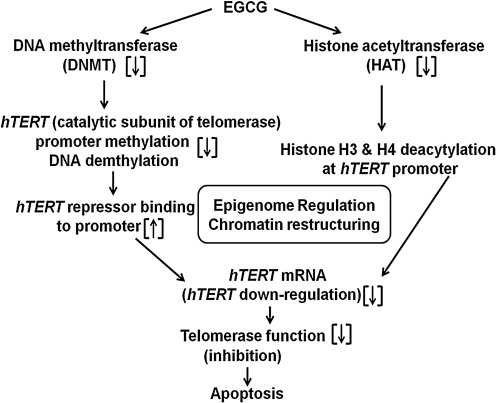
FIG. 3.
Mechanism of EGCG-induced apoptosis in cancer cells through epigenetic regulation of telomerase. According to Berletch et al. (11) and Meeran et al. (52), EGCG inhibits both deoxyribonucleic acid (DNA) methyltransferase (DNMT) and histone acetyltransferase (HAT), leading to the DNA demethylation and histones H3 and H4 deacetylation of the human telomerase– reverse transcriptase (hTERT) promoter, respectively. These events result in the epigenome regulation and chromatin restructuring involving hTERT messenger ribonucleic acid (mRNA) downregulation and inhibition of telomerase and ultimately cancer cell death (apoptosis). However, these studies have not provided any links between the antioxidant actions of EGCG and its epigenetic regulation of apoptosis of the cancer cells.
With intent to show the anticancer actions of EGCG, a study has been conducted on the epigenetic anticancer effects of EGCG on the UV-B radiation-induced skin cancer in vivo in a hairless mouse model (54). In this study, the green tea polyphenol, EGCG, has been applied on the affected skin as a topical cream. The results reveal that the EGCG as topical cream inhibits the UV-B-induced skin papillomas and carcinomas and also inhibits global DNA methylation. Furthermore, this study demonstrates that the EGCG topical application as a cream on the skin appears as a promising epigenetic therapeutic strategy for the treatment of skin photocarcinogenesis. The antimelanoma action of EGCG has been investigated in the human melanoma cell line (A-375 cells) with a focus on the epigenetic action of EGCG (58). In this study, the HDAC inhibitor drug, Vorinostat, has also been used in conjunction with EGCG. The results reveal that the antimelanoma effects of Vorinostat are more pronounced than those of EGCG, but the combined treatment of EGCG and Vorinostat has been more dramatic in causing the antimelanoma effects. Thus, it is clear from this study that EGCG can be used as a combination drug along with a known HDAC inhibitor for use in the epigenetic therapy of melanoma.
Dietary phytochemical antioxidants are also known to exert immunomodulatory effects, including upkeep of immunotolerance and autoimmunity suppression. Along those lines, a study has been launched to investigate modulation of the regulatory T cells by EGCG through epigenetic regulation (78). Regulatory T cells are crucial for the immunotolerance and autoimmunity suppression. The results of this study reveal that EGCG decreases DNMT expression and global DNA methylation through induction of forkhead box p3 (Foxp3) gene expression substantiating the epigenetic actions EGCG and modulation of immunity by EGCG. Thus, it appears that the green tea polyphenol, EGCG, also acts as an epigenetic immunomodulator. Overall, consumption of green tea has been attributed to the lower incidences of various cancers such as gastric, esophageal, ovarian, pancreatic, skin, and colorectal cancers (48). The cancer-preventive actions of green tea are linked to the presence of the bioactive natural product in the tea such as EGCG. Many studies have shown that EGCG directly inhibits the DNMT activity by directly interacting with the enzyme, causes demethylation of DNA, and reactivates methylation-silenced genes (48). This mechanism of action of EGCG in causing modulation of DNA methylation through the regulation of DNMTs and leading to epigenetic regulation of gene expression has been demonstrated in several cancer models, substantiating the epigenetic anticancer action of EGCG (48, 49).
Curcumin As a Modulator of Epigenome: Disease Prevention and Therapy
Curcumin (diferuloylmethane) is a polyphenolic ingredient of turmeric (the most popular and common Asian Indian curry spice; golden spice) obtained from the rhizomes of the plant, Curucuma longa (Fig. 2). Recently, curcumin has gained tremendous attention of the nutritionists, biomedical scientists, pharmacologists, drug discovery scientists, clinicians, and, above all, the common man worldwide as a preventive molecule or therapeutic agent for several disorders and diseases, which is substantiated by anecdotal accounts and scientific investigations. However, curcumin has long been known for its anticancer actions causing necrosis and apoptosis and arrest of division in cancer cells (33, 35). Curcumin has been identified as a pro-oxidant in generating ROS (causing oxidative stress) and as an antioxidant (protecting against oxidative stress) in cellular systems. The oxidative stress induced by curcumin has been considered as the probable mechanism of action of the compound to act as an anticancer agent. The biphasic action of curcumin depends upon its concentration that is used in experiments, for example, at higher concentrations (∼50 μM) curcumin acts as a pro-oxidant and at lower doses (∼10 μM), the same acts as an antioxidant (35). Although several studies have revealed multiple mechanisms/targets for the anticancer action of curcumin, the epigenetic regulation by curcumin in different disease states (models), including cancer, is sprouting (33). A study on the human hepatoma cells reveals that curcumin treatment lowers histone acetylation (hypoacetylation) by inhibiting the HAT activity and without an effect on the HDAC activity, which is linked with the enhancement of ROS production in cells (35). This study suggests that HAT is the target for curcumin, and its anticancer action could be attributed to its epigenome regulatory actions wherein ROS are apparently involved. On the other hand, curcumin has been shown to inhibit the HDAC activity in medulloblastoma (brain tumor) cells and to decrease medulloblastoma growth (tumor xenografts) in vitro and in vivo (44). In this study, curcumin has been observed to cause apoptosis and cell cycle arrest (G2/M phase) followed by the inhibition of HDAC activity in medulloblastoma cells. Also, the study reveals that curcumin enhances the survival of the mice that received the medulloblastoma tumor xenograft (44). Overall, this study features the importance of curcumin as an antimedulloblastoma agent with an epigenetic target in its path of action. Upon screening 33 carboxylate derivatives to identify potent HDAC inhibitors in HeLa cell nuclear extract an in vitro assay system, it has been identified that curcumin has the highest potency along with chlorogenic acid as compared to the established HDAC inhibitor, sodium butyrate (12). This study reveals that curcumin is a potent natural phytochemical polyphenol HDAC inhibitor with an ability to modulate the epigenome through regulation of histone acetylation.
Phosphodiesterases (PDEs) catalyze the hydrolysis of cyclic nucleotides such as cyclic adenosine-3′,5′-monophosphate and cyclic guanosine-3′,5′-monophosphate in cells and thus take crucial part in cell-signaling events responsible for cell division (1). Using the B16F10 melanoma cells, it has been shown that curcumin inhibits cell proliferation through PDE1-5 involvement. In this study, curcumin has also exerted epigenetic modulatory effectors such as inhibiting the expression of the epigenetic integrator ubiquitin-like containing PHD and ring finger domains 1 (UHRF1), and DNMT1 with upstream targeting of PDE1 and resulting in the antiproliferative effects in the melanoma cells (1). Thus, this study also demonstrates that curcumin exerts its epigenome-modulating effects in its action as an anticancer agent. The epigenetic regulation of HATs, HDACs, DNMTs, and miRNAs and associated modulation of gene expression by curcumin in conjunction with its anticancer actions and clinical utilization have been highlighted (66).
In addition to its anticancer actions, curcumin has been also shown to offer protection against inflammation, neurodegenerative diseases, autoimmune pathologies, and cardiovascular and lung disorders, while the epigenetic mechanisms of cardioprotective and lung-protective actions of curcumin are just budding (4, 8, 65, 79). Curcumin, as an antioxidant, has been reported to offer protection against several pathophysiological states (79). Especially, it has been demonstrated in studies with animal models that curcumin protects against cardiac hypertrophy and heart failure involving epigenetic regulation by HAT (79). One of the established mechanisms of epigenetic regulations of gene expression is maintaining a tight balance between the acetylation and deacetylation by a controlled regulation of the activities of HATs and HDACs. A specific transcription factor such as the hypertrophy-responsive transcription factor requires p300 (adenovirus E1A-associated protein), which also acts as a HAT bringing out the chromatin remodeling (79). p300-HAT has also been shown to be responsible for the cardiomyocyte growth and differentiation in the course of development (79). However, the activity of p300-HAT has been elevated during cardiac hypertrophy, and by inhibiting the p300-HAT activity, it has been possible to protect against the cardiac hypertrophy. Curcumin has been established as an inhibitor of p300-HAT (79). Thus, it is conclusive that curcumin protects against cardiac hypertrophy through modulation of epigenetic regulation that is mediated by p300-HAT. As histone acetylation mediated by HAT has been established as a critical player in the development of the heart, its inhibition or attenuation by curcumin has been studied in the cardiomyocytes (72). Curcumin has been revealed to inhibit p300-HAT in cardiomyocytes causing decreased acetylation of histone H3 in the promoter regions of certain cardiac-specific genes responsible for the cardiogenesis (72). This observation has been associated with curcumin-induced cell morphological alterations, inhibition of the HAT (p300-HAT) activity, decreased acetylation of histone H3, and suppression of cardiac-specific gene expression confirming curcumin-mediated modulation of epigenetic regulation of cardiomyocyte gene expression during cardiogenesis. This study also underscores the importance of curcumin as a therapeutic compound in alleviating congenital myocardial diseases and cardiac hypertrophy through epigenetic regulation.
Mechanisms behind several debilitating lung diseases in humans are complex, and specific targets in the onset and progression of those lung diseases such as chronic obstructive pulmonary diseases (COPDs) for pharmacological (drug) intervention are being constantly hunted. In this regard, specific and effective drugs are also highly warranted. Multiple mechanisms operating behind COPD have been documented, including the role of ROS, oxidative stress, membrane lipid deterioration by lipid peroxidation, loss of cellular thiol (GSH) redox, weakened antioxidant defense system, and other complex signaling mechanisms (65). Several therapeutic interventions, such as antioxidant therapy, thiol-redox and GSH boosting, enhancement of antioxidant defenses, enzyme activators, spin traps for reactive radicals have been sought after for the COPD therapy (65). Among those, polyphenols and curcumin have been described as therapeutic agents for COPD. More strikingly, the role of epigenome in the curcumin therapy of COPD has been brought to the lime light (8). HDAC, especially HDAC2, has been recognized to play an important role in the inflammatory gene expression and inflammatory lung pathologies, including COPD, asthma, and airway diseases, since histone acetylation upregulates the lung inflammatory genes and causes lung inflammation (8). Therefore, any pharmacological activation of HDAC appears to offer therapeutic intervention for lung inflammatory diseases. Curcumin has been suggested as a possible HDAC2 activator in protecting against the inflammatory lung diseases, but the challenge lies in its inhibitory action on HAT (8). However, the epigenetic mechanisms of protective action of curcumin on inflammatory lung diseases have to be thoroughly investigated prior to its clinical use.
Flavonoids As Modulators of Epigenome: Disease Prevention and Therapy
Flavonoids comprise a major group of phytochemicals, and they fall under polyphenol category with diverse chemical structures. Although their source of entry into humans is diet, there is a growing interest in flavonoids for their disease-preventive and therapeutic actions. Flavonoids have been established as potent and naturally available antioxidants with properties to relieve oxidative stress, disease prevention, and protection (60). However, the epigenome-regulating actions of flavonoids are just surfacing (27). Flavonoids—in particular, isoflavones, flavonols, and catechins—have been emphasized as the phytochemical polyphenol regulators of the epigenome with a focus on DNA methylation, histone acetylation, and chromatin alterations (27). Here, the findings of some selected studies dealing with the flavonoid-modulated epigenetic regulation with a relevance to diseases are discussed.
In the human leukemia-60 cells (HL-60 cells), quercetin (Fig. 2) has been shown to exert epigenetic modulations involving activation of HAT and inhibition of HDAC, leading to histone acetylation (45). This study revealed that quercetin induces Fas ligand-mediated apoptosis in HL-60 cells that is associated with the epigenetic regulation through HAT and HDAC (Fig. 4). Soybean isoflavones have been identified as phytochemical therapeutic flavonoids for the treatment of colorectal cancer wherein the isoflavones suppress metastasis of the tissue through epigenetic modulation of DNA methylation and histone modifications (47). In particular, this study highlights genistein, one of the soybean isoflavones, as an effective epigenetic modulator of the colorectal cancer metastasis, and dietary genistein may be beneficial for colorectal cancer (Fig. 2) (47). Genistein, through epigenetic modulations, including chromatin remodeling and DNA methylation, leads to the activation of tumor suppressor genes and suppression of the survival of cancer cells (82). Genistein has also been shown to inhibit the DNMT activity, which causes inhibition of DNA methylation and thus may be acting as an anticancer agent (48). Genistein has been observed to enhance the acetylation of histones H3 and H4 in the transcription sites of p21 and p16 with which the upregulation of the tumor suppressor genes in prostate cancer cells (82). The cell-cycle arrest results from the genistein-induced downregulation of cyclins from the upregulation of p21 and p16 in prostate cancer cells. From this study, genistein appears as a promising anticancer flavonoid that operates through epigenetic regulation of gene expression. An association between genistein consumption and the low mortality rate among Asian women with breast cancer has been documented (48). In various experimental cellular models of cancer such as the prostate, esophagus, and colon cancer cells, genistein has been shown to act as an anticancer flavonoid (48). Although multiple mechanisms of the anticancer actions of genistein, such as inhibition of DNA mutation, suppression of cancer cell division, antiangiogenic effect, and stimulation of cell differentiation have been reported, the mechanism of epigenetic regulation of gene expression by genistein while exerting its anticancer actions is becoming more evident. An important discovery that has led to the understanding of the estrogenic activity of genistein in which prenatal exposure to genistein permanently affects the erythropoiesis in fetus and alters the gene expression and DNA methylation in hematopoietic cells (75). Also, ligands for isoflavones in the ERα-mediated HAT activity have been identified, and genistein has been observed to cause modulation of the HAT activity and extent of histone acetylation (64). These studies underscore the estrogenic nature of the phytoestrogen and genistein and their effects on ERα, which may be seriously considered for use as an anticancer agent.
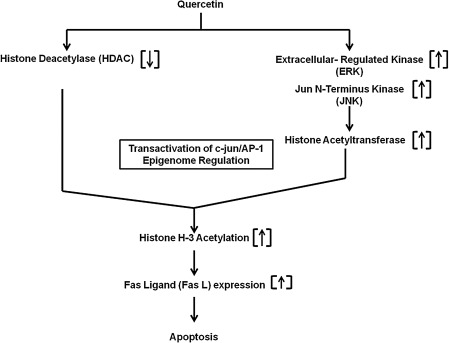
FIG. 4.
Mechanism of quercetin-induced apoptosis of cancer cells through epigenetic regulation of Fas ligand (Fas L) expression. As investigated by Lee et al. (45), quercetin inhibits the histone deacetylase (HDAC) activity and activates HAT through the upstream activation of the extracellular-regulated kinase (ERK) and Jun N-terminus kinase (JNK). Here, the transactivation of c-jun/AP-1 is involved. Thus, the epigenome is regulated by quercetin, leading to apoptosis of the cancer cells (medulloblastoma) through elevation of histone H3 acetylation, which causes downstream upregulation of Fas L. In this study, no attempt has been made to establish the connection between the antioxidant actions of quercetin and its epigenetic regulation of apoptosis of the medulloblastoma.
Isoflavones of dietary origin have been documented to offer vascular protection in different experimental models and humans through protection against oxidative stress and upregulation of the antioxidant-signaling mechanisms (69). Dietary isoflavones have been shown to elevate the production of nitric oxide and ROS in the vessel wall and enhance the activities of antioxidant enzymes in the vascular endothelial and smooth muscle cells, which are attributed to the estrogenic activities of isoflavones that upregulate the genes for antioxidant enzymes in those vascular cells. Although the isoflavones offer vasculoprotection and, in particular, are capable of inhibiting HAT and DNMT (69), the exact epigenetic mechanism of protection of vascular endothelial and smooth muscle cells needs to be established.
Conclusions: Critical Issues and Future Directions
Studies conducted so far have provided convincing evidence that phytochemical antioxidants (polyphenols and flavonoids such as quercetin and curcumin) do modulate epigenetic regulation of gene expression, which could stand out as a plausible target for the intervention of certain diseases by the phytochemicals of choice (Fig. 5). In cancer biology, the phytochemical-modulated epigenome actions have picked up the pace, and the clinical use of phytochemical polyphenols as epigenetic therapeutics is promising. However, with respect to other diseases and disorders, studies on the epigenome actions of phytochemical antioxidants are nascent. The most important aspects of phytochemical antioxidants that should be understood thoroughly are (i) the metabolism of the phytochemicals in mammalian and human cells in vitro and in vivo to identify the active metabolite of the phytochemical that is responsible for its pharmacological actions and (ii) precise cellular targets (receptors or proteins) for either the parent phytochemical molecule or its cellular metabolites under normal physiological and pathological states. In this regard, specific bioactive metabolites of a particular phytochemical antioxidant of choice, in target cells and their normal counterparts, have to be identified and characterized (Fig. 5). In addition, the redox biology of these phytochemical antioxidants in the cellular milieu has to be established as most of these natural compounds have a dual behavior of acting either as a pro-oxidant or as an antioxidant. The question that still remains to be answered is whether the redox-active phytochemical antioxidant that causes epigenetic regulation is either dependent upon the oxidants generated through the pro-oxidant actions or the antioxidant nature of the phytochemical polyphenol antioxidant (Fig. 6). In spite of the well-established fact that the phytochemical polyphenolics are among the most effective naturally occurring antioxidants, there is a void bridging their antioxidant actions and redox-signaling mediation to their epigenetic regulatory actions. It is high time to establish whether the epigenome regulatory actions of the phytochemical polyphenol antioxidants are related or unrelated to their antioxidative actions (Fig. 6). So far, laboratory studies conducted offer convincing evidence in favor of a connection between the chromatin remodeling and epigenetic regulation of gene expression, and this should be established or verified in the preclinical and clinical studies.
FIG. 5.
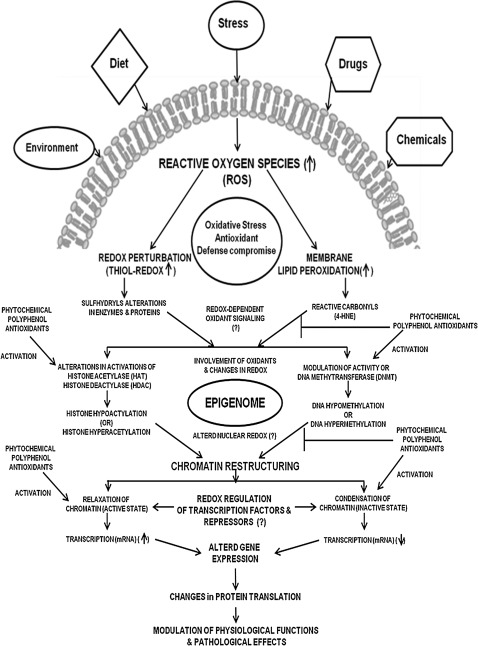
Proposed mechanisms of epigenetic regulation by phytochemical polyphenol antioxidants and the link between reactive oxygen species (ROS) and oxidative stress and antioxidative and nonantioxidative pathways in physiological and pathophysiological states. Stress, diet, chemicals, drugs, and environmental factors cause oxidative stress through ROS and thiol-redox dysregulation, which result in epigenome alterations resulting in chromatin restructuring through modulations in DNMT, HDACs, and HAT and subsequent changes in DNA methylation and histone acetylation. Thus, the regulation of gene expression is brought by the alterations in the epigenome. Phytochemical polyphenol antioxidants may act either as (i) antioxidants relieving the oxidative stress and/or (ii) direct modulators of DNMT, HDACs, and HATs.
FIG. 6.
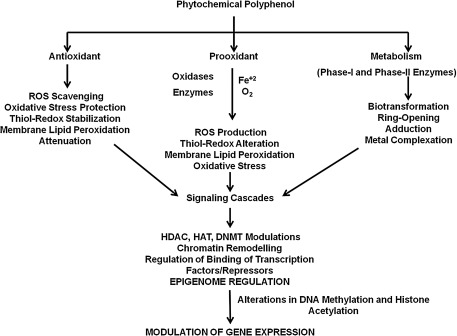
Proposed mechanisms of epigenome regulatory activities of phytochemical polyphenols through their antioxidant actions, pro-oxidant nature, and bioactive metabolite functions. As the phytochemical polyphenols are known to act as effective antioxidants, they may regulate the functions of cellular epigenome by acting as antioxidants. Phytochemical polyphenols also act as pro-oxidants in presence of iron (Fe+2) and oxygen or in concert with oxidases and induce oxidative stress, which may cause the modulation of the cellular epigenome through oxidant and thiol-redox signaling. Phytochemical polyphenols are also known to undergo biotransformation into metabolites that is catalyzed by the cellular phase-I and phase-II xenobiotic-metabolizing enzymes. It is possible that these metabolites regulate the cellular epigenome through signaling cascades.
Abbreviations Used
- 4-HNE
4-hydroxy-2-nonenal
- ADP
adenosine-3′,5′-diphosphate
- C
cytosine
- COPDs
chronic obstructive pulmonary diseases
- CpG
phosphodiester bond between cytosine and guanine
- CVD
cardiovascular disease
- DNA
deoxyribonucleic acid
- DNMT
DNA methyltransferase
- ECG
(−)-epicatechin-3-gallate
- EGCG
(−)-epigallocatechin-3-gallate
- ERK
extracellular-regulated kinase
- Erα
estrogen receptor-α
- Foxp3
forkhead box p3
- G
guanine
- GSH
glutathione
- H2A
histone 2A
- H2B
histone 2B
- H3
histone 3
- H4
histone 4
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HL-60 cells
human leukemia-60 cells
- hTERT
human telomerase reverse transcriptase
- JNK
Jun N-terminus kinase
- miRNA
micro-ribonucleic acid
- mRNA
messenger ribonucleic acid
- PDE
phosphodiesterase
- pro-EGCG
pro-drug of (−)-epigallocatechin-3-gallate
- RNA
ribonucleic acid
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SAM
S-adenosylmethionine
- SIRT
sirtuin
- UHRF1
ubiquitin-like containing PHD and ring finger domains 1
- UV
ultraviolet
Acknowledgments
Funding support from the International Academy of Oral Medicine and Toxicology (IAOMT); Dorothy M. Davis Heart & Lung Research Institute; the Division of Pulmonary, Allergy, Critical Care, and Sleep Medicine of the Ohio State University College of Medicine; and the National Institutes of Health (HL 093463) is acknowledged.
References
- 1.Abusnina A. Keravis T. Yougbare I. Bronner C. Lugnier C. Anti-proliferative effect of curcumin on melanoma cells is mediated by PDE1A inhibition that regulates the epigenetic integrator UHRF1. Mol Nutr Food Res. 2011;55:1677–1689. doi: 10.1002/mnfr.201100307. [DOI] [PubMed] [Google Scholar]
- 2.Acamovic T. Brooker JD. Biochemistry of plant secondary metabolites and their effects in animals. Proc Nutr Soc. 2005;64:403–412. doi: 10.1079/pns2005449. [DOI] [PubMed] [Google Scholar]
- 3.Adibhatla RM. Hatcher JF. Lipid oxidation and peroxidation in CNS health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2010;12:125–169. doi: 10.1089/ars.2009.2668. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal BB. Harikumar KB. Potential therapeutic effects of curcumin, the anti-inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int J Biochem Cell Biol. 2009;41:40–59. doi: 10.1016/j.biocel.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arts IC. Hollman PC. Polyphenols and disease risk in epidemiologic studies. Am J Clin Nutr. 2005;81:317S–325S. doi: 10.1093/ajcn/81.1.317S. [DOI] [PubMed] [Google Scholar]
- 6.Baker JE. Oxidative stress and adaptation of the infant heart to hypoxia and ischemia. Antioxid Redox Signal. 2004;6:423–429. doi: 10.1089/152308604322899495. [DOI] [PubMed] [Google Scholar]
- 7.Bao J. Sack MN. Protein deacetylation by sirtuins: delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell Mol Life Sci. 2010;67:3073–3087. doi: 10.1007/s00018-010-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes PJ. Histone deacetylase-2 and airway disease. Ther Adv Respir Dis. 2009;3:235–243. doi: 10.1177/1753465809348648. [DOI] [PubMed] [Google Scholar]
- 9.Barnes S. Prasain J. D'Alessandro T. Arabshahi A. Botting N. Lila MA. Jackson G. Janle EM. Weaver CM. The metabolism and analysis of isoflavones and other dietary polyphenols in foods and biological systems. Food Funct. 2011;2:235–244. doi: 10.1039/c1fo10025d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry SP. Townsend PA. What causes a broken heart—molecular insights into heart failure. Int Rev Cell Mol Biol. 2010;284:113–179. doi: 10.1016/S1937-6448(10)84003-1. [DOI] [PubMed] [Google Scholar]
- 11.Berletch JB. Liu C. Love WK. Andrews LG. Katiyar SK. Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103:509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bora-Tatar G. Dayangac-Erden D. Demir AS. Dalkara S. Yelekci K. Erdem-Yurter H. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: activity and docking studies. Bioorg Med Chem. 2009;17:5219–5228. doi: 10.1016/j.bmc.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 13.Bors W. Michel C. Chemistry of the antioxidant effect of polyphenols. Ann N Y Acad Sci. 2002;957:57–69. doi: 10.1111/j.1749-6632.2002.tb02905.x. [DOI] [PubMed] [Google Scholar]
- 14.Cave AC. Brewer AC. Narayanapanicker A. Ray R. Grieve DJ. Walker S. Shah AM. NADPH oxidases in cardiovascular health and disease. Antioxid Redox Signal. 2006;8:691–728. doi: 10.1089/ars.2006.8.691. [DOI] [PubMed] [Google Scholar]
- 15.Cheeseman KH. Mechanisms and effects of lipid peroxidation. Mol Aspects Med. 1993;14:191–197. doi: 10.1016/0098-2997(93)90005-x. [DOI] [PubMed] [Google Scholar]
- 16.Chen H. Yoshioka H. Kim GS. Jung JE. Okami N. Sakata H. Maier CM. Narasimhan P. Goeders CE. Chan PH. Oxidative stress in ischemic brain damage: mechanisms of cell death and potential molecular targets for neuroprotection. Antioxid Redox Signal. 2011;14:1505–1517. doi: 10.1089/ars.2010.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi DY. Lee YJ. Hong JT. Lee HJ. Antioxidant properties of natural polyphenols and their therapeutic potentials for Alzheimer's disease. Brain Res Bull. 2012;87:144–153. doi: 10.1016/j.brainresbull.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Chung S. Yao H. Caito S. Hwang JW. Arunachalam G. Rahman I. Regulation of SIRT1 in cellular functions: role of polyphenols. Arch Biochem Biophys. 2010;501:79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dell'Agli M. Busciala A. Bosisio E. Vascular effects of wine polyphenols. Cardiovasc Res. 2004;63:593–602. doi: 10.1016/j.cardiores.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Dinkova-Kostova AT. Phytochemicals as protectors against ultraviolet radiation: versatility of effects and mechanisms. Planta Med. 2008;74:1548–1559. doi: 10.1055/s-2008-1081296. [DOI] [PubMed] [Google Scholar]
- 21.Duthie SJ. Epigenetic modifications and human pathologies: cancer and CVD. Proc Nutr Soc. 2011;70:47–56. doi: 10.1017/S0029665110003952. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson LR. Schlothauer RC. The potential role of nutritional genomics tools in validating high health foods for cancer control: broccoli as example. Mol Nutr Food Res. 2012;56:126–146. doi: 10.1002/mnfr.201100507. [DOI] [PubMed] [Google Scholar]
- 23.Forester SC. Lambert JD. The role of antioxidant versus pro-oxidant effects of green tea polyphenols in cancer prevention. Mol Nutr Food Res. 2011;55:844–854. doi: 10.1002/mnfr.201000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraga CG. Plant polyphenols: how to translate their in vitro antioxidant actions to in vivo conditions. IUBMB Life. 2007;59:308–315. doi: 10.1080/15216540701230529. [DOI] [PubMed] [Google Scholar]
- 25.Fritz KS. Galligan JJ. Smathers RL. Roede JR. Shearn CT. Reigan P. Petersen DR. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem Res Toxicol. 2011;24:651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghiringhelli F. Rebe C. Hichami A. Delmas D. Immunomodulation and anti-inflammatory roles of polyphenols as anticancer agents. Anticancer Agents Med Chem. 2012 Jan 31; doi: 10.2174/187152012802650048. [Epub ahead of print] PubMed PMID: 22292769. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert ER. Liu D. Flavonoids influence epigenetic-modifying enzyme activity: structure—function relationships and the therapeutic potential for cancer. Curr Med Chem. 2010;17:1756–1768. doi: 10.2174/092986710791111161. [DOI] [PubMed] [Google Scholar]
- 28.Gill PS. Wilcox CS. NADPH oxidases in the kidney. Antioxid Redox Signal. 2006;8:1597–1607. doi: 10.1089/ars.2006.8.1597. [DOI] [PubMed] [Google Scholar]
- 29.Gravina GL. Festuccia C. Marampon F. Popov VM. Pestell RG. Zani BM. Tombolini V. Biological rationale for the use of DNA methyltransferase inhibitors as new strategy for modulation of tumor response to chemotherapy and radiation. Mol Cancer. 2010;9:305. doi: 10.1186/1476-4598-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo RF. Ward PA. Role of oxidants in lung injury during sepsis. Antioxid Redox Signal. 2007;9:1991–2002. doi: 10.1089/ars.2007.1785. [DOI] [PubMed] [Google Scholar]
- 31.Hodek P. Trefil P. Stiborova M. Flavonoids—potent and versatile biologically active compounds interacting with cytochromes P450. Chem Biol Interact. 2002;139:1–21. doi: 10.1016/s0009-2797(01)00285-x. [DOI] [PubMed] [Google Scholar]
- 32.Hollman PC. Cassidy A. Comte B. Heinonen M. Richelle M. Richling E. Serafini M. Scalbert A. Sies H. Vidry S. The biological relevance of direct antioxidant effects of polyphenols for cardiovascular health in humans is not established. J Nutr. 2011;141:989S–1009S. doi: 10.3945/jn.110.131490. [DOI] [PubMed] [Google Scholar]
- 33.Huang J. Plass C. Gerhauser C. Cancer chemoprevention by targeting the epigenome. Curr Drug Targets. 2011;12:1925–1956. doi: 10.2174/138945011798184155. [DOI] [PubMed] [Google Scholar]
- 34.Janssen LJ. Catalli A. Helli P. The pulmonary biology of isoprostanes. Antioxid Redox Signal. 2005;7:244–255. doi: 10.1089/ars.2005.7.244. [DOI] [PubMed] [Google Scholar]
- 35.Kang J. Chen J. Shi Y. Jia J. Zhang Y. Curcumin-induced histone hypoacetylation: the role of reactive oxygen species. Biochem Pharmacol. 2005;69:1205–1213. doi: 10.1016/j.bcp.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Khan SI. Aumsuwan P. Khan IA. Walker LA. Dasmahapatra AK. Epigenetic events associated with breast cancer and their prevention by dietary components targeting the epigenome. Chem Res Toxicol. 2012;25:61–73. doi: 10.1021/tx200378c. [DOI] [PubMed] [Google Scholar]
- 37.Kim HJ. Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res. 2011;3:166–179. [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J. Lee HJ. Lee KW. Naturally occurring phytochemicals for the prevention of Alzheimer's disease. J Neurochem. 2010;112:1415–1430. doi: 10.1111/j.1471-4159.2009.06562.x. [DOI] [PubMed] [Google Scholar]
- 39.Korkina L. Kostyuk V. De Luca C. Pastore S. Plant phenylpropanoids as emerging anti-inflammatory agents. Mini Rev Med Chem. 2011;11:823–835. doi: 10.2174/138955711796575489. [DOI] [PubMed] [Google Scholar]
- 40.Kulkarni AC. Kuppusamy P. Parinandi N. Oxygen, the lead actor in the pathophysiologic drama: enactment of the trinity of normoxia, hypoxia, and hyperoxia in disease and therapy. Antioxid Redox Signal. 2007;9:1717–1730. doi: 10.1089/ars.2007.1724. [DOI] [PubMed] [Google Scholar]
- 41.Kurdistani SK. Histone modifications in cancer biology and prognosis. Prog Drug Res. 2011;67:91–106. doi: 10.1007/978-3-7643-8989-5_5. [DOI] [PubMed] [Google Scholar]
- 42.Lambert JD. Elias RJ. The antioxidant and pro-oxidant activities of green tea polyphenols: a role in cancer prevention. Arch Biochem Biophys. 2010;501:65–72. doi: 10.1016/j.abb.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lappas M. Hiden U. Desoye G. Froehlich J. Hauguel-de Mouzon S. Jawerbaum A. The role of oxidative stress in the pathophysiology of gestational diabetes mellitus. Antioxid Redox Signal. 2011;15:3061–3100. doi: 10.1089/ars.2010.3765. [DOI] [PubMed] [Google Scholar]
- 44.Lee SJ. Krauthauser C. Maduskuie V. Fawcett PT. Olson JM. Rajasekaran SA. Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo. BMC Cancer. 2011;11:144. doi: 10.1186/1471-2407-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee WJ. Chen YR. Tseng TH. Quercetin induces FasL-related apoptosis, in part, through promotion of histone H3 acetylation in human leukemia HL-60 cells. Oncol Rep. 2011;25:583–591. doi: 10.3892/or.2010.1097. [DOI] [PubMed] [Google Scholar]
- 46.Li G. Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q. Chen H. Epigenetic modifications of metastasis suppressor genes in colon cancer metastasis. Epigenetics. 2011;6:849–852. doi: 10.4161/epi.6.7.16314. [DOI] [PubMed] [Google Scholar]
- 48.Li Y. Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y. Yuan YY. Meeran SM. Tollefsbol TO. Synergistic epigenetic reactivation of estrogen receptor-alpha (ERalpha) by combined green tea polyphenol and histone deacetylase inhibitor in ERalpha-negative breast cancer cells. Mol Cancer. 2010;9:274. doi: 10.1186/1476-4598-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Link A. Balaguer F. Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manach C. Mazur A. Scalbert A. Polyphenols and prevention of cardiovascular diseases. Curr Opin Lipidol. 2005;16:77–84. doi: 10.1097/00041433-200502000-00013. [DOI] [PubMed] [Google Scholar]
- 52.Meeran SM. Patel SN. Chan TH. Tollefsbol TO. A novel prodrug of epigallocatechin-3-gallate: differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev Res (Phila) 2011;4:1243–1254. doi: 10.1158/1940-6207.CAPR-11-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minko IG. Kozekov ID. Harris TM. Rizzo CJ. Lloyd RS. Stone MP. Chemistry and biology of DNA containing 1,N(2)-deoxyguanosine adducts of the alpha,beta-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem Res Toxicol. 2009;22:759–778. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mittal A. Piyathilake C. Hara Y. Katiyar SK. Exceptionally high protection of photocarcinogenesis by topical application of (-)-epigallocatechin-3-gallate in hydrophilic cream in SKH-1 hairless mouse model: relationship to inhibition of UVB-induced global DNA hypomethylation. Neoplasia. 2003;5:555–565. doi: 10.1016/s1476-5586(03)80039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nandakumar V. Vaid M. Katiyar SK. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicholson SK. Tucker GA. Brameld JM. Physiological concentrations of dietary polyphenols regulate vascular endothelial cell expression of genes important in cardiovascular health. Br J Nutr. 2010;103:1398–1403. doi: 10.1017/S0007114509993485. [DOI] [PubMed] [Google Scholar]
- 57.Nicholson SK. Tucker GA. Brameld JM. Effects of dietary polyphenols on gene expression in human vascular endothelial cells. Proc Nutr Soc. 2008;67:42–47. doi: 10.1017/S0029665108006009. [DOI] [PubMed] [Google Scholar]
- 58.Nihal M. Roelke CT. Wood GS. Anti-melanoma effects of vorinostat in combination with polyphenolic antioxidant (-)-epigallocatechin-3-gallate (EGCG) Pharm Res. 2010;27:1103–1114. doi: 10.1007/s11095-010-0054-5. [DOI] [PubMed] [Google Scholar]
- 59.Otani H. Oxidative stress as pathogenesis of cardiovascular risk associated with metabolic syndrome. Antioxid Redox Signal. 2011;15:1911–1926. doi: 10.1089/ars.2010.3739. [DOI] [PubMed] [Google Scholar]
- 60.Panickar KS. Anderson RA. Effect of polyphenols on oxidative stress and mitochondrial dysfunction in neuronal death and brain edema in cerebral ischemia. Int J Mol Sci. 2011;12:8181–8207. doi: 10.3390/ijms12118181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pereira CV. Nadanaciva S. Oliveira PJ. Will Y. The contribution of oxidative stress to drug-induced organ toxicity and its detection in vitro and in vivo. Expert Opin Drug Metab Toxicol. 2012;8:219–237. doi: 10.1517/17425255.2012.645536. [DOI] [PubMed] [Google Scholar]
- 62.Perron NR. Brumaghim JL. A review of the antioxidant mechanisms of polyphenol compounds related to iron binding. Cell Biochem Biophys. 2009;53:75–100. doi: 10.1007/s12013-009-9043-x. [DOI] [PubMed] [Google Scholar]
- 63.Perron NR. Wang HC. Deguire SN. Jenkins M. Lawson M. Brumaghim JL. Kinetics of iron oxidation upon polyphenol binding. Dalton Trans. 2010;39:9982–9987. doi: 10.1039/c0dt00752h. [DOI] [PubMed] [Google Scholar]
- 64.Piaz FD. Vassallo A. Rubio OC. Castellano S. Sbardella G. De Tommasi N. Chemical biology of histone acetyltransferase natural compounds modulators. Mol Divers. 2011;15:401–416. doi: 10.1007/s11030-010-9299-5. [DOI] [PubMed] [Google Scholar]
- 65.Rahman I. Antioxidant therapeutic advances in COPD. Ther Adv Respir Dis. 2008;2:351–374. doi: 10.1177/1753465808098224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reuter S. Gupta SC. Park B. Goel A. Aggarwal BB. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 2011;6:93–108. doi: 10.1007/s12263-011-0222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santos-Reboucas CB. Pimentel MM. Implication of abnormal epigenetic patterns for human diseases. Eur J Hum Genet. 2007;15:10–17. doi: 10.1038/sj.ejhg.5201727. [DOI] [PubMed] [Google Scholar]
- 68.Scalbert A. Johnson IT. Saltmarsh M. Polyphenols: antioxidants and beyond. Am J Clin Nutr. 2005;81:215S–217S. doi: 10.1093/ajcn/81.1.215S. [DOI] [PubMed] [Google Scholar]
- 69.Siow RC. Mann GE. Dietary isoflavones and vascular protection: activation of cellular antioxidant defenses by SERMs or hormesis? Mol Aspects Med. 2010;31:468–477. doi: 10.1016/j.mam.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 70.Spatafora C. Tringali C. Natural-derived polyphenols as potential anticancer agents. Anticancer Agents Med Chem. 2012 Jan 31; doi: 10.2174/187152012802649996. [Epub ahead of print] PubMed PMID 22292761. [DOI] [PubMed] [Google Scholar]
- 71.Stoclet JC. Chataigneau T. Ndiaye M. Oak MH. El Bedoui J. Chataigneau M. Schini-Kerth VB. Vascular protection by dietary polyphenols. Eur J Pharmacol. 2004;500:299–313. doi: 10.1016/j.ejphar.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 72.Sun H. Yang X. Zhu J. Lv T. Chen Y. Chen G. Zhong L. Li Y. Huang X. Huang G. Tian J. Inhibition of p300-HAT results in a reduced histone acetylation and down-regulation of gene expression in cardiac myocytes. Life Sci. 2010;87:707–714. doi: 10.1016/j.lfs.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 73.Tsao R. Chemistry and biochemistry of dietary polyphenols. Nutrients. 2010;2:1231–1246. doi: 10.3390/nu2121231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Usatyuk PV. Natarajan V. Hydroxyalkenals and oxidized phospholipids modulation of endothelial cytoskeleton, focal adhesion and adherens junction proteins in regulating endothelial barrier function. Microvasc Res. 2012;83:45–55. doi: 10.1016/j.mvr.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vanhees K. Coort S. Ruijters EJ. Godschalk RW. van Schooten FJ. Barjesteh van Waalwijk van Doorn-Khosrovani S. Epigenetics: prenatal exposure to genistein leaves a permanent signature on the hematopoietic lineage. FASEB J. 2011;25:797–807. doi: 10.1096/fj.10-172155. [DOI] [PubMed] [Google Scholar]
- 76.Visioli F. De La Lastra CA. Andres-Lacueva C. Aviram M. Calhau C. Cassano A. D'Archivio M. Faria A. Fave G. Fogliano V. Llorach R. Vitaglione P. Zoratti M. Edeas M. Polyphenols and human health: a prospectus. Crit Rev Food Sci Nutr. 2011;51:524–546. doi: 10.1080/10408391003698677. [DOI] [PubMed] [Google Scholar]
- 77.Waggoner D. Mechanisms of disease: epigenesis. Semin Pediatr Neurol. 2007;14:7–14. doi: 10.1016/j.spen.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Wong CP. Nguyen LP. Noh SK. Bray TM. Bruno RS. Ho E. Induction of regulatory T cells by green tea polyphenol EGCG. Immunol Lett. 2011;139:7–13. doi: 10.1016/j.imlet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wongcharoen W. Phrommintikul A. The protective role of curcumin in cardiovascular diseases. Int J Cardiol. 2009;133:145–151. doi: 10.1016/j.ijcard.2009.01.073. [DOI] [PubMed] [Google Scholar]
- 80.Wood LG. Wark PA. Garg ML. Antioxidant and anti-inflammatory effects of resveratrol in airway disease. Antioxid Redox Signal. 2010;13:1535–1548. doi: 10.1089/ars.2009.3064. [DOI] [PubMed] [Google Scholar]
- 81.Yao H. Rahman I. Current concepts on oxidative/carbonyl stress, inflammation and epigenetics in pathogenesis of chronic obstructive pulmonary disease. Toxicol Appl Pharmacol. 2011;254:72–85. doi: 10.1016/j.taap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y. Chen H. Genistein, an epigenome modifier during cancer prevention. Epigenetics. 2011;6:888–891. doi: 10.4161/epi.6.7.16315. [DOI] [PubMed] [Google Scholar]