Abstract
Significance: Epidemiological and animal studies have demonstrated a close link between maternal nutrition and chronic metabolic disease in children and adults. Compelling experimental results also indicate that adverse effects of intrauterine growth restriction on offspring can be carried forward to subsequent generations through covalent modifications of DNA and core histones. Recent Advances: DNA methylation is catalyzed by S-adenosylmethionine-dependent DNA methyltransferases. Methylation, demethylation, acetylation, and deacetylation of histone proteins are performed by histone methyltransferase, histone demethylase, histone acetyltransferase, and histone deacetyltransferase, respectively. Histone activities are also influenced by phosphorylation, ubiquitination, ADP-ribosylation, sumoylation, and glycosylation. Metabolism of amino acids (glycine, histidine, methionine, and serine) and vitamins (B6, B12, and folate) plays a key role in provision of methyl donors for DNA and protein methylation. Critical Issues: Disruption of epigenetic mechanisms can result in oxidative stress, obesity, insulin resistance, diabetes, and vascular dysfunction in animals and humans. Despite a recognized role for epigenetics in fetal programming of metabolic syndrome, research on therapies is still in its infancy. Possible interventions include: 1) inhibition of DNA methylation, histone deacetylation, and microRNA expression; 2) targeting epigenetically disturbed metabolic pathways; and 3) dietary supplementation with functional amino acids, vitamins, and phytochemicals. Future Directions: Much work is needed with animal models to understand the basic mechanisms responsible for the roles of specific nutrients in fetal and neonatal programming. Such new knowledge is crucial to design effective therapeutic strategies for preventing and treating metabolic abnormalities in offspring born to mothers with a previous experience of malnutrition. Antioxid. Redox Signal. 17, 282–301.
Introduction
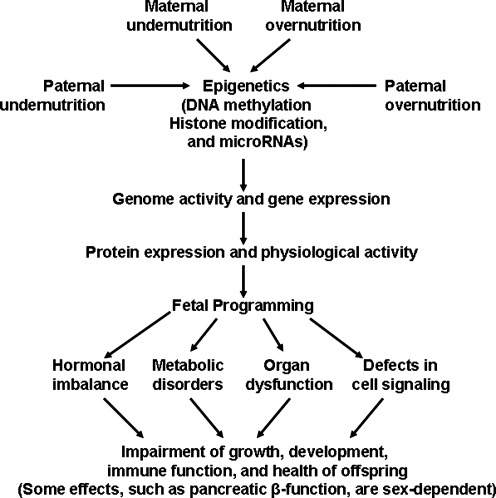
The United Nations Food and Agriculture Organization recently reported that 925 million people worldwide suffer from hunger (190). This population includes not only males and females of reproductive age, but also children who will grow to be adults and produce their own offspring. Currently, deficiencies in protein (amino acids), vitamin A, and iron remain major nutritional problems in developing nations (146), and a deficiency in folic acid can also occur when individuals have a low intake of fresh green vegetables or no consumption of folate-fortified foods (31). In both developed and developing countries, gestating women can experience undernutrition owing to: 1) severe nausea and vomiting known as hyperemesis gravidarum; 2) early or closely-spaced pregnancies; and 3) multi-fetal pregnancies resulting from assisted reproductive technologies (84, 104, 199). These conditions may result in intrauterine growth restriction (IUGR) and impaired health of the offspring (Fig. 1).
FIG. 1.
Impacts of maternal and paternal nutrition on fetal programming. Either undernutrition or overnutrition of the mother or father affects expression of the fetal genome, which may have lifelong consequences. Thus, alterations in fetal nutrition may result in developmental adaptations that permanently change the structure, physiology, and metabolism of offspring, thereby predisposing individuals to metabolic, endocrine, and cardiovascular diseases in adult life.
The other end of the nutrition spectrum is overnutrition, which can lead to overweight or obesity in human beings (115). One billion adults worldwide are overweight and more than 300 million are obese (126). Many overweight and obese women unknowingly enter pregnancy and continue overeating during gestation (39). These women usually gain more weight during the first pregnancy and accumulate more fat during subsequent pregnancies (128). Maternal obesity before or during gestation may result in adverse pregnancy outcomes, including maternal insulin resistance and hyperglycemia often associated with large-for-gestation age infants, IUGR, and impaired health of the offspring (Fig. 1). Thus, obesity is a global epidemic that negatively affects infants, children, and adults (48).
Extensive epidemiological studies have linked maternal undernutrition and overnutrition with the etiology of many chronic diseases in offspring when they reach adulthood (108). The metabolic syndrome can be defined as a cluster of disorders that include obesity, hyperglycemia (fasting serum glucose (>6.1 mM), hyperinsulinemia, hyperlipidemia, hypertension, and insulin resistance (an impaired response of cells or tissues to physiological concentrations of insulin) (78). These factors, independently or collectively, contribute to a high risk for metabolic disease, a major cause of death in developed nations and developing countries (126). This phenomenon led to the new concept of fetal programming that is defined as an adaptive process whereby nutrition and other environmental factors alter developmental pathways during the critical period of prenatal growth, thereby inducing changes in postnatal metabolism and susceptibility of adults to chronic disease (11). Such findings have prompted animal and human studies to identify the biological mechanisms responsible for the effects of intrauterine nutrition on long-term health consequences of the offspring (156, 200). Results of molecular studies indicate that fetal programming can be explained by epigenetics, which can be defined as stable and heritable alterations of gene expression through covalent modifications of DNA and core histones without changes in the DNA sequence (43). The present article highlights recent advances in this exciting area of biomedical research.
Impacts of Maternal Nutrition on Fetal Programming
Nutritional requirements for embryonic and fetal development
The conceptus (embryo/fetus and associated membranes) requires water, amino acids, lipids, carbohydrates, minerals, and vitamins for growth and development (13). In humans, the blastocyst stage of development is reached on about Day 5 after fertilization of the oocyte and then enters the uterus where the embryo differentiates into the inner cell mass (which will develop into the fetus) and the outer layer of cells (trophoblasts which will develop into the placenta and associated membranes). This is followed by implantation (Days 7 to 9 post-fertilization in humans), which is the first stage in sequence of events leading to placentation (the formation and growth of the placenta within the uterus). By Week 4 after fertilization, the basic structure of the mature placenta will has been established. Before placentation, the embryo receives nutrients from uterine secretions and oxygen from its surrounding environment (14). These secretions, including glucose and amino acids (e.g., arginine, leucine, proline, and glutamine), play crucial roles in activating cell signaling and metabolic pathways necessary for protein synthesis and cytoskeletal remodeling in the conceptus (196, 201). A deficiency of nutrients during this period or an inability of the conceptus to respond to the nutrients can result in abnormal development or even death of the conceptus (13).
The placenta transports nutrients, respiratory gases, and the products of their metabolism between the maternal and fetal circulations (186). Rates of uteroplacental blood flows depend on placental vascular growth (a result of angiogenesis) and placental vascularization that are greatly influenced by the availability of nitric oxide and polyamines (143, 198). To support increased uterine and placental blood flows, placental angiogenesis increases markedly from the first to the second third of gestation and continues to increase during late gestation. Uptake of nutrients by the uterus or the fetus is determined by both rate of blood flow and concentrations of nutrients in the arterial and venous blood. To support the rapid rate of placental and fetal growth, uptake of both macro- and micro-nutrients by the uterus is greater in pregnant women relative to that for nonpregnant women (143). Thus, impaired placental blood flow contributes to IUGR in mammalian pregnancies.
Energy is required for a variety of physiological processes in the fetus, including nutrient transport, cell motility, and biosynthetic pathways (64). Dietary macronutrients (glucose, amino acids, and fatty acids) are the ultimate sources of energy substrates during fetal growth (201). In the pregnant mother and her growing fetus, glucose is the major fuel for red blood cells, brain, retinal cells, and medullary cells of the kidney, while the maternal and fetal hearts utilize both glucose and lactate. Additionally, the small intestine of both the mother and the fetus oxidizes glutamate, aspartate, and glutamine to meet most of their energy requirements (78). Through β-oxidation, fatty acids are the major energy substrates for the maternal liver, skeletal muscle, heart, and kidneys (21). The fetal liver also oxidizes long-chain fatty acids (LCFA) to CO2 at relatively high rates. However, fetal muscle has a low capacity for oxidation of LCFA due to relatively low concentrations of carnitine and low carnitine palmitoyltransferase-I activity (78).
The human placenta is permeable to LCFA and transfers them to the fetus for metabolic utilization. In both maternal and fetal tissues (e.g., the liver), fatty acids are synthesized from acetyl-CoA which is a product of oxidation of glucose and amino acids (200). In most mammals (including humans), the fetus accumulates only a small amount of lipids prior to mid-gestation, but a relatively large amount of lipids accumulate exponentially thereafter (21). Because placental and fetal tissues have Δ9 desaturase, they are able to synthesize the ω9 (oleic acid) family of unsaturated fatty acids from palmitate (C16:0; a saturated fatty acid). Humans cannot synthesize linoleic acid or α-linolenic acid, which are ω6 and ω3 fatty acids, respectively. Therefore, these two nutritionally essential unsaturated fatty acids must be provided in the diet and used to synthesize the long-chain ω-6 and ω-3 polyunsaturated fatty acids. Physiologically and nutritionally important long chain ω-6 and ω-3 polyunsaturated fatty acids include arachidonic acid (20:4ω6), eicosapentaenoic acid (20:5ω3), and docosahexaenoic acid (22:6ω3).
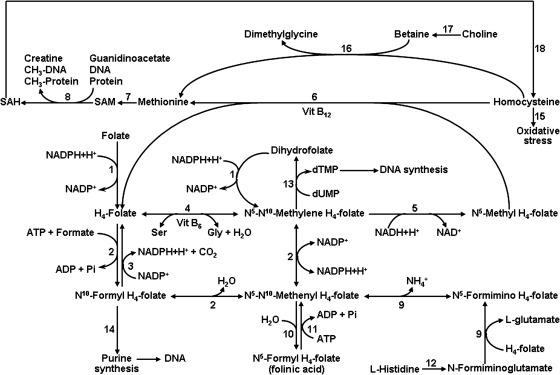
Amino acids serve not only as building blocks for proteins, but also as essential precursors for synthesis of a variety of physiologically important molecules, including hormones, small peptides (e.g., glutathione), neurotransmitters, nitric oxide, creatine, carnitine, and polyamines (196). Additionally, through multiple cell signaling pathways, amino acids regulate key metabolic pathways that are vital to human health, growth, development, and reproduction (197). Notably, some amino acids (histidine, methionine, glycine, serine) participate in one-carbon-unit metabolism, which is essential for DNA synthesis, as well as cell growth and development (Fig. 2). Based on nitrogen balance and growth, amino acids have been traditionally classified as either nutritionally essential (EAA) or nonessential (NEAA). The EAA are histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine. The NEAA are alanine, arginine, asparagine, aspartate, cysteine, glutamate, glutamine, glycine, proline, serine, taurine, and tyrosine. However, the basis for classification of an amino acid as a NEAA has conceptual limitations because there is no compelling evidence that certain NEAA (e.g., arginine and glutamine) can be adequately synthesized by the mother or fetus to support optimal survival, growth, and development of the conceptus (203, 204). The synthesis of proteins in the fetus depends on the balanced provision of all of the constituent amino acids, namely both EAA and NEAA.
FIG. 2.
One-carbon unit metabolism for provision of methyl donors in cells. Folate, histidine, methionine, glycine and serine participate in the transfer of one-carbon units for the synthesis of purines and DNA as well as methylation reactions in animals. The enzymes that catalyzed the indicated reactions are: 1) folate reductase; 2) N5-N10-methylene H4-folate dehydrogenase (a trifunctional enzyme possessing N10-formyl H4-folate synthetase, N5-N10-methylene H4-folate dehydrogenase, and N5-N10-methenyl H4-folate cyclohydrolase activities); 3) N10-formyl H4-folate dehydrogenase; 4) serine hydroxymethyl transferase; 5) N5-N10-methylene H4-folate reductase; 6) methionine synthase; 7) S-adenosylmethionine synthase; 8) S-adenosylmethionine as a major methyl group donor in methyltransferase reactions; 9) formiminotransferase-cyclodeaminase (a bifunctional enzyme possessing glutamate formiminotransferase and formimidoyl H4-folate cyclodeaminase activities); 10) serine hydroxymethyl transferase, N5-N10-methenyl H4-folate cyclohydrolase, and spontaneous reaction at pH 4 to 7.0; 11) N5-N10-methenyl H4-folate synthetase; 12) a sequential series of enzymes: histidase, urocanase, and imidazolone propionate hydrolase; 13) thymidylate synthase; 14) formyltransferase; 15) an oxidant for cells; 16) betaine:homocysteine methyltransferase; 17) choline dehydrogenase; and 18) S-adenosylhomocysteine hydrolase. dTMP, deoxythymidine 5'-monophosphate; dUMP, deoxyuridine 5'-monophosphate; Gly, glycine; H4-folate, tetrahydrofolate; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; Ser, serine; Vit, vitamin.
Nutritionally significant macro- and micro-minerals cannot be synthesized by the body and must, therefore, be provided in the diet (195, 213). The macro-minerals are sodium, potassium, chloride, calcium, phosphorus, magnesium, and sulfur (177). The micro-minerals are iodine, iron, zinc, selenium, manganese, copper, cobalt, chromium, molybdenum, silicon, fluoride, vanadium, and nickel (177). The inorganic nutrients are the backbone for development, maintenance, and function of the skeleton system, and they serve as second messengers in cell signaling, and maintain polarity of the plasma membrane. Minerals also regulate extracellular and intracellular osmolality, which is important for maintaining cell and blood volume as well as cell viability and shape (213). Most minerals are either cofactors for enzymes or components of metalloproteins, and also participate in gene expression, electron transport, and redox reactions (21).
Vitamins are organic nutrients that are required in small amounts for biochemical reactions. Water-soluble vitamins include vitamin C and members of the B complex that are absorbed by the small intestine into the portal circulation (127). Members of the B complex vitamins are vitamin B1 (thiamin), vitamin B2 (riboflavin), vitamin B3 (niacin), vitamin B5 (pantothenic acid), vitamin B6 (pyridoxal, pyridoxine, and pyridoxamine), vitamin B12 (cobalamin), biotin, and folate (21). Because of their water solubility, there is limited storage of these vitamins in the body (127). As a result, these micronutrients must be provided regularly in the diet. Lipid-soluble vitamins include vitamin A, vitamin D, vitamin E, and vitamin K. Most vitamins, except niacin and vitamin D, cannot be synthesized by humans and must be supplied in the diet (177). Vitamins serve as cofactors for enzymes involved in gene expression, metabolism of nutrients (e.g., fatty acids, glucose, and amino acids), one-carbon-unit transfer (Fig. 2), and ATP production.
Impacts of maternal undernutrition on offspring metabolism and health
In both humans and animals, birth weight is the predominant indicator of prior nutrient availability in utero. Epidemiological studies of the Dutch famine (November 1944 and May 1945) have shown that undernutrition of women during the second or third trimester, but not the first trimester, reduced weight, length, and head circumference of infants at birth (102). The growth-stunting effects were more pronounced for third trimester exposure than second trimester exposure (132) because most growth of the fetus occurs during late gestation (201). However, this does not necessarily mean that a severe deficiency in specific nutrients (e.g., micronutrients and amino acids) during the first trimester of pregnancy had little impact on maternal health or overall fetal growth and development. Indeed, maternal malnutrition during early gestation has a great effect on adversely influencing metabolism and health of offspring (200). Of note, in response to maternal undernutrition, the fetus develops mechanisms for adapting to the extrauterine environment that may confer an evolutionary advantage for survival of the species (152, 162). However, when environmental cues during prenatal life inappropriately program offspring, there are adverse consequences, including the increased prevalence of adult onset of disease (21, 156).
Maternal undernutrition increases risk for adult-onset of metabolic syndrome (11, 32). This notion is supported by four lines of evidence. First, at 4 years of age, children with low birth weight have elevated levels of glucose and insulin in response to a glucose challenge (206). Second, individuals exposed to the Dutch winter famine of 1944–1945 in utero had higher rates of insulin resistance, vascular disease, morbidity, and mortality in adulthood (102). Indeed, exposure of pregnant women to famine during any stage of gestation was associated with insulin resistance in adult offspring (102, 206). Of particular interest, maternal undernutrition during early gestation had greater effects in increasing the incidence of cardiovascular and metabolic disorders in the offspring in later life, compared with maternal undernutrition during mid- or late-gestation (102). Third, maternal undernutrition was correlated with the incidences of coronary heart disease and hypertension in a cohort of 4630 adult men (10, 42) and 3447–4130 women (10, 51) born in Helsinki, Finland between 1924 and 1933 (Table 1). Moreover, among adult men and women, small size at birth (an indicator of IUGR) was associated with increased mortality rates from cardiovascular causes (80). Fourth, a cohort study of 15,000 Swedish men and women born between 1915 and 1929 perhaps provides by far the most convincing evidence for the close association between reduced fetal growth rate and increased risk of death from ischemic heart disease (94). These results highlight the importance of birth weight as a simple measure on predicting and potentially managing potential health risks in adults. Thus, in terms of long-term health, the embryo/fetus is most vulnerable to nutritional insults during early gestation.
Table 1.
Effects of Birth Weight on Hazard Ratios for Coronary Heart Disease and the Cumulative Incidence of Hypertension in Adult Men and Women
| |
Adult men |
Adult women |
||
|---|---|---|---|---|
| Birth weight (g) | Hazard ratios for coronary heart diseasea | Cumulative incidence of hypertension (%)b | Hazard ratios for coronary heart diseasec | Cumulative incidence of hypertension (%)d |
| ≤ 2500 | 3.63 | — | 1.34 | — |
| 2501–3000 | 1.86 | 19.0 | 1.38 | 21.1 |
| 3001–3500 | 1.99 | 17.0 | 1.24 | 16.3 |
| 3501–4000 | 2.08 | 14.1 | 1.17 | 13.0 |
| > 4000 | 1.00 | 12.5 | 1.00 | 12.1 |
| P value for trend | 0.006 | < 0.001 | 0.007e | <0.001 |
Experimental evidence from a variety of species and animal models has provided a wealth of knowledge regarding effects of reduced nutrient availability in utero on the development of adult disease (3). For example, global undernutrition increases the risk for dyslipidemia, obesity, and type-2 diabetes mellitus in rats (156). Also, maternal protein restriction induces hypertension and vascular dysfunction in adult female offspring of rats (65, 149). Extensive research with large animals (e.g., sheep and nonhuman primates) provides further evidence for fetal programming of vascular dysfunction and metabolic abnormalities. For example, in sheep, maternal undernutrition induces left ventricular hypertrophy and alters gene expression in the left ventricle of the fetal heart (62, 180). Additionally, maternal nutrient restriction results in increased myocardial lipid and altered gene expression in offspring at 1 year of age (28). Moreover, in sheep, maternal nutrient restriction impairs renal function, increases the development of glomerulosclerosis, and enhances apoptosis in kidneys, while altering the expression of proteins involved in regulating inflammatory processes (155, 192). In both pigs and sheep, maternal undernutrition also results in decreased skeletal muscle fiber number, increased deposition of adipose tissue, and increased connective tissue content (15, 58). Collectively, these alterations in normal development result in: a) reduced postnatal growth, including both whole-body and skeletal muscle growth; b) reduced efficiency of nutrient utilization; and c) suboptimal function of multiple organs.
Impacts of maternal overnutrition on metabolism and health of offspring
Maternal obesity has been linked to metabolic perturbations, obesity, and cardiovascular disorders in the offspring of a number of mammalian species, including humans, mice, rats, and sheep (6, 83, 85, 156). In nonhuman primates, a maternal high fat diet promotes the development of nonalcoholic fatty liver and atherosclerosis (110) and alteration of seven metabolites in the fetus related to one-carbon unit metabolism (33). Notably, changes in the fetal metabolome result, at least in part, from an altered fetal hepatic chromatin structure that leads to aberrant gene expression in the liver (1) and other insulin-sensitive tissues (162).
Feeding a high-fat diet during pregnancy or the neonatal period stimulates adipogenesis, induces cardiovascular dysfunction characterized by elevated systolic blood pressure, and impairs endothelium-dependent relaxation in rats (3, 61, 83). Importantly, the response to maternal dietary treatment differed between male and female offspring in that female offspring were more susceptible to elevated systolic and diastolic blood pressure when their mothers were fed a high-fat diet during gestation and/or lactation (145). This interesting phenomenon may be explained by imprinting of genes and sex-related differences in hormone secretion. In contrast, endothelium-dependent relaxation of blood vessels in response to acetylcholine was consistently blunted in both males and females exposed to a high-fat diet either in utero or during lactation (137). A major underlying mechanism for the high fat-induced vascular dysfunction is likely the reduced release of nitric oxide (a major vasodilator) from endothelial cells (198).
Maternal obesity due to a high-fat diet reduced mitochondrial copy number in the kidney in 1-year-old rats, while altering expression of a number of mitochondrial genes in the rat aorta (166). These findings suggest that mitochondria play a central role in developmental programming of vascular function. Similar results were observed in the offspring of pregnant mice fed a high-fat diet, including increased adiposity, dyslipidemia, hypertension, and insulin resistance (38, 148). In sheep, maternal obesity downregulates expression of genes involved in myogenesis and placental angiogenesis (212) and fetal skeletal muscle (173), while reducing the phosphorylation of AMP-activated kinase in the fetal and neonatal liver (134) as well as energy metabolism (185). Importantly, these changes in gene and protein expression in response to maternal overnutrition result in increased fetal and/or neonatal adiposity (50, 121) and increased expression of the leptin gene in peri-renal and subcutaneous adipose tissue depots (121). The consequences of maternal overnutrition continue to manifest in offspring as evidenced by impaired insulin sensitivity and reduced glucose utilization on postnatal Day 210 despite growing in an identical nutritional environment from birth (150). Similarly, a maternal high-fat diet during pregnancy results in impaired glucose homeostasis in rat offspring characterized by elevated concentrations of insulin in plasma at 1 year of age (166).
Impact of Maternal Nutrition on Gene Expression in Offspring
Maternal nutrition and gene expression in offspring
As noted previously, studies of offspring born to women during the Dutch famine in the 1940s suggested a link between the fetal environment (including nutrition) and postnatal health, particularly cardiovascular function or dysfunction, in humans (9, 30, 69, 133). Interestingly, 60 years after birth, offspring with early prenatal experience of the famine exhibited less DNA methylation of the imprinted IGF2 gene, as compared to the same-sex siblings without exposure to prenatal malnutrition (67). Furthermore, individuals with periconceptional exposure to the famine had lower methylation of the INSIGF gene, but higher methylation levels for several other genes (IL10, LEP, ABCA1, GNASAS, and MEG3) (171). These genes are closely linked with nutrient metabolism, cardiovascular function, and inflammation. Interactions between undernutrition and sex also affect methylation of the INSIGF, LEP, and GNASAS genes (171).
In monkeys, a 30% reduction in maternal nutrient intake did not alter fetal weight at stages 0.5 and 0.9 of gestation (full term being 1.0) (176). However, tissue-specific global methylation status (176) and the availability of methyl group donors (153) were altered in the kidneys of nonhuman primates at both 0.5 and 0.9 of gestation. Furthermore, emerging evidence indicates modifications of hepatic phosphoenolpyruvate carboxykinase (a key enzyme in gluconeogenesis) after exposure of fetal baboons to moderately reduced nutrient availability (124). These results indicate that even a relatively mild nutrient restriction can induce alterations in gene expression and provide a potential mechanism responsible for the increased incidence of endothelial dysfunction, renal dysfunction, and hypertension in offspring with previous experience of malnutrition during the period of fetal growth. Such an effect of fetal programming on gene expression in offspring has been experimentally tested in a number of species, including nonhuman primates (124), rats (5), mice (38), cattle (23), and sheep (162). Thus, effects of changes in nutrition or endocrine status during the fetal can negatively impact subsequent development of the offspring.
Transgenerational effects of maternal nutrition
Epidemiological studies have indicated the presence of transgenerational effects of maternal malnutrition in humans (209). For example, starvation in young boys (7–11 years of age) resulted in increased incidence of metabolic syndrome in them as adult men, and in their children and grandchildren (79). Likewise, food deprivation in female fetuses for 7–12 weeks was associated with increased prevalence of metabolic syndrome in them as adult women, and in their children and grandchildren (22). Results of animal studies also indicated that the effect of maternal undernutrition on hormone secretion and metabolism can be passed transgenerationally from mother (F0) to daughter (F1) and to the F2 progeny (16). For example, maternal calorie restriction in rats caused: a) decreases in the mass and number of pancreatic β-cells in the first generation of female offspring; and b) impaired responses of F2 pancreatic β-cells to pregnancy, leading to insulin resistance and gestational hyperglycemia (107, 168). Similar results were obtained for female rats fed a low-protein diet (210) or female mice subjected to caloric undernutrition (76) during gestation. In guinea pigs, even modest maternal undernutrition (70% of normal food intake) resulted in alterations in the cardiac structure (e.g., increases in wall thickness and mass of the left ventricle), hypothalamo-pituitary-adrenal function (e.g., increased basal levels of cortisol), and elevated blood pressure in adult male F1 and F2 offspring through the maternal line (4, 16). Interestingly, some of the transgenerational effects of maternal undernutrition-induced IUGR and developmental defects on diabetes and cardiovascular function are sex-dependent (168, 187), which can be explained well by differential genomic imprinting (35). Likewise, chronic high-fat feeding in fathers resulted in hypomethylation of the II13ra2 gene and β-cell dysfunction in female, but not male, offspring (123).
Fetal Programming
The mechanism of fetal programming: Epigenetics
The genetic code established by the DNA sequence exhibits minimal rates of change after the formation of the diploid chromatin state at fertilization (43). Therefore, an extensive search for alternative mechanisms of gene regulation resulted in the now widely accepted notion that changes in gene expression can be manifest by mitotically and/or meiotically heritable alterations in the DNA–protein complex without any change in the DNA sequence (156). This phenomenon was originally termed “epigenetics” by Conrad Waddington in 1940 (182) based on the Greek prefix “epi” meaning over or above. Molecular mechanisms about epigenetics and genomic imprinting have evolved over the past 40 years since the first identification in the 1970s of DNA methylation as a regulatory factor (70). The 1980s (109, 183, 193) and 1990s (12, 18, 63) witnessed pioneering research on the maintenance and regulation of DNA methylation patterns in mammals. In the mid 1990s, histone modifications were discovered as an epigenetic determinant of chromatin structure and, therefore, genome activity (29, 174). The identification of noncoding microRNAs and their functions early in the 2000s further expanded the field of epigenetic research (4, 95). Excitingly, the first whole epigenome analysis was completed for yeast in 2005 (136). It is now known that epigenetic modifications can be carried forward from one generation of cells to the next (mitotic inheritance) and between generations in a species (meiotic inheritance) (156).
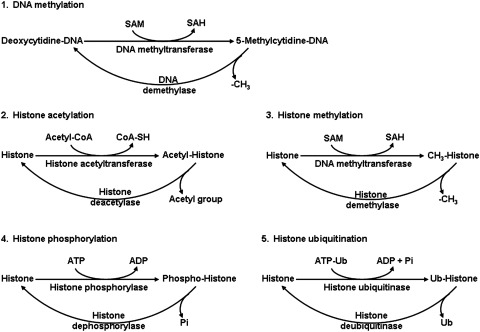
At present, the four mechanisms responsible for mediating epigenetic effects are: a) chromatin modifications, b) DNA methylation (occurring at the 5’-position of cytosine residues within CpG dinucleotides throughout the mammalian genome), c) histone modifications (acetylation, methylation, phosphorylation, ubiquitination, and sumoylation), and d) RNA-based mechanisms such as small noncoding RNAs or inhibitory RNAs (Fig. 3). The enzymes involved in these reactions include specific DNA and protein methyltransferases, DNA demethylases, histone acetylase (lysine acetyltransferase), GCN5-related N-acetyltransferase (a super family of acetyltransferase), and histone deacetyltransferase (18, 26, 30).
FIG. 3.
Biochemical reactions involving DNA methylation and histone modifications. These reactions are localized in specific compartments of the cell. SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; Ub, ubiquitin.
DNA methylation
The methylation of DNA is catalyzed by DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) and is a reversible physiological process in the eukaryotic genome (45). This biochemical event involves the donation of a methyl group from S-adenosylmethionine to the 5'-position of a cytosine nucleotide linked to a guanine nucleotide (CpG dinucleotide) by a phosphodiester bond. Regions with a high CpG dinucleotide content form CpG islands, which are located in the regulatory regions of many genes, including promoters and enhancers (37). Due to their presence in these regulatory regions, changes in methylation status can either facilitate (hypomethylation) or inhibit (hypermethylation) the expression of a gene (106). In normal mammalian cells, over 85% of CpG dinucleotides are methylated, which plays a role in maintaining the integrity of chromatin structure and transcriptional regulation (56). Except for imprinted genes, CpG islands present in the promoter regions of genes are usually unmethylated. Approximately 1% to 2% of the human genome consists of CpG dinucleotide whose methylation is inversely related to transcriptional activity in cells (187).
Epigenetic modifications are erased and re-established in a tissue-specific manner during embryonic development (184). The haploid genomes of the sperm and oocyte possess different patterns of DNA methylation that arise from sex-specific patterns of de novo methylation. After fertilization, the paternal genome is rapidly demethylated prior to the first cell division of the zygote. In contrast, the maternal genome is protected from this demethylation event, and, instead, is gradually demethylated during the development of the blastocyst. At the blastocyst stage, most methyl marks are removed, remaining present at elements regulating genomic imprinting and retroviral elements (43, 56). After implantation, de novo DNA methylation occurs, giving rise to tissue-specific patterns of DNA methylation (36). The process of demethylation and remethylation during the development of germ cells is referred to as “reprogramming” and this physiological event is regulated, in part, by the phosphatidylinositol 3-kinase pathway (161).
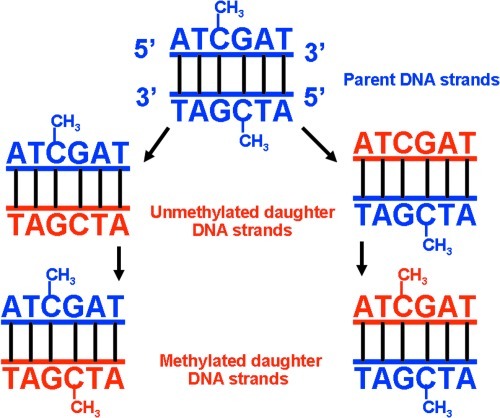
Epigenetic information is heritable between cell generations through the action of the methyltransferase, DNMT1 (184). In somatic cells, this enzyme recognizes and methylates CpG dinucleotides present on the newly synthesized strand. As DNA synthesis occurs, the parental strand remains methylated, whereas the newly synthesized daughter strand is not. DNMT1 “reads” the parental strand and methylates CpG dinucleotides on the daughter strand that are complementary to methylated CpG dinucleotides on the parental DNA strand (Fig. 4). The importance of DNA methylation for successful pregnancy outcome is evident by the fact that all homozygous DNMT1 knockout mice die at the morula stage of embryonic development (20).
FIG. 4.
Passage of epigenetic information during DNA replication in germlines. When double-stranded DNA is divided into daughter DNA, a specifc DNA methyltransferase can methylate the cytosine in CpG islands of the daughter DNA at the blastocyst stage of conceptus development. Tissue-specific gene expression results from differences in DNA methylation and promoter activity, leading to differentiated somatic cells.
Histone modifications
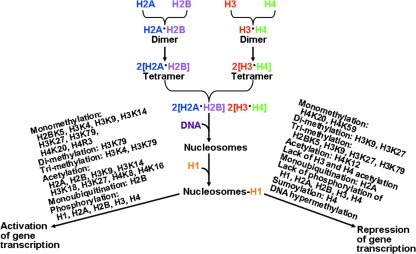
Histones are basic proteins that facilitate the packaging of DNA in the nucleus and the regulation of gene expression in cells. The histone proteins are H1, H5, H2A, H2B, H3, and H4. Two copies of the H2A, H2B, H3, and H4 histones assemble to form one octameric histone protein core (Fig. 5). DNA is wrapped around the core twice in a left-handed super-helical turn to make a structural unit called the nucleosome. In the histone protein core, a heterotetramer of histones H3 and H4 is flanked by two heterodimers of histones H2A and H2B (59). Histones H1 and H5 are linker histones (131). Histone H1 binds the nucleosome as well as the entry and exit sites of the DNA. Through changes in phosphorylation and O-glycosylation, linker histones modify chromatin structure (2). Post-translational modifications imparted onto the N-terminal tails of each histone protein alter the properties and structures of chromatin (131). These post-translational modifications include methylation of arginine or lysine, acetylation of lysine, phosphorylation of serine and threonine, ubiquitination of lysine, ADP-ribosylation, sumoylation of lysine, and glycosylation (147). The biochemical changes regulate gene expression by modulating: a) the accessibility of the transcriptional machinery to the DNA binding elements; and b) DNA replication, recombination, and chromosomal organization. At present, more than 100 specific modifications of chromatin are associated with either actively transcribed or silenced genes (45). Therefore, histone proteins play an important role in gene expression (Fig. 5).
FIG. 5.
Roles of histone modifications in the regulation of gene transcription. Methylation of histones, acetylation, or ubiquitination can either activate or repress gene expression, depending on specific histone proteins and the sites of modifications. In general, phosphorylation of histones promotes transcription, DNA repair, and apoptosis. H, histone; K, lysine residue; R, arginine residue.
Histone methylation is catalyzed by histone methyltransferases (HMT), which transfer one to three methyl groups from S-adenosylmethionine to lysine or arginine residues of histone proteins (7). There are four known HMTs, including SUV1, SUV2, SUV3, and RIZ. Examples of the enzymes are H3K4-, H3K9-, and H3K27-specific methyltransferases, which target lysine residues 4, 9, and 27 of histone H3, respectively. The H3K27-specific methyltransferase is also known as the enhancer of zeste homologue 2 (EZH2). Other significant histone methylases are H3K79- and H4H20-specific (131). Silencing of differentiation genes is associated with the methylated H3K27 chromatin mark. In a variety of cell types, histone methylation is generally associated with transcriptional repression, but methylation of some lysine (e.g., lysine 4 of histone 3) and arginine residues (e.g., those on H3 and H4) of histones can result in transcriptional activation (7). While histone methylation was previously thought to be a permanent modification, recent studies uncovered histone demethylating enzymes (45). It is now known that demethylation of histone proteins is performed by histone demethylases. Lysine-specific demethylase 1 (a flavin-dependent monoamine oxidase) can demethylate mono- and di-methylated lysines, specifically lysines 4 and 9 of histone 3 (H3K4 and H3K9) (54). This enzyme cannot demethylate tri-methylated lysines. However, the Jumonji domain-containing (JmjC) histone demethylases can demethylate mono-, di-, or tri-methylated lysines (187). It is possible that arginine-specific demethylase exists in cells to demethylate mono- and di-methylated arginines in histone proteins (19). Alternatively, methylated arginine residues can be converted into citrulline by peptidyl arginine deiminase 4, which acts on unmodified and mono-methylated arginine residues in proteins (7).
Histone acetylation also influences gene expression (43). Acetylation of lysine residues at the N terminus of histone proteins is catalyzed by histone acetyltransferases (HAT) (59). Superfamilies of HAT include GNAT, CBP/p300, and MYST. Examples of histone acetyltransferases are HAT1, HATp300, CDY1, and CLOCK. The source of the acetyl group in histone acetylation is acetyl-CoA. Thus, metabolism of energy substrates or the energy status of the cell can modulate histone acetylation (184). Notably, acetylation of lysine residues neutralizes the positive charge on histones, thereby decreasing the interaction of the N termini of histones with the negatively charged phosphate groups of DNA (187). Such a modification of histone proteins transforms the condensed chromatin into a more relaxed structure and makes RNA polymerase and transcription factors easier to access the promoter region of the DNA, leading to enhanced gene transcription. Thus, histone acetylation is usually associated with activation of gene transcription (59). Histone deacetylation is catalyzed by histone deacetylases (HDAC; also known as lysine deacetylases). In this reaction, the acetyl group is transferred from the acetylated histone protein to coenzyme A. Based on intracellular localization, three classes of HDAC are I: HDAC 1, 2, 3, and 8 (exclusively in the nucleus); II: HDAC 4, 5, 6, 7, 9, and 10 (predominantly residing in the cytoplasm and shuttling in and out of the nucleus); and III: surtuin enzymes (SIRT) 1, 2, 3, 4, 5, 6, and 7 (localized in either nucleus, cytoplasm, or mitochondria) (187). The histone deacetylase CBP/p300 can interact with numerous transcription regulators and likely plays a major role in repressing gene transcription (57).
The biological activity of histones is also regulated by phosphorylation. Histone H3 phosphorylation, which occurs during both mitosis and meiosis, is catalyzed by ATP-dependent protein kinases (26, 207). This event is coordinated in both space and time as a signaling mechanism to modulate nucleosome structure and thus DNA accessibility (73). Phosphorylation of H2A (at serine 1 and threonine T119), H3 (at serine 10 and 28; threonine 3 and 11), and H4 (at serine 1) results in transcriptional activation, as well as mitosis and meiosis (26, 131). Phosphorylation of H2A also enhances DNA repair, while phosphorylation of H4 promotes spermatogenesis. Likewise, histone H1 phosphorylation is associated with decondensation of chromatin and activation of transcription (2), and histone-3 phosphorylation is critical for cytokine-induced gene expression (207). In contrast, phosphorylation of H2B (at serine 14) stimulates apoptosis, which plays an important role in cell metabolism and growth (101). Thus, histone phosphorylation can allow cells to rapidly respond and adapt to changes in maternal nutritional status by regulating numerous processes, including transcription, DNA repair, and apoptosis (Fig. 5).
The monoubiquitination of histones H2A and H2B plays a critical role in regulating DNA repair, gene transcription, and mRNA translation (188). In this process, ubiquitin (a 76-amino acid polypeptide) is conjugated to histones H2A and H2B in a series of reactions involving three separate enzymatic activities: a) ATP-dependent ubiquitin activating enzyme (E1; activation of ubiquitin), b) ubiquitin-conjugating enzyme (E2; conjugation via a thioester bond to a cysteine residue); and c) ubiquitin-protein isopeptide ligase (E3; transfer of ubiquitin from the E2 enzyme to a target lysine residue in a histone) (135). Monoubiquitination of H2B promotes transcription initiation and elongation through efficient reassembly of nucleosomes (49). There is evidence that H2B monoubiquitination is a prerequisite for Lys-4 H3 and Lys-79 H3 methylation, whereas H2A monoubiquitination is associated with transcriptional silencing (187). Although H2A and H2B ubiquitination appear to perform separate functions, there is cross talk between ubiquitinated H2A and H2B proteins to modulate Lys-4 H3 methylation and RNA polymerase II activity during transcription elongation (49, 169).
RNA-based epigenetic mechanisms
Epigenetic regulation of gene expression by RNA-based mechanisms can occur at both the post-transcriptional level and the level of chromatin (187). These mechanisms are mediated by small-interfering RNAs or small non-coding RNAs which act through their respective pathways to induce DNA methylation or histone modifications to silence or enhance gene expression (82, 169). In mammals, the small non-coding RNAs include Piwi RNA and microRNA (miRNA) (74). Piwi RNA is restricted to the germ line (e.g., testes in males and oocytes in females), and regulates gene expression possibly through sequence-specific targeting of heterochromatin formation factors to the mobile elements in the genome and the degradation of mobile element transcripts (172). In contrast, miRNAs are widespread in cells and tissue to regulate gene expression at the post-transcriptional level (44). Since the discovery of lin-4 as the first miRNA in 1993 (93), approximately 1000 human miRNAs have been identified, annotated and catalogued (103, 113).
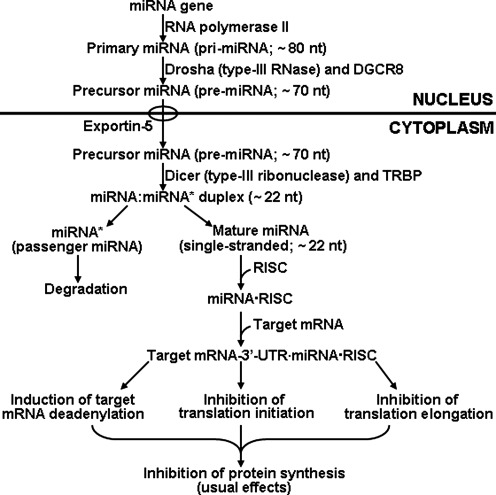
The biogenesis of mature (functional) miRNA from its miRNA gene involves both the nucleus and cytoplasm in mammalian cells (Fig. 6). In the nucleus, an miRNA gene is transcribed by RNA polymerase II to produce primary miRNA with ∼80 nucleotides (nt) in length (44). The primary miRNA, which is a long capped and polyadenylated transcript, is processed by Drosha (type-III RNase) and its associated protein DGCR8 (Pasha) to generate precursor miRNA (∼70 nt). After being transported to the cytoplasm by exportin-5, the precursor miRNA is cleaved by Dicer (type-III ribonuclease) in association with its binding partner TRBP (transacting RNA-binding protein) to generate a small imperfect miRNA:miRNA* duplex (∼ 22 nt) (74). This duplex is unwound to form a) a passenger strand miRNA (miRNA*) for degradation; and b) a mature (functional) single-stranded miRNA with approximately 22 nt (72). The mature miRNA is loaded into an RNA-induced silencing complex (RISC) containing several components, including Dicer, TRBP, Argonaute proteins, GW182 protein, and fragile X mental retardation protein (FMRP1) (44). The mature miRNA then pairs with the 3’-untranslated region of the target mRNA, which usually results in: a) induction of target mRNA deadenylation and, thus, degradation; b) inhibition of the assembly of the 80S ribosome complex and, thus, translation initiation; and c) inhibition of translation elongation possibly through premature termination and subsequent ribosome drop-off (44). This leads to decreased synthesis of proteins which are either inhibitors or activators of metabolic pathways depending on cell types, tissues, and developmental stage. Therefore, miRNAs have versatile functions in physiological processes and play crucial roles in growth, development, and health.
FIG. 6.
Biogenesis of microRNA and its role in the regulation of gene expression. The biogenesis of mature (functional) miRNA from its miRNA gene involves both the nucleus and cytoplasm in mammalian cells. A single-stranded mature (functional) miRNA with approximately 22 nucleotides (nt) binds the 3’-untranslated region of the target mRNA. This usually results in decreased protein synthesis through induction of mRNA deadenylation, reduction of translation initiation, and inhibition of translation elongation. DGCR8, associated protein DGCR8 of Drosha (also known as Pasha); TRBP (transacting RNA-binding protein); miRNA*, passenger strand miRNA for degradation; RISC, RNA-induced silencing complex (RISC) containing several components, including Argonaute proteins, PW182 protein, and fragile X mental retardation protein.
Aberrant Epigenetic Expression in the Metabolic Syndrome
Aberrant DNA methylation
Since the early report that dietary deficiency of methionine caused abnormal cell metabolism through DNA hypomethylation (194), there has been growing evidence indicating that disruption of epigenetic mechanisms results in metabolic disorders related to aging, obesity, and diabetes in animals and humans (46, 101). Aging is associated with decreased expression of DNMT1 and DNMT3, global hypomethylation of DNA, and genomic instability in mammalian tissues, although gene-specific hypermethylation can occur (193). For example, increased DNA methylation in the NDUFB6 promoter with aging is attributable to decreased expression of the NDUFB6 gene and whole-body energy metabolism in humans (99). There is evidence that increased DNA methylation of the glucokinase promoter plays a role in decreasing expression of the glucokinase protein in the liver of aged rats (75), thereby increasing the risk for hyperglycemia.
Epigenetic defects (epimutations) can play a role in the development of obesity (3). Of interest, loss-of-function of the histone demethylase, Jhdm2a, is associated with reduced expression of peroxisome proliferator-activated receptor-α and medium-chain acyl-CoA dehydrogenase in skeletal muscle, impairment of cold-induced expression of uncoupling protein-1, and obesity in mice (165). Accordingly, hypomethylation increases risk of obesity in humans and animals (71). For example, maternal high-fat diet increased global and gene (dopamine and opioid) specific hypomethylation of DNA in the brains of offspring that resulted in alterations in gene expression and behavior (181). In addition, secretion of leptin by white adipose tissue is controlled by epigenetic mechanisms. Specifically, the leptin promoter that contains a CpG island has a high degree of methylation in preadipocytes (116, 208), and the promoter is demethylated in association with induction of leptin expression in differentiated adipocytes (208). Furthermore, a high-fat diet increases DNA methylation of one CpG site in the leptin promoter of adult rat adipocytes (119). Finally, NAD-dependent sirtuins (class III HDAC), which target both histones and nonhistone proteins, regulate nutrient metabolism, mitogenesis, adipogenesis, and insulin secretion (154). Additionally, defects in orchestration of circadian rhythm by HDAC3 lead to abnormal lipid metabolism in the liver (46).
Diabetes may also have an epigenetic basis (99, 100). First, recent work revealed that COX7A1 (part of complex 4 of the respiratory chain) is a target of DNA methylation (144). Interestingly, methylation of the DNA for the COX7A1 promoter is increased in skeletal muscle of diabetic animals, and its gene transcription is reduced in association with reduced glucose uptake by muscle (144). Second, genetic mutation in the Pdx1/insulin promoter factor-1, which is a transcription factor regulating development of the pancreas and β-cell differentiation, results in a monogenic form of diabetes (178). Interestingly, IUGR resulting from uteroplacental insufficiency is associated with the silencing of Pdx1 and the development of type 2 diabetes in adult offspring of rats (130). Third, DNA methylation of the PPARGC1A gene is elevated in pancreatic islets from patients with type 2 diabetes and the degree of DNA methylation is inversely correlated with PPARGC1A expression (100). Fourth, hypomethylation at chromosome 6q24 is associated with transient neonatal diabetes in humans, and these subjects develop type 2 diabetes as adults (167).
Aberrant histone modifications
Abnormal modifications of histone proteins contribute to metabolic disorders (101). GLUT4 is a major glucose transporter in insulin-sensitive tissues; therefore, intensive research on epigenetic regulation of diabetes has focused on this protein. In animal studies, reduced expression of GLUT4 is associated with elevated HDAC activity and hyperglycemia (112). Normally, GLUT4 expression is regulated by the transcription factor myocyte enhancer factor 2 (MEF2), which interacts with HDAC5 in the nucleus. However, when the histone tails that are colocalized with the GLUT4 gene are deacetylated by HDAC5 in skeletal muscle, the formation of condense chromatin structures impairs GLUT4 gene expression (111). In contrast, enhanced HAT activity to acetylate histones is positively correlated with oxygen consumption in human skeletal muscle (129). Similarly, Class II HDAC enzymes limit GLUT4 gene expression during adipocyte differentiation (187). Likewise, histone code modifications by HDAC1 and HDAC4 repress GLUT4 expression in IUGR offspring (140). Furthermore, phosphorylation of HDAC5 has a stimulatory effect on GLUT4 transcription mediated by AMP-activated protein kinase in skeletal muscle (111).
A novel finding in epigenetic research is the regulation of insulin secretion by histone modifications (125, 156). In normal pancreatic β-cells, the insulin gene exhibits hyperacetylation of H4 and hypermethylation of H3K4. This pattern is not observed for the insulin gene in other cells (e.g., HeLa cells) that do not produce insulin (27). Recruitment of histone acetyltransferase and the histone methyltransferase SET7/9 to the insulin promoter is necessary to activate the gene for insulin (27). HDAC may inhibit pancreatic development in the fetus (66). Also, reduced levels of histone H3K27 trimethylation and the histone methyltransferase Ezh2, as well as the loss of H2A ubiquitination are associated with elevated expression of Ink4a/Arf and reduced beta-cell regeneration in aging mice (88). Thus, reduced activity of HAT (e.g., CBP/p300) is associated with a monogenic form of diabetes (8).
Another exciting new development in histone modifications is their crucial roles in cellular antioxidative reactions and cellular signaling. Poor glycemic control increases NF-kB activity in monocytes and expression of genes encoding inflammatory cytokines through an interaction between NF-kB and HAT (e.g., CBP/p300) (117). The H3K4 methyltransferase SET7/9 affects the recruitment of NF-kB p65 to gene promoters and thus expression of NF-kB-dependent inflammatory genes (98). Histone methylase (SET7) and histone demethylase (LSD1) can regulate epigenetic changes in the NF-kB p65 promoter induced by hyperglycemia (19). Vascular smooth muscle cells from diabetic mice display reduced activities of H3K9 methyltransferase and lysine-specific histone demethylase-1, as well as reduced levels of H3K9 trimethylation and elevated levels of H3K4 dimethylation (179). Abnormal H3K9 dimethylation has also been reported for lymphocytes from patients with type-2 diabetes (118).
Aberrant expression of small non-coding RNAs
Aberrant expression of miRNAs has been implicated in obesity, insulin resistance, diabetes, cardiovascular complications, and placental dysfunction (72, 120). For example, high fat feeding results in upregulation of miR-143 but downregulation of primary miR-27a in mouse mesenteric adipose tissue via mechanisms involving expression of the PPAR-family of genes (113). Also, enhanced levels of miR-335 have been reported for the liver and white adipose tissues of three murine models of obesity (leptin-deficient ob/ob mice, leptin receptor-deficient db/db mice, and KKAy 44 mice) (122). Similarly, type-2 diabetes in humans is associated with down expression of miR-27a and miR-378, which promotes the formation of lipid droplets, adipocyte differentiation, and adipogenesis in white adipocytes (113). Furthermore, miR-17-5P and miR-132 in omental fat and blood are positively correlated with human obesity (68). Interestingly, emerging evidence indicates that miR-17-92, miR-21, miR-103, miR-143, miR-371, and miR-378 promote, but let-7, miR-27, miR-130, miR-138, miR 369-5P, and miR-448 inhibit, oxidative stress and inflammation in animals with obesity and atherosclerosis (72). Finally, mis-expression of miRNAs contributes to inflammatory processes in placental dysfunction (120), cardiovascular diseases (e.g., coronary artery disease, ischemic injury, hypertension, and atherosclerosis) (158), the metabolic syndrome (103), and aging (158). Besides microRNAs, little is known about effects of nutrition on expression of other small non-coding RNAs in animal tissues.
Inhibition to Regulate Epigenetics and Ameliorate Metabolic Syndrome
Environmental cues appear to be the primary trigger to induce changes in the epigenome and include such factors as nutrients, stress, environmental pollutants, and toxins (55). The extent to which cells are able to respond to these cues lies in large part with the plasticity of cells. Thus, the developing embryo/fetus is a prime target for epigenetic modifications (43) and interventions during pregnancy may be an important strategy for ameliorating or preventing metabolic disorders (including vascular insulin resistance) in adult life (Fig. 7). Despite a recognized role for epigenetics in fetal programming for metabolic syndrome (175), work in this field remains very limited.
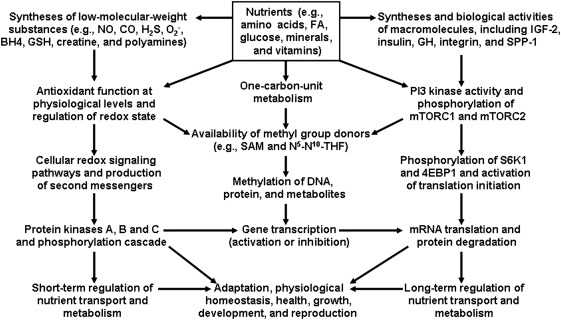
FIG. 7.
Roles of macro- and micro-nutrients in epigenetics and physiological responses. Nutrients, particularly amino acids, regulate cellular redox state, the secretion of hormones (e.g., insulin and insulin-like growth factors), physiological functions, and whole-body homeostasis in humans and animals through three mechanisms: (1) the expression of genes; (2) the production of signaling gases and other metabolites; and (3) MTOR activation. S-Adenosylmethionine is the major methyl group donor in cells and its synthesis is affected by amino acids (e.g., methionine, serine, glycine, and histidine), B vitamins (including folate, vitamin B12, and vitamin B6), choline, and creatine. Methylation of DNA and protein contributes to epigenetics, which results in transcriptional activation or inhibition of select genes. Changes in intracellular protein turnover (protein synthesis and degradation) and protein kinase cascades can alter physiological responses in the fetus and offspring. CO, carbon monoxide; 4EBP1, eIF4E-binding protein-1; GH, growth hormone; H2S, hydrogen sulfide; IGF, insulin-like growth factor; MTOR, mechanistic target of rapamycin; NO, nitric oxide; SAM, S-adenosylmethionine; S6K1, ribosomal protein S6 kinase-1; SPP1, secreted phosphoprotein 1.
Inhibition of DNA methylation and histone deacetylation
Based on epigenetic research, chemically synthesized drugs have been used to target deleterious effects of DNA methylation and histone deacetylation to reverse metabolic disorders (164). The classic DNMT inhibitor is 5-azacytidine, a derivative of cytidine. Results of both in vitro and in vivo studies indicate that 5-azacytidine increases expression of the PPARγ gene in white adipocytes in a dose-dependent manner and the metabolic pattern of the cells (53). Also of particular interest is the application of isoform-specific HDAC inhibitors (e.g., short-chain fatty acids, hydroxamic acids, cyclic tetrapeptides, and benzamides) to treat vascular complications associated with diabetes (17, 60). For example, myocardial ischemia-reperfusion injury is associated with increased HDAC activity, but decreased acetylation of histones H3 and H4 in the heart of mice (57). Notably, administration of synthetic HDAC inhibitors (trichostatin A and Scriptaid) to these animals reduced myocardial infarction and cell death (57). Similarly, HDAC inhibitors attenuated load-and agonist-induced cardiac hypertrophy and abolished the associated activation of autophagy in cardiomyocytes (24).
Inhibition of miRNA expression and activity
Because abnormally high levels of some miRNAs cause translational repression in tissues (particularly insulin-sensitive tissues) and are associated with the development of chronic diseases (72, 113), modulation of their expression and activity may be a potentially effective therapeutic means for prevention and treatment of the metabolic syndrome. At present, the expression or activity of a specific miRNA can be inhibited or silenced using: a) a locked nucleic acid oligonucleotide (a Morpholino oligonucleotide); b) an antisense 2'-O-methyl RNA oligonucleotide; c) an antagomir (a small synthetic RNA oligonucleotide that is perfectly complementary to the specific miRNA target), or d) a steric-blocking oligonucleotide to prevent the maturation of miRNA or block the miRNA-binding site of a target mRNA (40, 89). Although research in this area is still limited, results from both in vitro and in vivo studies have been promising. For example, specific inhibition of miR-375 (a pancreatic islet-specific microRNA) by transfection with an antisense 2’-O-methyl oligonucleotide complementary to miR-375 decreased the expression of the myotrophin and PDK1 genes and increased glucose-stimulated insulin secretion in cultured murine pancreatic β-cells (138). The use of a similar technology led to the discovery that miR-143 plays a key role in the differentiation of cultured human pre-adipocytes by inhibiting expression of the ERK5 gene (41). Based on the in vitro studies, intravenous administration of an antagomir against miR-122 three times within 24 h (80 mg/kg body weight/day) to mice reduced expression of 11 genes involved in cholesterol synthesis (including hydroxy-3-methylglutaryl-CoA reductase) in the liver, thereby decreasing concentrations of cholesterol in plasma (89). Likewise, subcutaneous injection of an anti-miR-33 oligonucleotide to mice weekly for 4 weeks promoted the reversal transport of cholesterol from peripheral tissues to the plasma, liver, and feces, as well as regression of established atherosclerosis, and ameliorated dyslipidemia (as indicated by reduced concentrations of triacyglycerols and cholesterol in plasma) (141). Employing the similar technology, Rayner et al. (142) recently reported that inhibition of miR-33a/b increased the expression of: a) the sterol-response-element-binding protein genes SREBF1 and SREBF2; b) the cholesterol transporter gene ABCA1; and c) genes involved in fatty acid oxidation (CROT, CPT1A, HADHB, and PRKAA1), while reducing the expression of genes involved in fatty acid synthesis (SREBF1, FASN, ACLY, and ACACA) in nonhuman primates. This anti-microRNA strategy resulted in increased concentrations of high-density lipoproteins and reduced concentrations of very-low-density lipoproteins in plasma to reduce risk for cardiovascular disease.
Targeting epigenetically disturbed metabolic pathways
Maternal malnutrition may affect growth and health of F1 and subsequent offspring (200). It is now known that IUGR is associated with reduced expression of the imprinted gene IGF2 in animals (52). Interestingly, loss of imprinting of the IGF2 gene results in enhanced signaling of IGF2/IGF1 at the receptor level due to a positive feedback mechanism (35) which leads to abnormal metabolism of nutrients (e.g., uncontrolled utilization of glucose via glycolysis) (170). Inhibition of the IGF2 receptor can reverse the epigenetically disturbed metabolic pathways in animals with loss of imprinting of the IGF2 gene (81). Additionally, results of recent studies indicate that alterations in DNA methylation and histone modifications in cardiovascular diseases all result in elevated levels of hyperhomocysteinemia due to reduced expression of the enzymes involved in the utilization of homocysteine (25, 47, 87). Accordingly, reducing circulating levels of homocysteine through dietary supplementation with folic acid can improve function of the cardiovascular system (139, 191).
Nutritional interventions with amino acids and vitamins
Besides serving as building blocks of proteins, amino acids are signaling molecules, regulators of gene expression and the protein phosphorylation cascade, and they are key precursors for syntheses of hormones and low-molecular weight nitrogenous substances with enormous biological importance (Fig. 7). Amino acids also modulate cellular redox state and the secretion of hormones from endocrine organs (e.g., insulin, growth hormone, lactogen, and insulin-like factors) (197). Physiological concentrations of metabolites (e.g., nitric oxide, polyamines, glutathione, taurine, thyroid hormones, and serotonin) of amino acids are required for the functions of cells and whole-body homeostasis (197). It is noteworthy that maintenance and regulation of the epigenetic state, which depends on one-carbon unit metabolism, require adequate provision of methionine, serine, glycine, histidine, choline, creatine, and B vitamins (including folate, vitamin B12, and vitamin B6) (Fig. 2). These nutrients play an important role in regulating the availability of S-adenosylmethionine, a major methyl donor for DNA and protein methylation by specific DNA and protein methytransferases (200). Thus, restriction of essential B vitamins, folate, and methionine during the peri-conceptual period in sheep resulted in altered DNA methylation, insulin resistance, and elevated blood pressure observed most notably in adult male offspring (157). Conversely, supplementing the maternal diet with folate, vitamin B12, choline, or betaine increases DNA methylation of the agouti gene in the offspring, thereby leading to low agouti expression and preventing the development of obesity (77). Interestingly, the effect of maternal supplementation with methyl donors can be inherited in the F2 generation through germline epigenetic modifications (34).
S-Adenosylmethionine is synthesized from methionine by S-adenosylmethionine synthase (also known as methionine adenosyltransferase) (197). In addition to its role as a methyl donor, S-adenosylmethionine is required for the synthesis of polyamines, cysteine, taurine, and creatine. Polyamines are required for the proliferation of endothelial cells and the remodeling of the vasculature (96). Importantly, the nutritional and physiological state of the animal will alter the production of these bioactive substances, thus regulating the availability of methyl donors. When cysteine or taurine is deficient in the diet, their synthesis from methionine will be increased in vivo, thus decreasing total S-adenosylmethionine availability for DNA or protein methylation. Inadequate synthesis of glycine and serine, coupled with low supplies from the diet, can also impair one-carbon unit metabolism (200). Therefore, an amino acid deficiency can alter the epigenetic code through changes in DNA methylation as well as histone modifications (127).
Both epidemiological and experimental evidence indicate that IUGR contributes to a plethora of metabolic disorders and chronic diseases in adults (Fig. 1). These problems include: 1) hormonal imbalances (e.g., increased plasma levels of glucocorticoids and renin; decreased plasma levels of insulin, growth hormone, and insulin-like growth factor-I); 2) metabolic disorders (e.g., insulin resistance, β-cell dysfunction, dyslipidemia, glucose intolerance, impaired energy homeostasis, obesity, type-II diabetes, oxidative stress, mitochondrial dysfunction, and aging); 3) organ dysfunction and abnormal development (e.g., testes, ovaries, brain, heart, skeletal muscle, liver, thymus, and small intestine); and 4) cardiovascular disorders (e.g., coronary heart disease, hypertension, stroke, and atherosclerosis) (3, 156, 162, 199). These novel findings led to the design of nutritional means (including supplementation with functional amino acids) to enhance fetal growth and development (200).
Enteral feeding or intravenous administration of amino acids (e.g., arginine and citrulline) is effective in increasing their circulating concentrations in the mother and her fetus(es) (90–92). Therefore, these nutrients may provide an effective solution to fetal growth restriction in underfed and overfed dams and postnatal metabolic disorders. The importance of amino acids in supporting fetal growth, development, and health is an emerging area of investigation and will undoubtedly shape the future of nutritional management in medicine and animal production (201). The translation of basic research on amino acid nutrition into practice has yielded fruitful outcomes. For example, in pigs, arginine supplementation to pregnant sows during gestation increases placental vascularity (97) and embryonic/fetal survival (105). Interestingly, piglets from gilts that received arginine supplementation during gestation have higher rates of neonatal survival and growth (203). Additionally, supplementing the gestational diet for gilts with 0.4% L-arginine plus 0.6% L-glutamine between Days 30 and 114 of gestation enhanced the efficiency of nutrient utilization, reduced variation in piglet birth weight, and increased litter birth weight (203). In sheep, maternal arginine administration during late gestation increased the development of fetal peri-renal brown adipose tissue (152), which is enriched with endothelial cells. The increased mass of brown adipose tissue may lead to enhancement of blood flow to tissues and the ability of neonates to combat cold exposure at birth. Moreover, intravenous administration of L-arginine-HCl (3 x 27 mg/kg body weight per day) enhanced fetal growth in ovine models of both undernutrition-induced and naturally-occurring IUGR (91). Finally, during late (Week 33) gestation, daily intravenous infusion of L-arginine (20 g/day) for 7 days to women with unknown causes of IUGR increased birth weight at term (Week 39) by 6.4% (205). The beneficial effects of arginine likely result from increases in angiogenesis, the number and size of blood vessels, and ultimately the transfer of nutrients from the mother to the conceptus through utero-placental blood flows (151, 202). Future studies are required to determine cardiovascular function and health of offspring from mothers supplemented with arginine during pregnancy.
Physiological effects of supplemental nutrients depend on many factors, including species, developmental stage, nutritional status, dose, duration, window of receptivity, and the content of dietary components. Thus, too much of a good thing can be detrimental to health and care must be taken to ensure the safety of use of nutrients in epigenetic interventions. For example, a high dose of supplemental arginine can disturb acid-base balance and cause antagonism among basic amino acids to yield undesirable effects (e.g., reduced growth, skin lesions, and organ dysfunction) on animals and humans (196). For this reason, it is important that arginine be administered in divided doses on each day to: a) prevent gastrointestinal tract disorders due to abrupt production of large amounts of NO; b) increase the availability of circulating arginine over a longer period of time; and c) avoid a potential imbalance among dietary amino acids. Likewise, dietary supplementation with high doses of folate may not benefit all the people and rather may cause harm to some subjects. For example, high intake of dietary folate by animals and humans may reduce the cytotoxicity of natural killer cells, impair the response to antifolate-based drugs, and result in phenotypic changes in subsequent generations (159). Additionally, because folate interferes with the actions of vitamin B12-dependent enzymes, excess folate in diet can worsen vitamin B12 deficiency in subjects whose diets are inadequate in vitamin B12, which may increase risks for: a) cognitive impairment and anemia in the elderly; and b) in pregnant mothers, adverse fetal programming on insulin resistance and obesity in their offspring (159).
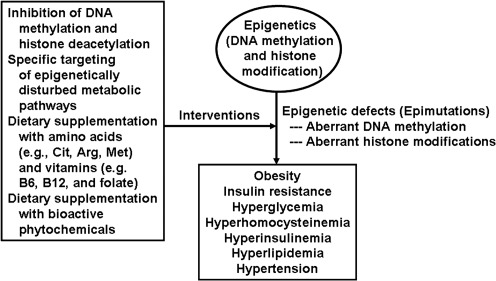
Nutritional interventions with bioactive phytochemicals
Besides specific nutrients, botanicals have been identified as possible epigenetic modulators to ameliorate metabolic syndrome by regulating expression and activity of DNMT, HMT, HAT, and HDAC (86). For example, PMI 5011 (A dracunculus L), which has hypoglycemic activity in diabetic animals, reduced DNMT1 and DNMT3b gene expression in NIH 3T3 fibroblasts. Similar results were obtained for ALT-S (A tuberosum L) (86). There are also reports that phytochemicals (including epigallocatechingallate, resveratrol, genistein, curcumin, and isothiocyanates) can interfere with enzymatic activities of DNMT, Class I, II, IV HDAC, HAT, and Class III HDAC sirtuins (SIRT), thereby beneficially modulating inflammatory responses and immunological senescence (163). Plant extracts may also affect interactions among histone proteins or between nucleosomes and binding factors. Thus, dietary ingredients in food can be epigenetic medicine to remedy abnormalities in DNA methylation, histone modifications, chromatin remodeling, and microRNA patterns. Based on current knowledge of epigenetics, we propose pharmacological and nutritional interventions to prevent and treat metabolic disorders caused by epigenetic defects (Fig. 8). However, as noted above for folate supplementation, cautions (e.g,, timing, dose, frequency, and length of administration) should be taken when using epigenetic compounds to treat human disorders.
FIG. 8.
Pharmacological and nutritional interventions of epigenetic defects. Epigenetic defects can occur in response to environmental stress (e.g., malnutrition and heat stress), abnormal endocrine status, and abnormal levels of metabolites in the body. Chemically synthesized drugs (e.g., 5'-azacytidine as an inhibitor of DNA methyltransferase), physiological metabolites (e.g., butyrate as an inhibitor of histone deacetylase), specific nutrients (e.g., amino acids and vitamins), or bioactive phytochemicals may be used to prevent and treat epimutations.
Conclusion
Either undernutrition or overnutrition during pregnancy (particularly the periconception period), remains a significant problem in both medicine and animal agriculture as it can result in epigenetic changes of some genes in both animals and humans. These changes are affected by multiple factors (e.g., sex, gestational period, and the severity of malnutrition) and may persist in offspring throughout postnatal life, and may carry over to the next generation. A unified explanation for impairment of fetal growth and development in response to both maternal undernutrition and overnutrition may be reduced utero-placental blood flow and, therefore, the reduced transfer of nutrients from mother to fetus (Fig. 9). This hypothesis is gaining support from results of studies with animal models, including rats, pigs, and sheep (114, 145, 160, 211). Nutrients, particularly amino acids and B vitamins, are essential for the regulation of epigenetics and vascular function. Compelling evidence indicates that the fetal and early neonatal periods of development are extremely sensitive to environmental cues, which have long-lasting consequences to postnatal growth, health, and likely athletic performance. Much research is needed with both cell and animal models to understand basic mechanisms responsible for effects mediated by specific nutrients in fetal and neonatal programming. Such new knowledge is expected to aid in designing effective therapeutic means for metabolic abnormalities in offspring that experience an adverse intrauterine environment.
FIG. 9.
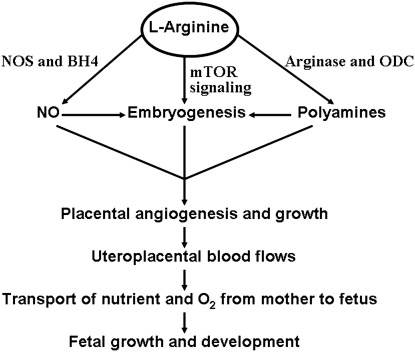
Role of L-arginine in enhancing fetal growth and development in response to maternal undernutrition or overnutrition. Through the tetrahydrobiopterin (BH4)-dependent synthesis of nitric oxide (NO) and production of polyamines from arginine-derived ornithine as well as activation of the mechanistic target of rapamycin (MTOR) signaling pathway, L-arginine can promote embryogenesis and placental vascular growth. Antioxidants are essential to maintain sufficient levels of BH4 in cells for NO synthase (NOS). Enhancement of uteroplacental blood flow ensures adequate transport of nutrients and oxygen from mother to fetus to support fetal growth and development under the conditions of maternal malnutrition. ODC, ornithine decarboxylase.
Abbreviations Used
- DNMT
DNA methyltransferase
- EAA
essential amino acids
- H3K4
histone 3 lysine residue 4
- HAT
histone acetyltransferase
- HDAC
histone deacetyltransferase
- HMT
histone methyltransferase
- IUGR
intrauterine growth restriction
- LCFA
long-chain fatty acids
- miR
microRNA
- NEAA
nonessential amino acids
Acknowledgments
This work was supported, in part, by grants from the Thousand-People-Talent program at China Agricultural University, National Natural Science Foundation of China (3081010390, 30972156, 31129006, and 31172217), National Institutes of Health (1R21 HD049449), American Heart Association (10GRNT4480020), National Research Initiative Competitive Grants from the Animal Reproduction Program (2008-35203-19120), and Animal Growth & Nutrient Utilization Program (2008-35206-18764 and 2009-35206-05211) of the USDA National Institute of Food and Agriculture, and Texas AgriLife Research Hatch Project (H-8200).
Author Disclosure Statement
For all authors, there are no financial or other contractual agreements that might cause conflicts of interest or be perceived as causing conflicts of interest.
References
- 1.Aagaard-Tillery KM. Grove K. Bishop J. Ke X. Fu Q. McKnight R. Lane RH. Developmental origins of disease and determinants of chromatin structure: Maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad W. Shabbiri K. Nazar N. Nazar S. Qaiser S. Mughal MAS. Human linker histones: Interplay between phosphorylation and O-β-GlcNAc to mediate chromatin structural modifications. Cell Div. 2011;6:15. doi: 10.1186/1747-1028-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ainge H. Thompson C. Ozanne SE. Rooney KB. A systematic review on animal models of maternal high fat feeding and offspring glycaemic control. Int J Obes. 2011;35:325–335. doi: 10.1038/ijo.2010.149. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson CM. Lopez F. Zimmer A. Benoit JN. Placental insufficiency leads to developmental hypertension and mesenteric artery dysfunction in two generations of Sprague-Dawley rat offspring. Biol Reprod. 2006;74:538–544. doi: 10.1095/biolreprod.105.045807. [DOI] [PubMed] [Google Scholar]
- 6.Armitage JA. Lakasing L. Taylor PD. Balachandran AA. Jensen RI. Dekou V. Ashton N. Nyengaard JR. Poston L. Developmental programming of aortic and renal structure in offspring of rats fed fat-rich diets in pregnancy. J Physiol. 2005;565:171–184. doi: 10.1113/jphysiol.2005.084947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister AJ. Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 8.Barbacci E. Chalkiadaki A. Masdeu C. Haumaitre C. Lokmane L. Loirat C. Cloarec S. Talianidis I. Bellanne-Chantelot C. Cereghini S. HNF1beta/TCF2 mutations impair transactivation potential through altered coregulator recruitment. Hum Mol Genet. 2004;13:3139–3149. doi: 10.1093/hmg/ddh338. [DOI] [PubMed] [Google Scholar]
- 9.Barker DJP. Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;8489:1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 10.Barker DJP. Growth and living conditions in childhood and hypertension in adult life: A longitudinal study. J Hypertens. 2002;20:1951–1956. doi: 10.1097/00004872-200210000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Barker DJP. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 12.Barlow DP. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 13.Bazer FW. Wu G. Spencer TE. Johnson GA. Burghardt RC. Bayless K. Novel pathways for implantation and establishment and maintenance of pregnancy in mammals. Mol Human Reprod. 2010;16:135–152. doi: 10.1093/molehr/gap095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazer FW. Wu G. Johnson GA. Kim JY. Song GW. Uterine histotroph and conceptus development: Select nutrients and secreted phosphoprotein 1 affect MTOR cell signaling in ewes. Biol Reprod. 85:1094–1107. doi: 10.1095/biolreprod.111.094722. [DOI] [PubMed] [Google Scholar]
- 15.Berard J. Bee G. Effects of dietary L-arginine supplementation to gilts during early gestation on foetal survival, growth and myofiber formation. Animal. 2010;4:1680–1687. doi: 10.1017/S1751731110000881. [DOI] [PubMed] [Google Scholar]
- 16.Bertram C. Khan O. Ohri S. Phillips DI. Matthews SG. Hanson MA. Transgenerational effects of prenatal nutrient restriction on cardiovascular and hypothalamic-pituitary- adrenal function. J Physiol. 2008;586:2217–2229. doi: 10.1113/jphysiol.2007.147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieliauskas AV. Pflum MK. Isoform-selective histone deacetylase inhibitors. Chem Soc Rev. 2008;37:1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird A. Molecular biology. DNA methylation de novo. Science. 1999;286:2287–2288. doi: 10.1126/science.286.5448.2287. [DOI] [PubMed] [Google Scholar]
- 19.Brasacchio D. Okabe J. Tikellis C. Balcerczyk A. George P. Baker EK. Calkin AC. Brownlee M. Cooper ME. El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown KD. Robertson KD. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat Genet. 2007;39:289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 21.Burdge GC. Lillycrop KA. Nutrition, epigenetics, and developmental plasticity: Implications for understanding human disease. Annu Rev Nutr. 2010;30:315–339. doi: 10.1146/annurev.nutr.012809.104751. [DOI] [PubMed] [Google Scholar]
- 22.Bygren LO. Kaati G. Edvinsson S. Longevity determined by paternal ancestors' nutrition during their slow growth period. Acta Biotheor. 2001;49:53–59. doi: 10.1023/a:1010241825519. [DOI] [PubMed] [Google Scholar]
- 23.Cafe LM. Hennessy DW. Hearnshaw H. Morris SG. Greenwood PL. Consequences of prenatal and preweaning growth for feedlot growth, intake and efficiency of Piedmontese- and Wagyu-sired cattle. Anim Prod Sci. 2009;49:461–467. [Google Scholar]
- 24.Cao DJ. Wang ZV. Battiprolu PK. Jiang N. Morales CR. Kong YL. Rothermel BA. Gillette TG. Hill JA. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro R. Rivera I. Blom HJ. Jakobs C. Tavares de Almeida I. Homocysteine metabolism, hyperhomocysteinaemia and vascular disease: An overview. J Inherit Metab Dis. 2006;29:3–20. doi: 10.1007/s10545-006-0106-5. [DOI] [PubMed] [Google Scholar]
- 26.Cerutti H. Casas-Mollano JA. Histone H3 phosphorylation: Universal code or lineage specific dialects? Epigenetics. 2009;4:71–75. doi: 10.4161/epi.4.2.7781. [DOI] [PubMed] [Google Scholar]
- 27.Chakrabarti SK. Francis J. Ziesmann SM. Garmey JC. Mirmira RG. Covalent histone modifications underline the developmental regulation of insulin gene transcription in pancreatic beta cells. J Biol Chem. 2003;278:23617–23623. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- 28.Chan LL. Sébert SP. Hyatt MA. Stephenson T. Budge H. Symonds ME. Gardner DS. Effect of maternal nutrient restriction from early to midgestation on cardiac function and metabolism after adolescent-onset obesity. Am J Physiol Regul Integr Comp Physiol. 2009;296:R1455–R1463. doi: 10.1152/ajpregu.91019.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen ZJ. Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation in nucleolar dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chmurzynska A. Fetal programming: Link between early nutrition, DNA methylation, and complex diseases. Nutr Rev. 2010;68:87–98. doi: 10.1111/j.1753-4887.2009.00265.x. [DOI] [PubMed] [Google Scholar]
- 31.Christian P. Micronutrients, birthweight, and survival. Annu Rev Nutr. 2010;30:83–104. doi: 10.1146/annurev.nutr.012809.104813. [DOI] [PubMed] [Google Scholar]
- 32.Cleal JK. Poore KR. Boullin JP. Khan O. Chau R. Hambidge O. Torrens C. Newman JP. Poston L. Noakes DE. Hanson MA. Green LR. Mismatched pre- and postnatal nutrition leads to cardiovascular dysfunction and altered renal function in adulthood. Proc Natl Acad Sci USA. 2007;104:9529–9533. doi: 10.1073/pnas.0610373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J. Williams S. Grove K. Lane RH. Aagaard-Tillery KM. A maternal high-fat diet is accompanied by alterations in the fetal primate metabolome. Am J Obstet Gynecol. 2009;201:281.e281–289. doi: 10.1016/j.ajog.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cropley JE. Suter CM. Beckman KB. Martin DI. Germline epigenetic modification of the murine A vy allele by nutritional supplementation. Proc Natl Acad Sci USA. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui H. Cruz-Correa M. Giardiello FM. Hutcheon DF. Kafonek DR. Brandenburg S. Wu Y. He X. Powe NR. Feinberg AP. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 36.Dindot SV. Kent KC. Evers B. Loskutoff N. Womack J. Piedrahita JA. Conservation of genomic imprinting at the XIST, IGF2 and GTL2 loci in the bovine. Mamm Genome. 2004;15:966–974. doi: 10.1007/s00335-004-2407-z. [DOI] [PubMed] [Google Scholar]
- 37.Dindot SV. Person R. Strivens M. Garcia R. Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19:1374–1383. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn GA. Bale TL. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009;150:4999–5009. doi: 10.1210/en.2009-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards LE. Hellerstedt WL. Alton IR. Story M. Himes JH. Pregnancy complications and birth outcomes in obese and normal-weight women: Effects of gestational weight change. Obstet Gynecol. 1996;87:389–394. doi: 10.1016/0029-7844(95)00446-7. [DOI] [PubMed] [Google Scholar]
- 40.Elmen J. Lindow M. Schutz S. Lawrence M. Petri A. Obad S. Lindholm M. Hedtjarn M. Hansen HF. Berger U. Gullans S. Kearney P. Sarnow P. Straarup EM. Kauppinen S. LNA-mediated microRNA silencing in nonhuman primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 41.Esau C. Kang X. Peralta E. Hanson E. Marcusson EG. Ravichandran LV. Sun Y. Koo S. Perera RJ. Jain R. Dean NM. Freier SM. Bennett CF. Lollo B. Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 42.Eriksson JG. Forsen T. Tuomilehto J. Osmond C. Barker DJP. Early growth and coronary heart disease in later life: Longitudinal study. Br Med J. 2001;322:949–953. doi: 10.1136/bmj.322.7292.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evertts AG. Zee BM. Garcia BA. Modern approaches for investigating epigenetic signaling pathways. J Appl Physiol. 2010;109:927–933. doi: 10.1152/japplphysiol.00007.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabian MR. Sonenberg N. Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 45.Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299:1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- 46.Feng D. Liu T. Sun Z. Bugge A. Mullican SE. Alenghat T. Liu XS. Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez AZ. Siebel AL. El-Osta A. Atherogenic factors and their epigenetic relationships. Int J Vasc Med. 2010;2010:437809. doi: 10.1155/2010/437809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flegal KM. Carroll MD. Ogden CL. Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 49.Fleming AB. Kao CF. Hillyer C. Pikaart M. Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Ford SP. Zhang L. Zhu M. Miller MM. Smith DT. Hess BW. Moss GE. Nathanielsz PW. Nijland MJ. Maternal obesity accelerates fetal pancreatic beta- cell but not alpha-cell development in sheep: Prenatal consequences. Am J Physiol Regul Integr Comp Physiol. 2009;297:R835–843. doi: 10.1152/ajpregu.00072.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forsen T. Eriksson JG. Tuomilehto J. Osmond C. Barker DJP. Growth in utero and during childhood among women who develop coronary disease: Longitudinal study. Br Med J. 1999;319:1403–1407. doi: 10.1136/bmj.319.7222.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fowden AL. Ward JW. Wooding FP. Forhead AJ. Constancia M. Programming placental nutrient transport capacity. J Physiol. 2006;572:5–15. doi: 10.1113/jphysiol.2005.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujiki K. Kano F. Shiota K. Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godmann M. Auger V. Ferraroni-Aguiar V. Di Sauro A. Sette C. Behr R. Kimmins S. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol Reprod. 2007;77:754–764. doi: 10.1095/biolreprod.107.062265. [DOI] [PubMed] [Google Scholar]
- 55.Godfrey KM. Lillycrop KA. Burdge GC. Gluckman PD. Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr Res. 2007;61:5R–10R. doi: 10.1203/pdr.0b013e318045bedb. [DOI] [PubMed] [Google Scholar]
- 56.Golding MC. Williamson GL. Stroud TK. Westhusin ME. Long CR. Examination of DNA methyltransferase expression in cloned embryo reveals an essential role for Dnmt1 in bovine development. Mol Reprod Dev. 2011;78:306–317. doi: 10.1002/mrd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Granger A. Abdullah I. Huebner F. Stout A. Wang T. Huebner T. Epstein JA. Gruber PJ. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 2008;22:35–49. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenwood PL. Hunt AS. Hermanson JW. Bell AW. Effects of birth weight and postnatal nutrition on neonatal sheep: II. Skeletal muscle growth and development. J Anim Sci. 2000;78:50–61. doi: 10.2527/2000.78150x. [DOI] [PubMed] [Google Scholar]
- 59.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 60.Haberland M. Montgomery RL. Olsen EN. The many roles of histone deacetylation in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hales CN. Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 62.Han HC. Austin KJ. Nathanielsz PW. Ford SP. Nijland MJ. Hansen TR. Maternal nutrient restriction alters gene expression in the ovine fetal heart. J Physiol. 2004;558:111–121. doi: 10.1113/jphysiol.2004.061697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen RS. Gartler SM. Scott CR. Chen SH. Laird CD. Methylation analysis of CGG sites in the CPG island of the human FMR1 gene. Hum Mol Genet. 1992;1:571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- 64.Hardie DG. Hawley SA. Scott JW. AMP-activated protein kinase: Development of the energy sensor concept. J Physiol. 2006;574:7–15. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrison M. Langley-Evans SC. Intergenerational programming of impaired nephrogenesis and hypertension in rats following maternal protein restriction during pregnancy. Br J Nutr. 2009;101:1020–1030. doi: 10.1017/S0007114508057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haumaitre C. Lenoir O. Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endogenous progenitors. Mol Cell Biol. 2008;28:6373–6383. doi: 10.1128/MCB.00413-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heijmans BT. Tobi EW. Stein AD. Putter H. Blauw GJ. Susser ES. Slagboom PE. Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heneghan FM. Miller N. McAnena OJ. O'Brien T. Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab. 2011;96:E846–850. doi: 10.1210/jc.2010-2701. [DOI] [PubMed] [Google Scholar]
- 69.Hill DJ. Duville B. Pancreatic development and adult diabetes. Pediatr Res. 2000;48:269–274. doi: 10.1203/00006450-200009000-00002. [DOI] [PubMed] [Google Scholar]
- 70.Holliday R. Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 71.Hosoki K. Ogata T. Kagami M. Tanaka T. Saitoh S. Epimutation (hypomethylation) affecting the chromosome 14q32.2 in a girl with upd914)mat-like phenotype. Eur J Hum Genet. 2008;16:1019–1023. doi: 10.1038/ejhg.2008.90. [DOI] [PubMed] [Google Scholar]
- 72.Hulsmans M. De Keyzer D. Holvoet P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB J. 2011;25:2515–2527. doi: 10.1096/fj.11-181149. [DOI] [PubMed] [Google Scholar]
- 73.Hurd PJ. Bannister AJ. Halls K. Dawson MA. Vermeulen M. Olsen JV. Ismail H. Somers J. Mann M. Owen-Hughes T. Gout I. Kouzarides T. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J Biol Chem. 2009;284:16575–16583. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ilnytskyy Y. Kovalchuk O. Non-targeted radiation effects—An epigenetic connection. Mutat Res. 2011;714:113–25. doi: 10.1016/j.mrfmmm.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 75.Jiang MH. Fei J. Lan MS. Lu ZP. Liu M. Fan WW. Gao X. Lu DR. Hypermethylation of hepatic Gck promoter in ageing rats contributes to diabetogenic potential. Diabetologia. 2008;51:1525–1533. doi: 10.1007/s00125-008-1034-8. [DOI] [PubMed] [Google Scholar]
- 76.Jimenez-Chillaron JC. Isganaitis E. Charalambons M. Gesta S. Pentinat-Pelegrin T. Faucette RR. Otis JP. Chow A. Diaz R. Ferguson-Smith A. Patti ME. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jirtle RL. Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jobgen WS. Fried SK. Fu WJ. Meininger CJ. Wu G. Regulatory role for the arginine-nitric oxide pathway in metabolism of energy substrates. J Nutr Biochem. 2006;17:571–588. doi: 10.1016/j.jnutbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 79.Kaati G. Bygren LO. Pembrey M. Sjostrom M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 80.Kajantie E. Osmond C. Barker DJP. Forsen T. Phillips DIW. Eriksson JG. Size at birth as a predictor of mortality in adulthood: A follow-up of 350 000 person- years. Int J Epidemiol. 2005;34:655–663. doi: 10.1093/ije/dyi048. [DOI] [PubMed] [Google Scholar]
- 81.Kaneda A. Wang CJ. Cheong R. Timp W. Onyango P. Wen B. Iacobuzio-Donahue CA. Ohlsson R. Andraos R. Pearson MA. Sharov AA. Longo DL. Ko MS. Levchenko A. Feinberg AP Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation, tumor risk. Proc Natl Acad Sci USA. 2007;104:20926–20931. doi: 10.1073/pnas.0710359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kawasaki H. Taira K. Morris KV. siRNA induced transcriptional gene silencing in mammalian cells. Cell Cycle. 2005;4:442–448. doi: 10.4161/cc.4.3.1520. [DOI] [PubMed] [Google Scholar]
- 83.Khan IY. Dekou V. Douglas G. Jensen R. Hanson MA. Poston L. Taylor PD. A high-fat diet during rat pregnancy or suckling induces cardiovascular dysfunction in adult offspring. Am J Physiol Regul Integr Comp Physiol. 2005;288:R127–133. doi: 10.1152/ajpregu.00354.2004. [DOI] [PubMed] [Google Scholar]
- 84.Khan KS. Wojdyla D. Say L. Gulmezoglu AM. Van Look PF. WHO analysis of causes of maternal death: A systematic review. Lancet. 2006;367:1066–1074. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 85.Kiel DW. Dodson EA. Artal R. Boehmer TK. Leet TL. Gestational weight gain and pregnancy outcomes in obese women. Obstet Gynecol. 2007;110:752–758. doi: 10.1097/01.AOG.0000278819.17190.87. [DOI] [PubMed] [Google Scholar]
- 86.Kirk H. Cefalu WT. Ribnicky D. Liu Z. Eilertsen KJ. Botanicals as epigenetic modulators for mechanisms contributing to development of metabolic syndrome. Metabolism. 2008;57:S16–S23. doi: 10.1016/j.metabol.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 87.Krishna SM. Dear AE. Norman PE. Golledge J. Genetic and epigenetic mechanisms and their possible role in abdominal aortic aneurysm. Atherosclerosis. 2010;212:16–29. doi: 10.1016/j.atherosclerosis.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 88.Krishnamurthy J. Ramsey MR. Ligon KL. Torrice C. Koh A. Bonner-Weir S. Sharpless NE. p16INK4a induces an age-dependent decline in islet regeneration potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 89.Krutzfeldt M. Rajewsky N. Braich R. Rajeev KG. Tuschl T. Manoharan M. Stoffel M. Silencing of miRNAs in vivo with ‘antagomirs”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 90.Lassala A. Bazer FW. Cudd TA. Li P. Li XL. Satterfield MC. Spencer TE. Wu G. Intravenous administration of L-citrulline to pregnant ewes is more effective than L-arginine for increasing arginine availability in the fetus. J Nutr. 2009;139:660–665. doi: 10.3945/jn.108.102020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lassala A. Bazer FW. Cudd TA. Datta S. Keisler DH. Satterfield MC. Spencer TE. Wu G. Parenteral administration of L-arginine prevents fetal growth restriction in undernourished ewes. J Nutr. 2010;140:1242–1248. doi: 10.3945/jn.110.125658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lassala A. Bazer FW. Cudd TA. Datta S. Keisler DH. Satterfield MC. Spencer TE. Wu G. Parenteral administration of L-arginine enhances fetal survival and growth in sheep carrying multiple pregnancies. J Nutr. 2011;141:849–855. doi: 10.3945/jn.111.138172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee RC. Feinbaum RL. Ambross V. The C. elegan heterochronic gene lin-4 encodes small RNAs with antisense complementary to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 94.Leon DA. Lithell HO. Vagero D. Koupilova I. Mohsen R. Berglund L. Lithell UB. McKeigue PM. Reduced fetal growth rate and increased risk of death from ischaemic heart disease: Cohort study of 15000 Swedish men and women born 1915–29. Br Med J. 1998;317:241–245. doi: 10.1136/bmj.317.7153.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewis BP. Shih IH. Jones-Rhoades MW. Bartel DP. Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 96.Li H. Meininger CJ. Hawker JR. Kelly KA. Morris SM. Wu G. Activities of arginase I and II are limiting for endothelial cell proliferation. Am J Physiol Regul Integr Comp Physiol. 2002;282:R64–R69. doi: 10.1152/ajpregu.2002.282.1.R64. [DOI] [PubMed] [Google Scholar]
- 97.Li XL. Bazer FW. Johnson GA. Burghardt RC. Erikson DW. Frank JW. Spencer TE. Shinzato I. Wu G. Dietary supplementation with 0.8% L-arginine between days 0 and 25 of gestation reduces litter size in gilts. J Nutr. 2010;140:1111–1116. doi: 10.3945/jn.110.121350. [DOI] [PubMed] [Google Scholar]
- 98.Li Y. Reddy MA. Miao F. Shanmugam N. Yee JK. Hawkins D. Ren B. Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kB-dependent inflammatory genes. J Biol Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ling C. Poulsen P. Simonsson S. Ronn T. Holmkvist J. Almgren P. Hagert P. Nilsson E. Mabey AG. Nilsson P. Vaag A. Groop L. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest. 2007;117:3427–3435. doi: 10.1172/JCI30938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ling C. Del Guerra S. Lupi R. Ronn T. Granhall C. Luthman H. Masiello P. Marchetti P. Groop L. Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ling C. Groop L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lumey LH. Reproductive outcome in women prenatally exposed to undernutrition: a review of findings from the Dutch famine birth cohort. Proc Nutr Soc. 1998;57:129–135. doi: 10.1079/pns19980019. [DOI] [PubMed] [Google Scholar]
- 103.Lynn FC. Meta-regulation: MicroRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452–459. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 104.Marsal K. Intrauterine growth restriction. Curr Opin Obstet Gynecol. 2002;14:127–135. doi: 10.1097/00001703-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 105.Mateo RD. Wu G. Bazer FW. Park JC. Shinzato I. Kim SW. Dietary L- arginine supplementation enhances the reproductive performance of gilts. J Nutr. 2007;137:652–656. doi: 10.1093/jn/137.3.652. [DOI] [PubMed] [Google Scholar]
- 106.Matouk CC. Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 107.Martin JF. Johnston CS. Han CT. Benyshek DC. Nutritional origins of insulin resistance: A rat model for diabetes-prone human populations. J Nutr. 2000;130:741–744. doi: 10.1093/jn/130.4.741. [DOI] [PubMed] [Google Scholar]
- 108.Maulik N, editor; Maulik G, editor. Nutrition, Epigenetic Mechanisms, and Human Disease. New York: CRC Press; 2011. p. 426. [Google Scholar]
- 109.McClelland M. Nelson M. The effect of site-specific DNA methylation on restriction endonucleases and DNA modification methyltransferases. Gene. 1988;74:291–304. doi: 10.1016/0378-1119(88)90305-8. [DOI] [PubMed] [Google Scholar]
- 110.McCurdy CE. Bishop JM. Williams SM. Grayson BE. Smith MS. Friedman JE. Grove KL. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119:323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McGee SL. van Denderen BJ. Howlett KF. Mollica J. Schertzer JD. Kemp BE. Hargreaves M. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 112.McGee SL. Hargreaves M. Histone modifications and exercise adaptations. J Appl Physiol. 2011;110:258–263. doi: 10.1152/japplphysiol.00979.2010. [DOI] [PubMed] [Google Scholar]
- 113.McGregor RA. Choi MS. MicroRNAs in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11:304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McKnight JR. Satterfield MC. Jobgen WS. Smith SB. Spencer TE. Meininger CJ. McNeal CJ. Wu G. Beneficial effects of L-arginine on reducing obesity: Potential mechanisms and important implications for human health. Amino Acids. 2010;39:349–357. doi: 10.1007/s00726-010-0598-z. [DOI] [PubMed] [Google Scholar]
- 115.McKnight JR. Satterfield MC. Li XL. Gao HJ. Wang JJ. Li DF. Wu G. Obesity in pregnancy: Problems and potential solutions. Front Biosci. 2011;E3:442–452. doi: 10.2741/e259. [DOI] [PubMed] [Google Scholar]
- 116.Melzner I. Scott V. Dorsch K. Fischer P. Wabitsch M. Bruderlein S. Hasel C. Moller P. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J Biol Chem. 2002;277:45420–45427. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- 117.Miao F. Gonzalo IG. Lanting L. Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 118.Miao F. Smith DD. Zhang L. Min A. Feng W. Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: An epigenetic study in diabetes. Diabetes. 2008;57:3189–3198. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Milagro FI. Campion J. Garcia-Diaz DF. Goyenechea E. Paternain L. Martinez JA. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem. 2009;65:1–9. doi: 10.1007/BF03165964. [DOI] [PubMed] [Google Scholar]
- 120.Mouillet JF. Chu T. Sadovsky Y. Expression patterns of placental microRNAs. Birth Defects Res A. 2011;91:737–743. doi: 10.1002/bdra.20782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Muhlhausler BS. Duffield JA. McMillen IC. Increased maternal nutrition increases leptin expression in perirenal and subcutaneous adipose tissue in the postnatal lamb. Endocrinology. 2007;148:6157–6163. doi: 10.1210/en.2007-0770. [DOI] [PubMed] [Google Scholar]
- 122.Nakanishi N. Nakagawa Y. Tokushige N. Aoki N. Matsuzaka T. Ishii K. Yahagi N. Kobayashi K. Yatoh S. Takahashi A. Suzuki H. Urayama O. Yamada N. Shimano H. The upregulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385:492–496. doi: 10.1016/j.bbrc.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 123.Ng SF. Lin RC. Laybutt DR. Barres R. Owens JA. Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 124.Nijland MJ. Mitsuya K. Li C. Ford SP. McDonald TJ. Nathanielsz PW. Cox LA. Epigenetic modification of fetal baboon hepatic phosphoenolpyruvate carboxykinase following exposure to moderately reduced nutrient availability. J Physiol. 2010;588:1349–1359. doi: 10.1113/jphysiol.2009.184168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nolan CJ. Damm P. Prentki M. Type 2 diabetes across generations: From pathophysiology to prevention and management. Lancet. 2011;378:169–181. doi: 10.1016/S0140-6736(11)60614-4. [DOI] [PubMed] [Google Scholar]
- 126.Ogden CL. Yanovski SZ. Carroll MD. Flegal KM. The epidemiology of obesity. Gastroenterology. 2007;132:2087–2102. doi: 10.1053/j.gastro.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 127.Oommen AM. Griffin JB. Sarath G. Zempleni J. Roles for nutrients in epigenetic events. J Nutr Biochem. 2005;16:74–77. doi: 10.1016/j.jnutbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 128.Ovesen P. Rasmussen S. Kesmodel U. Effect of prepregnancy maternal overweight and obesity on pregnancy outcome. Obstet Gynecol. 2011;118:305–312. doi: 10.1097/AOG.0b013e3182245d49. [DOI] [PubMed] [Google Scholar]
- 129.Parikh H. Nilsson E. Ling C. Poulsen P. Almgren P. Nittby H. Eriksson KF. Vaag A. Croop LC. Molecular correlates for maximal oxygen uptake and type 1 fibers. Am J Physiol Endocrinol Metab. 2008;294:E1152–E1159. doi: 10.1152/ajpendo.90255.2008. [DOI] [PubMed] [Google Scholar]
- 130.Park JH. Stoffers DA. Nicholls RD. Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Peterson CL. Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 132.Philbin VJ. Levy O. Developmental biology of the innate immune response: Implications for neonatal and infant vaccine development. Pediatr Res. 2009;65:98R–105R. doi: 10.1203/PDR.0b013e31819f195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Phillips DIW. Programming of the stress response: A fundamental mechanism underlying the long-term effects of the fetal environment. J Intern Med. 2007;261:453–460. doi: 10.1111/j.1365-2796.2007.01801.x. [DOI] [PubMed] [Google Scholar]
- 134.Philp LK. Muhlhausler BS. Janovska A. Wittert GA. Duffield JA. McMillen IC. Maternal overnutrition suppresses the phosphorylation of 5'-AMP-activated protein kinase in liver, but not skeletal muscle, in the fetal and neonatal sheep. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1982–1990. doi: 10.1152/ajpregu.90492.2008. [DOI] [PubMed] [Google Scholar]
- 135.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 136.Pokholok DK. Harbison CT. Levine S. Cole M. Hannett NM. Lee TI. Bell GW. Walker K. Rolfe PA. Herbolsheimer E. Zeitlinger J. Lewitter F. Gifford DK. Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 137.Poston L. Harthoorn LF. Van Der Beek EM. Contributors to the ILSI Europe Workshop. Obesity in pregnancy: Implications for the mother and lifelong health of the child. A consensus statement. Pediatr Res. 2011;69:175–180. doi: 10.1203/PDR.0b013e3182055ede. [DOI] [PubMed] [Google Scholar]
- 138.Poy MN. Eliasson L. Krutzfeldt J. Kuwajima S. Ma X. Macdonald PE. Pfeffer S. Tuschl T. Rajewsky N. Rorsman P. Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 139.Qipshidze N. Metreveli N. Lominadze D. Tyagi SC. Folic acid improves acetylcholine- induced vasoconstriction of coronary vessels isolated from hyperhomocysteinemic mice: An implication to coronary vasospasm. J Cell Physiol. 2011;226:2712–2720. doi: 10.1002/jcp.22621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raychaudhuri N. Raychaudhuri S. Thamotharan M. Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283:13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rayner KJ. Sheedy FJ. Esau CC. Hussain FN. Temel RE. Parathath S. van Gils JM. Rayner AJ. Chang AN. Suarez Y. Fernandez-Hernando C. Fisher EA. Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rayner KJ. Esau CC. Hussain FN. McDaniel AL. Marshall SM. van Gils JM. Ray TD. Sheedy FJ. Goedeke L. Liu X. Khatsenko OG. Kaimal V. Lees CJ. Fernandez-Hernando C. Fisher EA. Temel RE. Moore KJ. Inhibition of miR-33a/b in nonhuman primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reynolds LP. Caton JS. Redmer DA. Grazul-Bilska AT. Vonnahme KA. Borowicz PB. Luther JS. Wallace JM. Wu G. Spencer TE. Evidence for altered placental blood flow and vascularity in compromised pregnancies. J Physiol. 2006;572:51–58. doi: 10.1113/jphysiol.2005.104430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ronn T. Poulsen P. Hansson O. Holmkvist J. Almgren P. Nilsson P. Tuomi T. Isomaa B. Groop L. Vaag A. Ling C. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51:1159–1168. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- 145.Rooney K. Ozanne SE. Maternal over-nutrition and offspring obesity predisposition: Targets for preventative interventions. Int J Obes. 2011;35:883–890. doi: 10.1038/ijo.2011.96. [DOI] [PubMed] [Google Scholar]
- 146.Rush D. Nutritional and maternal mortality in the developing world. Am J Clin Nutr. 2000;72:212S–240S. doi: 10.1093/ajcn/72.1.212S. [DOI] [PubMed] [Google Scholar]
- 147.Sakabe K. Wang ZH. Hart GW. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Samuelsson AM. Matthews PA. Argenton M. Christie MR. McConnell JM. Jansen EH. Piersma AH. Ozanne SE. Twinn DF. Remacle C. Rowlerson A. Poston L. Taylor PD. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: A novel murine model of developmental programming. Hypertension. 2008;51:383–392. doi: 10.1161/HYPERTENSIONAHA.107.101477. [DOI] [PubMed] [Google Scholar]
- 149.Sathishkumar K. Elkins R. Yallampalli U. Yallampalli C. Protein restriction during pregnancy induces hypertension and impairs endothelium-dependent vascular function in adult female offspring. J Vasc Res. 2009;46:229–239. doi: 10.1159/000166390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Satterfield MC. McKnight JR. Li XL. Wu G. Nutrition, epigenetics, and vascular function. In: Maulik N, editor; Maulik G, editor. Nutrition, Epigenetic Mechanisms, and Human Disease. New York: CRC Press; 2011. pp. 125–139. [Google Scholar]
- 151.Satterfield MC. Bazer FW. Spencer TE. Wu G. Sildenafil citrate treatment enhances amino acid availability in the conceptus and fetal growth in an ovine model of intrauterine growth restriction. J Nutr. 2010;140:251–258. doi: 10.3945/jn.109.114678. [DOI] [PubMed] [Google Scholar]
- 152.Satterfield MC. Wu G. Growth and development of brown adipose tissue: Significance and nutritional regulation. Front Biosci. 2011;16:1589–1608. doi: 10.2741/3807. [DOI] [PubMed] [Google Scholar]
- 153.Schlabritz-Loutsevitch N. Farley D. Tejero ME. Comuzzie AG. Higgins PB. Cox L. Werner SL. Jenkins SL. Li C. Choj J. Dick EJ. Hubbard GB. Frost P. Dudley DD. Wu G. Nathanielsz PW. Feto-placental adaptations to maternal obesity in the baboon. Placenta. 2009;30:752–760. doi: 10.1016/j.placenta.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schwer B. Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 155.Sharkey D. Gardner DS. Symonds ME. Budge H. Maternal nutrient restriction during early fetal kidney development attenuates the renal innate inflammatory response in obese young adult offspring. Am J Physiol Renal Physiol. 2009;297:F1199–1207. doi: 10.1152/ajprenal.00303.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Simmons R. Epigenetics and maternal nutrition: Nature v. nurture. Proc Nutr Soc. 2011;70:73–81. doi: 10.1017/S0029665110003988. [DOI] [PubMed] [Google Scholar]
- 157.Sinclair KD. Allegrucci C. Singh R. Gardner DS. Sebastian S. Bispham J. Thurston A. Huntley JF. Rees WD. Maloney CA. Lea RG. Craigon J. McEvoy TG. Young LE. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci USA. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schroen B. Heymans S. Small but smart—MicroRNAs in the center of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and aging. Cardiovasc Res. 2011 doi: 10.1093/cvr/cvr268. [DOI] [PubMed] [Google Scholar]
- 159.Smith AD. Kim YI. Refsum H. Is folic acid good for everyone? Am J Clin Nutr. 2008;87:517–533. doi: 10.1093/ajcn/87.3.517. [DOI] [PubMed] [Google Scholar]
- 160.Sullivan EL. Smith MS. Grove KL. Perinatal exposure to high-fat diet programs energy balance, metabolism and behavior in adulthood. Neuroendocrinol. 2011;93:1–8. doi: 10.1159/000322038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sun LD. Zhao HB. Xu ZB. Liu QL. Liang W. Wang LT. Cai X. Zhang L. Hu L. Wang G. Zha X. Phosphatidylinositol 3- kinase/protein kinase B pathway stabilizes DNA methyltransferase I protein and maintains DNA methylation. Cell Signal. 2007;19:2255–2263. doi: 10.1016/j.cellsig.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 162.Symonds ME. Sebert SP. Hyatt MA. Budge H. Nutritional programming of the metabolic syndrome. Nat Rev Endocrinol. 2009;5:604–610. doi: 10.1038/nrendo.2009.195. [DOI] [PubMed] [Google Scholar]
- 163.Szic KSV. Ndlovu MN. Haegeman G. Vanden Berghe W. Nature or nurture: Let food be your epigenetic medicine in chronic inflammatory disorders. Biochem Pharmacol. 2010;80:1816–1832. doi: 10.1016/j.bcp.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 164.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- 165.Tateishi K. Okada Y. Kallin EM. Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Taylor PD. McConnell J. Khan IY. Holemans K. Lawrence KM. Asare-Anane H. Persaud SJ. Jones PM. Petrie L. Hanson MA. Poston L. Impaired glucose homeostasis and mitochondrial abnormalities in offspring of rats fed a fat-rich diet in pregnancy. Am J Physiol Regul Integr Comp Physiol. 2005;288:R134–R139. doi: 10.1152/ajpregu.00355.2004. [DOI] [PubMed] [Google Scholar]
- 167.Temple IK. Shield JP. Transient neonatal diabetes, a disorder of imprinting. J Med Genet. 2002;39:872–875. doi: 10.1136/jmg.39.12.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Thamotharan M. Garg M. Oak S. Rogers LM. Pan A. Sangiorgi F. Lee PN. Devaskar SU. Transgenerational inheritance of the insulin resistant phenotypes in embryo- transferred intrauterine growth-restricted adult female rat offspring. Am J Physiol Endocrinol Metab. 2007;292:E1270–E1279. doi: 10.1152/ajpendo.00462.2006. [DOI] [PubMed] [Google Scholar]
- 169.Thambirajah AA. Li A. Ishibashi T. Ausio J. New developments in post- translational modifications and functions of histone H2A variants. Biochem Cell Biol. 2009;87:7–17. doi: 10.1139/O08-103. [DOI] [PubMed] [Google Scholar]
- 170.Timp W. Levchenko A. Feinberg AP. A new link between epigenetic progenitor lesions in cancer and the dynamics of signal transduction. Cell Cycle. 2009;8:383–390. doi: 10.4161/cc.8.3.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tobi EW. Lumey LH. Talens RP. Kremer D. Putter H. Stein AD. Slagboom PE. Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Todeschini AL. Teysset L. Delmarre V. Ronsseray S. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS One. 2010;5:e11032. doi: 10.1371/journal.pone.0011032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tong JF. Yan X. Zhu MJ. Ford SP. Nathanielsz PW. Du M. Maternal obesity downregulates myogenesis and beta-catenin signaling in fetal skeletal muscle. Am J Physiol Endocrinol Metab. 2009;296:E917–E924. doi: 10.1152/ajpendo.90924.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Turunen MP. Aavik E. Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 176.Unterberger A. Szyf M. Nathanielsz PW. Cox LA. Organ and gestational age effects of maternal nutrient restriction on global methylation in fetal baboons. J Med Primatol. 2009;38:219–227. doi: 10.1111/j.1600-0684.2008.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.USDA. Dietary Guidance. 2010. DRI Tables. http://fnic.nal.usda.gov. [Jul 26;]. 2011. http://fnic.nal.usda.gov.
- 178.Vaxillaire M. Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr Rev. 2008;29:254–264. doi: 10.1210/er.2007-0024. [DOI] [PubMed] [Google Scholar]
- 179.Villeneuve LM. Reddy MA. Lanting LL. Wang M. Meng L. Natarajan R. Epigenetic histine H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vonnahme KA. Hess BW. Hansen TR. McCormick RJ. Rule DC. Moss GE. Murdoch WJ. Nijland MJ. Skinner DC. Nathanielsz PW. Ford SP. Maternal undernutrition from early- to mid-gestation leads to growth retardation, cardiac ventricular hypertrophy, and increased liver weight in the fetal sheep. Biol Reprod. 2003;69:133–140. doi: 10.1095/biolreprod.102.012120. [DOI] [PubMed] [Google Scholar]
- 181.Vucetic Z. Kimmel J. Totoki K. Hollenbeck E. Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151:4756–4764. doi: 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Waddington CH. Organizers and Genes. Cambridge University Press; 1940. [Google Scholar]
- 183.Waechter DE. Baserga R. Effect of methylation on expression of microinjected genes. Proc Natl Acad Sci USA. 1982;79:1106–1110. doi: 10.1073/pnas.79.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wallace DC. Fan WW. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wallace JM. Milne JS. Aitken RP. The effect of overnourishing singleton-bearing adult ewes on nutrient partitioning to the gravid uterus. Br J Nutr. 2005;94:533–539. doi: 10.1079/bjn20041398. [DOI] [PubMed] [Google Scholar]
- 186.Wallace JM. Luther JS. Milne JS. Aitken RP. Redmer DA. Reynolds LP. Hay WW. Nutritional modulation of adolescent pregnancy outcome. A review. Placenta. 2006;27:S61–S68. doi: 10.1016/j.placenta.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 187.Warner MJ. Ozanne SE. Mechanisms involved in the developmental programming of adulthood disease. Biochem J. 2010;427:333–347. doi: 10.1042/BJ20091861. [DOI] [PubMed] [Google Scholar]
- 188.Weake VM. Workman JL. Histone ubiquitination: Triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 189.Weems J. Olson AL. Class II histone deacetylases limit GLUT4 gene expression during adipocyte differentiation. J Biol Chem. 2011;286:460–468. doi: 10.1074/jbc.M110.157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.WHO. United Nations Food and Agriculture Organization. http://www.fao.org/publications/sofi/en.2010. [Jul 20;2011 ]. http://www.fao.org/publications/sofi/en.2010
- 191.Williams KT. Schalinske KL. Homocysteine metabolism and its relation to health and disease. Biofactors. 2010;36:19–24. doi: 10.1002/biof.71. [DOI] [PubMed] [Google Scholar]
- 192.Williams PJ. Kurlak LO. Perkins AC. Budge H. Stephenson T. Keisler D. Symonds ME. Gardner DS. Hypertension and impaired renal function accompany juvenile obesity: The effect of prenatal diet. Kidney Int. 2007;72:279–289. doi: 10.1038/sj.ki.5002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wilson VL. Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 194.Wilson MJ. Shivapurkar N. Poirier LA. Hypomethylation of hepatic nuclear DNA in rats fed with a carcinogenic methyl-deficient diet. Biochem J. 1984;218:987–990. doi: 10.1042/bj2180987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Winzerling JJ. Law JH. Comparative nutrition of iron and copper. Annu Rev Nutr. 1997;17:501–526. doi: 10.1146/annurev.nutr.17.1.501. [DOI] [PubMed] [Google Scholar]
- 196.Wu G. Amino acids: Metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 197.Wu G. Functional amino acids in growth, reproduction and health. Adv Nutr. 2010;1:31–37. doi: 10.3945/an.110.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wu G. Meininger CJ. Nitric oxide and vascular insulin resistance. BioFactors. 2009;35:21–27. doi: 10.1002/biof.3. [DOI] [PubMed] [Google Scholar]
- 199.Wu G. Bazer FW. Cudd TA. Meininger CJ. Spencer TE. Maternal nutrition and fetal development. J Nutr. 2004;134:2169–2172. doi: 10.1093/jn/134.9.2169. [DOI] [PubMed] [Google Scholar]
- 200.Wu G. Bazer FW. Wallace JM. Spencer TE. Board-invited review: Intrauterine growth retardation: iImplications for the animal sciences. J Anim Sci. 2006;84:2316–2337. doi: 10.2527/jas.2006-156. [DOI] [PubMed] [Google Scholar]
- 201.Wu G. Bazer FW. Datta S. Johnson GA. Li P. Satterfield MC. Spencer TE. Proline metabolism in the conceptus: Implications for fetal growth and development. Amino Acids. 2008;35:691–702. doi: 10.1007/s00726-008-0052-7. [DOI] [PubMed] [Google Scholar]
- 202.Wu G. Bazer FW. Davis TA. Kim SW. Li P. Rhoads JM. Satterfield MC. Smith SB. Spencer TE. Yin YL. Arginine metabolism and nutrition in growth, health and disease. Amino Acids. 2009;37:153–168. doi: 10.1007/s00726-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu G. Bazer FW. Burghardt RC. Johnson GA. Kim SW. Li XL. Satterfield MC. Spencer TE. Impacts of amino acid nutrition on pregnancy outcome in pigs: Mechanisms and implications for swine production. J Anim Sci. 2010;88:E195–E204. doi: 10.2527/jas.2009-2446. [DOI] [PubMed] [Google Scholar]
- 204.Wu G. Bazer FW. Burghardt RC. Johnson GA. Kim SW. Knabe DA. Li P. Li XL. McKnight JR. Satterfield MC. Spencer TE. Proline and hydroxyproline metabolism: Implications for animal and human nutrition. Amino Acids. 2011;40:1053–1063. doi: 10.1007/s00726-010-0715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xiao XM. Li LP. L-arginine treatment for asymmetric fetal growth restriction. Int J Gynecol Obstet. 2005;88:15–18. doi: 10.1016/j.ijgo.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 206.Yajnik CS. Deshmukh US. Maternal nutrition, intrauterine programming and consequential risks in the offspring. Rev Endocr Metab Disord. 2008;9:203–211. doi: 10.1007/s11154-008-9087-z. [DOI] [PubMed] [Google Scholar]
- 207.Yamamoto Y. Verma UN. Prajapati S. Kwak YT. Gaynor RB. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 208.Yokomori N. Tawata M. Onaya T. DNA demethylation modulates mouse leptin promoter activity during the differentiation of 3T3-L1 cells. Diabetologia. 2002;45:140–148. doi: 10.1007/s125-002-8255-4. [DOI] [PubMed] [Google Scholar]
- 209.Youngson NA. Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 210.Zambrano E. Martnez-Samayoa PM. Bautista CJ. Deas M. Guillen L. Rodriguez- Gonzales GL. Guzmán C. Larrea F. Nathanielsz PW. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J Physiol. 2005;566:225–236. doi: 10.1113/jphysiol.2005.086462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zeng XF. Wang FL. Fan X. Yang WJ. Zhou B. Li PF. Yin YL. Wu G. Wang JJ. Dietary arginine supplementation during early pregnancy enhances embryonic survival in rats. J Nutr. 2008;138:1421–1425. doi: 10.1093/jn/138.8.1421. [DOI] [PubMed] [Google Scholar]
- 212.Zhu MJ. Du M. Nijland MJ. Nathanielsz PW. Hess BW. Moss GE. Ford SP. Down- regulation of growth signaling pathways linked to a reduced cotyledonary vascularity in placentomes of over-nourished, obese pregnant ewes. Placenta. 2009;30:405–410. doi: 10.1016/j.placenta.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zimmermann MB. The influence of iron status on iodine utilization and thyroid function. Annu Rev Nutr. 2006;26:367–389. doi: 10.1146/annurev.nutr.26.061505.111236. [DOI] [PubMed] [Google Scholar]