Abstract
The mir-51 family of microRNAs (miRNAs) in C. elegans are part of the deeply conserved miR-99/100 family. While loss of all six family members (mir-51-56) in C. elegans results in embryonic lethality, loss of individual mir-51 family members results in a suppression of retarded developmental timing defects associated with the loss of alg-1. The mechanism of this suppression of developmental timing defects is unknown. To address this, we characterized the function of the mir-51 family in the developmental timing pathway. We performed genetic analysis and determined that mir-51 family members regulate the developmental timing pathway in the L2 stage upstream of hbl-1. Loss of the mir-51 family member, mir-52, suppressed retarded developmental timing defects associated with the loss of let-7 family members and lin-46. Enhancement of precocious defects was observed for mutations in lin-14, hbl-1, and mir-48(ve33), but not later acting developmental timing genes. Interestingly, mir-51 family members showed genetic interactions with additional miRNA-regulated pathways, which are regulated by the let-7 and mir-35 family miRNAs, lsy-6, miR-240/786, and miR-1. Loss of mir-52 likely does not suppress miRNA-regulated pathways through an increase in miRNA biogenesis or miRNA activity. We found no increase in the levels of four mature miRNAs, let-7, miR-58, miR-62 or miR-244, in mir-52 or mir-52/53/54/55/56 mutant worms. In addition, we observed no increase in the activity of ectopic lsy-6 in the repression of a downstream target in uterine cells in worms that lack mir-52. We propose that the mir-51 family functions broadly through the regulation of multiple targets, which have not yet been identified, in diverse regulatory pathways in C. elegans.
Introduction
microRNAs (miRNAs) are ∼22 nucleotide, non-coding RNAs that post-transcriptionally regulate the expression of their downstream targets. miRNAs bind to sites with imperfect complementarity in target mRNAs [1], which, in most cases, results in lower target protein levels due to the inhibition of translation and the reduced stability of target mRNAs [2], [3]. The effects of miRNA regulation on target protein levels can vary widely. In some cases, miRNA binding to a target can function as a ‘switch’ by directing the nearly complete suppression of target mRNA translation. For example, the lin-4 and let-7 miRNAs function as developmental switches to strongly down-regulate their respective targets, lin-14 and lin-41 [4]-[7]. In other cases, miRNA binding to a target can function as a ‘fine tuner’ to direct modest repression of target mRNA translation. For example, in flies, miR-8 maintains the levels of atrophin in an optimal range [8]. However, in recent work, Mukherji et al. [9] demonstrate that the effect of miRNA regulation upon target protein levels is not an inherent property of the miRNA but rather depends on the stoichiometry and binding affinity of a miRNA and its associated target mRNAs. At low levels of target mRNA, a miRNA can act to strongly repress translation, whereas at high levels of the target mRNA, a miRNA can act to modestly repress translation [9].
While penetrant mutant phenotypes are observed in lin-4 and let-7 mutants, defects were not identified for most individual miRNA mutants in C. elegans [10], though progress in identifying functions for miRNAs is being made [11]. For some miRNA mutants, like let-7 or mir-35 family mutants, the lack of observed defects is a result of functional redundancy among miRNA family members, which share a six nucleotide 5′ seed sequence [12], [13]. For other miRNA mutants, like lsy-6, the lack of obvious defects reflects highly specialized functions for individual miRNAs that were not observed in broad-based phenotypic analyses [14]. Furthermore, since some miRNAs function to modestly regulate, or fine tune, target gene expression, the loss of these miRNAs may not result in obvious defects during normal growth conditions. Approaches that have examined miRNA mutant worms under conditions of stress, such as altered environmental conditions [15], [16] or genetic backgrounds [17], have been successful in identifying mutant phenotypes associated with the loss of individual miRNAs.
Using a sensitized genetic background, we characterized defects in 80% of the individual miRNA mutants analyzed [17]. For that analysis, we used strains that lack one of two Argonaute proteins that function in the miRNA pathway in C. elegans, ALG-1, as a sensitized background in which to identify mutant phenotypes. In alg-1 mutants, overall miRNA levels are reduced, including the lin-4 and let-7 miRNAs, which leads to observable defects in the developmental timing pathway [18]. This pathway controls the appropriate temporal execution of stage-specific developmental programs through the four larval stages, L1–L4 [19]. Loss of alg-1 activity results in developmental timing defects including incomplete alae formation at the L4 to adult transition, an increased number of hypodermal seam cells, and a failure to exit the molting cycle [17], [18], [20]–[23]. Loss of mir-51 family members partially suppresses these developmental timing defects in alg-1 worms [17].
The mir-51 family is part of the larger miR-99/100 family, a miRNA family that shows deep conservation from cnidarians through humans [24]. In C. elegans, the mir-51 family comprises six miRNAs, miR-51 through miR-56. Loss of the entire mir-51 family in C. elegans results in embryonic lethality, due to a failure of pharyngeal attachment [25]. Loss of multiple members causes several mutant phenotypes including larval lethality and slow growth [13], [25]. These pleiotropic phenotypes indicate that mir-51 family members likely function to regulate multiple downstream targets and pathways. The mechanism whereby loss of individual mir-51 family members suppresses alg-1 developmental timing defects is unclear. Unlike other genes that regulate developmental timing, mir-51 family members are expressed broadly and abundantly throughout the life of the worm [25]–[29]. We therefore wanted to determine the function of the mir-51 family members in the regulation of the developmental timing pathway.
Here, we have defined the genetic interactions of mir-51 family members with components of the developmental timing pathway. Additionally, we report that the mir-51 family interacts with multiple, diverse, miRNA regulated genetic pathways, including pathways regulated by the let-7 and mir-35 family miRNAs, as well as lsy-6, miR-240/786, and miR-1. We provide evidence that is inconsistent with the model that the mir-51 family regulates miRNA biogenesis or miRNA activity. Instead, we propose that the mir-51 family functions to regulate multiple targets in diverse developmental pathways in C. elegans.
Results
Loss of mir-51 family members partially suppresses retarded developmental timing phenotypes
The loss of mir-51 family members suppresses alg-1 developmental timing defects [17], suggesting a possible direct role in the regulation of the developmental timing pathway. However, mutants lacking individual mir-51 family members did not display developmental timing abnormalities such as defects in alae formation or defects in seam cell divisions (Table 1 and Table 2). Further, worms that are multiply mutant for 5 out of 6 members of the mir-51 family, mir-52/53/54/55/56, also do not display alae formation defects (Table 1 and Table 2), despite displaying other mutant phenotypes including slow growth and larval lethality [13], [25]. Because the alg-1 developmental timing defects are similar to those associated with the loss of the let-7 family miRNAs [18], we determined if loss of individual mir-51 family members was sufficient to suppress let-7 timing defects. To do this, we used a temperature sensitive let-7 allele, n2853. At 25°C, these let-7(ts) mutants display a repetition of a late larval program with failure to form complete alae and lethality due to bursting at the vulva at the L4 to adult transition ([5]; Table 1). Loss of mir-51 family members did not suppress either of these phenotypes in let-7 mutants (Table 1).
Table 1. Genetic interactions of the mir-51 family with retarded developmental timing mutants.
| Alae at L4 to Adult transition | Lethality | |||||||
| Straina | seam cellsb | complete | gapped | none | n | % burst | % bag of worms | n |
| RG733 wild type | 16.0 | 100 | 0 | 0 | 20 | 0 | 0 | 208 |
| RF481 wild type | 16.1 | 100 | 0 | 0 | 20 | 0 | 0 | 109 |
| RF491 mir-51 | 16.2 | 100 | 0 | 0 | 20 | 0 | 0 | 151 |
| RF499 mir-52 | 15.9 | 100 | 0 | 0 | 20 | 0 | 0 | 181 |
| RF483 mir-53 | 16.1 | 100 | 0 | 0 | 20 | 0 | 0 | 176 |
| RF399 mir-54/55/56 | 16.1 | 99 | 1 | 0 | 98 | 0 | 0 | 228 |
| RF692 mir-52/53/54/55/56 | –c | 100 | 0 | 0 | 16 | – | – | – |
| MT7626 let-7ts @25°C | – | 0 | 50d | 50 | 16 | 100 | – | 103 |
| RF447 mir-51; let-7ts @25° | – | 0 | 80d | 20 | 20 | 100 | - | 119 |
| RF448 mir-52; let-7ts @25° | – | 7 | 73d | 20 | 15 | 96 | – | 114 |
| RF449 mir-53; let-7ts @25° | – | 0 | 53d | 47 | 17 | 99 | 1 | 92 |
| RF442 mir-54/55/56; let-7ts @25° | – | 7 | 21d | 71 | 14 | 99 | 1 | 91 |
| RF554 mir-48/84/241 | 22.6 | 0 | 100 | 0 | 40 | 56 | 37 | 111 |
| RF556 mir-52; mir-48/84/241 | 17.7e | 49f | 51 | 0 | 39 | 3f | 77 | 90 |
| RF553 mir-48/84/241 | 22.7 | 0 | 100 | 0 | 37 | 66 | 26 | 125 |
| RF555 mir-51; mir-48/84/241 | 21.8 | 0 | 100 | 0 | 37 | 42g | 41 | 112 |
| RF557 mir-53; mir-48/84/241 | 22.2 | 0 | 100 | 0 | 38 | 49g | 39 | 134 |
| RF558 mir-54/55/56; mir-48/84/241 | 20.6h | 21g | 79 | 0 | 38 | 25g | 57 | 141 |
| VT1064 mir-48/84 | – | – | – | – | – | 0 | 69 | 236 |
| RF451 mir-51; mir-48/84 | – | – | – | – | – | 0 | 30i | 101 |
| RF469 mir-52; mir-48/84 | – | – | – | – | – | 0 | 5i | 148 |
| RF454 mir-53; mir-48/84 | – | – | – | – | – | 0 | 62 | 106 |
| RF415 mir-54/55/56; mir- 48/84 | – | – | – | – | – | 0 | 2i | 93 |
| RF619 mir-48/241 | 19.1 | 5 | 95 | 0 | 21 | 31 | 49 | 144 |
| RF730 mir-48/241; mjEx160[mir-54/55/56] | 22.1j | 9 | 91 | 0 | 32 | 66k | 24k | 136k |
| RF568 lin-46 @15° | 19.4 | 5 | 95 | 0 | 40 | – | – | – |
| RF569 mir-52; lin-46 @15° | 17.8l | 23m | 77 | 0 | 39 | – | – | – |
| VC894 puf-9 | – | 29 | 71 | 0 | 34 | – | – | – |
| RF578 mir-52; puf-9 | – | 34 | 66 | 0 | 50 | – | – | – |
| RF620 mir-52; mir-48/241 | 16.6n | 85 | 15 | 0 | 20 | – | – | – |
| RF625 mir-48/241; puf-9 | 19.2 | 0 | 100 | 0 | 19 | – | – | – |
| RF626 mir-52; mir-48/241; puf-9 | 16.2o | 0 | 100 | 0 | 17 | – | – | – |
Full genotype information can be found in Table S1.
seam cells counted in L4-stage worms using wIs78 or wIs79[scm::gfp], n≥18 (range 19–30).
indicates results not determined.
alae scored categorized as partially or faintly visible rather than gapped as elsewhere.
indicates significant difference compared to RF554 mir-48/84/241 (student's t-test, p<0.05), which contained wIs79.
indicates significant difference compared to RF554 mir-48/84/241 (χ2, p<0.05) which contained wIs79.
indicates significant difference compared to RF553 mir-48/84/241 (χ2, p<0.05) which contained wIs78.
indicates significant difference compared to RF553 mir-48/84/241 (student's t-test, p<0.05), which contained wIs78.
indicates significant difference compared to VT1064 mir-48/84 (χ2, p<0.05).
indicates significant difference comparing worms from the same strain, but +/− for mjEx160[mir-54/55/56] (student's t-test, p<0.05).
population scored for lethality is a mix of worms +/− for mjEx160[mir-54/55/56].
indicates significant difference compared to RF568 lin-46 (student's t-test, p<0.05).
indicates significant difference compared to RF568 lin-46 (χ2, p<0.05).
indicates significant difference compared to RF619 mir-48/241 (student's t-test, p<0.05).
indicates significant difference compared to RF625 mir-48/241; puf-9 (student's t-test, p<0.05).
Table 2. Genetic interactions of mir-51 family with precocious developmental timing mutants.
| Precocious Alaeb | ||||
| Straina | complete | gapped | none | n |
| RG733 wild type | 0 | 0 | 100 | 9 |
| RF481 wild type | 0 | 0 | 100 | 12 |
| RF491 mir-51 | 0 | 0 | 100 | 14 |
| RF499 mir-52 | 0 | 0 | 100 | 13 |
| RF483 mir-53 | 0 | 0 | 100 | 15 |
| RF399 mir-54/55/56 | 0 | 0 | 100 | 13 |
| RF692 mir- 52/53/54/55/56 | 0 | 0 | 100 | 15 |
| RG490 mir-48(ve33) | 0 | 55 | 45 | 47 |
| RF583 mir-52; mir-48(ve33) | 0 | 88d | 12 | 34 |
| RF534 hbl-1 | 0 | 76 | 24 | 41 |
| RF535 mir-52; hbl-1 | 2d | 95 | 2 | 44 |
| RF563 lin-14ts @25°C | 34 | 66 | 0 | 29 |
| RF588 mir-52; lin-14ts @25°C | 76d | 20 | 4 | 25 |
| RF536 lin-41 | 0 | 11 | 89 | 38 |
| RF537 mir-52; lin-41 | 0 | 14 | 86 | 36 |
| RF538 lin-42 | 0 | 89 | 11 | 37 |
| RF541 mir-52; lin-42 | 3 | 93 | 3 | 29 |
| VT517 lin-28 c | 5 | 90 | 5 | 20 |
| RF573 mir-52; lin-28 c | 0 | 100 | 0 | 20 |
full genotype information can be found in Table S1.
alae were scored in L3 molt or early L4-stage worms, except where otherwise noted.
alae were scored in the L2 molt.
indicates significant difference between strains of same genotype +/− mir-52 (χ2, p<0.05).
The let-7 family members, mir-48, mir-84, and mir-241, function together to control the timing of the L3 stage program through down-regulation of their target, hbl-1 [12]. In the L2 stage, a subset of hypodermal seam cells undergo two rounds of cell division resulting in an increase in the number of seam cells from 10 to 16. In mutants lacking mir-48, mir-84 and mir-241 (hereafter referred to as mir-48/84/241), the L3 stage program is not executed properly and the L2 stage program is reiterated. This reiteration of the L2 stage program results in an increased number of seam cells [12]. mir-48/84/241 mutant worms often display defects in alae formation at the L4 to adult transition. In addition, many of these mutants burst at the L4 to adult transition or execute an extra adult-stage molt, which leads to the “bag-of-worms” phenotype [12]. mir-52; mir-48/84/241 had fewer seam cells than mir-48/84/241 worms, indicating a suppression of the L2 reiteration phenotype (Table 1). Additionally, loss of mir-52 suppressed the alae formation defects and bursting phenotypes of mir-48/84/241: 100% of mir-48/84/241 displayed incomplete alae formation and 56% of mir-48/84/241 worms burst at the L4 to adult transition compared to 51% and 3% of mir-52;mir-48/84/241 mutants, respectively (Table 1). However, 77% of mir-52; mir-48/84/241 worms showed the bag of worms phenotype, indicating an extra adult-stage molt. This likely reflects a partial suppression of mir-48/84/241 phenotypes, rather than an inability to suppress molting since loss of mir-52 strongly suppresses the ectopic molting phenotype of alg-1 worms [17] as well as mir-48/84 double mutant worms (Table 1).
Next, we examined the effect of elevated expression of mir-51 family members on the retarded development of mir-48 mir-241 (mir-48/241) mutant worms. To accomplish this, we used mjEx160, an extrachromosomal array with the genomic fragment for mir-54/55/56 that was previously shown to rescue the embryonic lethality of mir-51 family mutant worms [25] and the developmental timing phenotypes in mir-54/55/56 alg-1 mutant worms [17]. mjEx160 enhanced developmental timing defects of mir-48/241 mutant worms (Table 1). mir-48/241 worms have 19.1 seam cells on average. This is increased to 22.1 in mir-48/241; mjEx160 worms (Table 1). This indicates elevated expression of mir-51 family members enhances the L2 repetition phenotype.
We determined if the loss of mir-51 family members can suppress the phenotypes of lin-46 and puf-9, mutants that display retarded developmental timing defects [30], [31]. lin-46 functions in parallel to the let-7 family to control the timing of the L3 stage program [12], [30]. lin-46 mutants fail to properly execute the L3 stage program and show reiteration of the L2 program at 15°C. lin-46 mutants display extra seam cells and incomplete alae formation [30]. Loss of mir-52 partially suppressed lin-46 developmental timing defects: mir-52; lin-46 double mutant worms had fewer seam cells and displayed weaker alae defects compared to lin-46 mutant worms (Table 1). Loss of the other mir-51 family members had no significant effect on lin-46 developmental timing defects (data not shown). puf-9 encodes a pumilio family protein that acts to negatively regulate hbl-1 through its 3′UTR [31]. puf-9 mutant worms fail to form complete alae at the L4 to adult transition. Loss of mir-52 did not suppress the puf-9 alae defects (Table 1). This suggests that puf-9 may function downstream of the mir-51 family to regulate developmental timing.
To determine if puf-9 is necessary for mir-52-mediated suppression of the let-7 family developmental timing defects, we examined worms multiply mutant for mir-52, puf-9, and let-7 family miRNAs, mir-48 and mir-241 (mir-48/241). Loss of mir-52 suppressed the seam cell and alae formation defects in mir-48/241 mutants (Table 1). Loss of puf-9 did not affect the mir-52 mediated suppression of the extra seam cell phenotype of mir-48/241 mutant worms (Table 1). However, no suppression of alae formation defects was observed in mir-52; mir-48/241; puf-9 worms relative to mir-52; mir-48/241 (Table 1). This is consistent with a function for puf-9 later in development, after the L2 to L3 transition. Together, these data indicate that the mir-51 family functions to regulate the execution of the L3 stage program, acting either downstream or in parallel to the let-7 family miRNAs and lin-46 and may have additional activity in the control of alae formation in late larval development.
Loss of mir-52 enhances precocious developmental timing phenotypes
We next characterized genetic interactions between mir-52 and a set of precocious developmental timing genes. Loss of mir-52 enhanced the developmental timing defects observed in three precocious mutants: mir-48(ve33), hbl-1(ve18), and lin-14(n179). (Table 2). First, mir-48(ve33) worms display early accumulation of miR-48 and precocious formation of adult-specific alae one stage early at the L3 to L4 transition [32]. Loss of mir-52 significantly enhanced this precocious alae formation in the mir-48(ve33) background (Table 2). We found that 55% of mir-48(ve33) mutants displayed precocious alae formation compared to 88% of mir-52;mir-48(ve33) worms. Next, hbl-1 is a central regulator of the L2 versus L3 cell fate decision [33], [34]. Loss of mir-52 enhanced the precocious alae phenotype of hbl-1(ve18) mutants: 76% of hbl-1(ve18) worms displayed either complete or gapped precocious alae in the L4 stage compared to 97% of mir-52; hbl-1 double mutant worms (Table 2). Enhancement of the precocious phenotype of hbl-1(ve18) worms may reflect reduced activity of hbl-1 itself, since ve18 is a reduced function, not a null, allele [33]. Finally, lin-14 functions to regulate the timing of both L1 versus L2 and L2 versus L3 cell fate decisions [35]. To analyze genetic interactions with lin-14, we used the temperature sensitive allele, n179. At 25°C, 34% of lin-14(ts) worms form complete precocious alae at the L3 to L4 transition compared to 76% of mir-52; lin-14(n179) worms (Table 2). Enhancement was not observed for the lin-41, lin-42, or lin-28 phenotypes (Table 2). The enhancement of the precocious developmental timing defects observed in mir-48(ve33), hbl-1(ve18), and lin-14(n179ts) mutant worms is consistent with a role for the mir-51 family in the regulation of L2 versus L3 cell fate decisions.
mir-51 family members function upstream of hbl-1, but not lin-28, to suppress developmental timing defects in let-7 family mutants
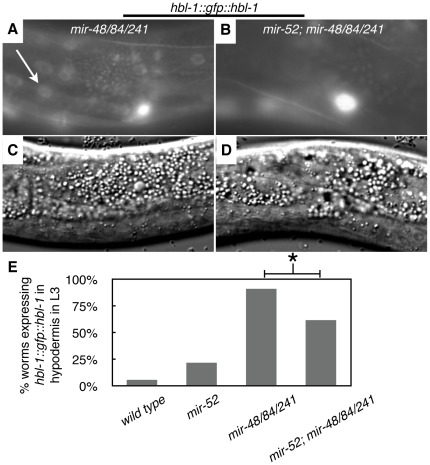
Genetic interactions between mir-52 and let-7 family members as well as hbl-1(ve18) suggest that mir-52 may act upstream of hbl-1 to promote its activity. hbl-1 is robustly expressed in the hypodermis during embryonic and early larval development and then is subsequently down-regulated through its 3′ UTR by the early L3 stage [33], [34], [36]. The down-regulation of hbl-1 in the hypodermis requires the let-7 family members, mir-48, mir-84, and mir-241 [12]. We therefore determined whether the observed suppression of developmental timing defects in mir-52; mir-48/84/241 reflects a suppression of hbl-1 misregulation. Indeed, loss of mir-52 partially suppressed the hbl-1 misexpression phenotype of mir-48/84/241 mutant worms: in 91% of mir-48/84/241 worms hbl-1::gfp::hbl-1 transgene expression remained high in L3, whereas only 62% of mir-52; mir-48/84/241 displayed high hbl-1::gfp::hbl-1 expression (Figure 1). This indicates that mir-52 acts upstream of hbl-1 expression in opposition to let-7 family activity.
Figure 1. Loss of mir-52 .
suppresses hbl-1 misregulation in mir-48/84/241 mutants. Representative fluorescent micrographs of hbl-1::gfp::hbl-1 transgene expression in (A) mir-48/84/241 and (B) mir-52; mir-48/84/241 mutant worms in the L3 stage with corresponding DIC images (C and D, respectively). White arrow in A indicates a hyp7 nucleus. (E) Percentage of worms with hbl-1::gfp::hbl-1 expression in hypodermis of L3 stage worms, n≥33 (range 33–37). * indicates significant difference (χ2, p<0.01).
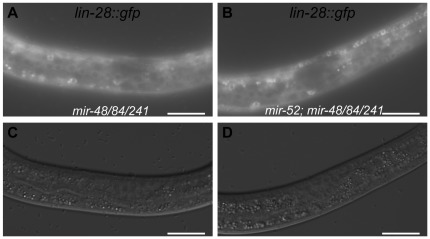
Like hbl-1, lin-28 is also a critical regulator of L2 versus L3 cell fate decisions. We used a lin-28::gfp::lin-28 transgene to determine whether mir-52 suppression is the result of a misregulation of lin-28. However, no difference was observed in lin-28::gfp::lin-28 expression between mir-48/84/241 and mir-52; mir-48/84/241 in L2 molt stage worms (Figure 2). Thus, misregulation of lin-28 does not account for the observed suppression of developmental timing defects in mir-52; mir-48/84/241 worms. Together, these data are consistent with the mir-51 family functioning downstream or in parallel to lin-28, lin-46 and the let-7 family, but upstream of hbl-1 to regulate the L2 versus L3 cell fate decisions.
Figure 2. Loss of mir-52 .
does not result in increased expression of lin-28::gfp::lin-28 . (A, B) Representative fluorescent micrographs of lin-28::gfp::lin-28 transgene expression at in (A) mir-48/84/241 and (B) mir-52; mir-48/84/241 worms in the L2 molt stage with corresponding DIC images, (C and D, respectively). Strains were scored for expression of lin-28::gfp::lin-28 at the L2 molt (n = 17). No significant difference was observed between strains (χ2, p>0.05).
Loss of mir-51 family members suppresses additional miRNA-dependent regulatory pathways in C. elegans
Genetic interactions with the developmental timing pathway may reflect a specific function for the mir-51 family miRNAs in the regulation of targets in this pathway. Alternatively, these interactions may reflect a broader function for the mir-51 family in the regulation of miRNA biogenesis or activity. For example, the developmental timing defects observed in alg-1 or ain-1 mutants [18], [20] are due to lower overall miRNA activity, including the lin-4 and let-7 family miRNAs, rather than a specific function in the developmental timing pathway. Therefore, we tested whether the mir-51 family interacted with additional miRNA-regulated pathways by determining if loss of mir-51 family members could suppress other miRNA mutant phenotypes that are distinct from developmental timing, including lsy-6 regulation of neuronal asymmetry, let-7 family regulation of vulva cell fate specification, mir-240/786 regulation of defecation, mir-35 family regulation of embryonic development and mir-1 regulation of neuromuscular function.
lsy-6
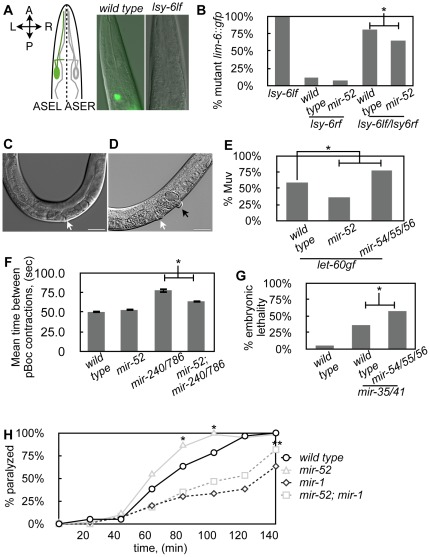
The lsy-6 miRNA specifies the ASEL cell fate through the down-regulation of its target, cog-1 [14]. lsy-6 repression of cog-1 is necessary for lim-6::gfp expression in the ASEL [14]. To achieve a genetic background with optimally compromised lsy-6 activity, we used heterozygous worms that carry a loss of function allele, ot149, and a reduced function allele, ot150. 85% of these lsy-6(ot149lf)/lsy-6(ot150rf) heterozygous worms fail to express lim-6::gfp in the ASEL neuron compared to 100% of lsy-6(ot149lf) and 14% of lsy-6(ot150rf) worms (Figure 3B; [14]). Loss of mir-52 partially suppressed mutant lim-6::gfp expression in lsy-6(ot149lf)/lsy-6(ot150rf): 85% of lsy-6rf/lsy-6lf worms displayed mutant lim-6::gfp expression compared to 61% of mir-52; lsy-6rf/lsy-6lf (Figure 3B).
Figure 3. The mir-51 family members, mir-52 and mir-54/55/56 .
, function in multiple miRNA-dependent developmental pathways. (A, B) mir-52 suppresses ASEL specification defects of lsy-6(rf)/lsy-6(lf) worms. (A) Cartoon of lim-6::gfp expression in wild-type and lsy-6(lf) worms. A, anterior; P, posterior; L, left; R, right. (B) Worms of indicated genotypes were scored for lim-6::gfp expression in late larval and young adult stages, n≥169. * indicates significant difference (χ2, p<0.01). (C–E) Loss of mir-52 partially suppresses, while loss of mir-54/55/56 enhances, the multivulva (Muv) phenotype of let-60gf worms. (C) A wild type worm with one normal vulva, white arrow. (D) A let-60gf worm with one normal vulva, white arrow, and one ectopic vulva, black arrow. Bars represent 100 µm. (E) Synchronized L1 worms of the indicated genotype were allowed to develop at 25°C for 2–3 days and then scored as young adults for the Muv phenotype. n≥100. * indicates significant difference (χ2, p<0.01). (F) Loss of mir-52 reduces the average defecation cycle time of mir-240/786 mutant worms. Average time between consecutive pBoc contractions for n≥5 worms. * indicates significant difference (student's t-test, p<0.01). Error bars indicate SEM values. (G) Loss of mir-54/55/56 enhances the embryonic lethality of mir-35 through 41 mutant worms. L4 worms of the indicated genotypes were shifted to 25° and the next day embryos from these worms were collected. After 24 hours, unhatched embryos were counted to determine the percentage of embryonic lethality (n≥148). * indicates significant difference (χ2, p<0.01). (H) Loss of mir-52 modestly suppresses the resistance to levamisole of mir-1 worms. mir-52 mutants show weakly enhanced sensitivity to levamisole. * indicates significant difference compared to wild type at the indicated time point (χ2, p<0.05). ** indicates significant difference compared to mir-1 at the indicated time point (χ2, p<0.05).
let-7 family regulation of vulva development
The let-7 family miRNAs repress let-60/RAS in the regulation of vulva development [37]. Worms with a gain-of-function mutation in let-60 display defects in cell fate specification, which often results in a ‘Muv’ phenotype with multiple vulva structures produced [38]. Overexpression of let-7 family members partially suppresses the let-60gf Muv phenotype [37]. If the mir-51 family opposes let-7 activity in vulva development, as it did in the developmental timing pathway, then it would be expected that loss of mir-51 family members should suppress the let-60gf Muv phenotype. This is observed in mir-52;let-60gf worms (Figure 3E). Interestingly, loss of mir-54/55/56 enhanced the Muv phenotype of let-60gf (Figure 3E). This may reflect distinct activities of individual mir-51 family members in the control of vulva development. Identification of mir-51 family targets in the vulva specification pathway is required to elucidate the functions of individual mir-51 family members.
mir-240/786
mir-240/786 is necessary for the normal rhythmicity of the defecation motor program [10]. In wild type worms, a defecation motor program occurs every ∼50 seconds [39]. In mir-240/786 mutant worms, the average defecation cycle time is increased [10]. We found that loss of mir-52 significantly reduced the average defecation cycle time of mir-240/786 worms (Figure 3F). Loss of mir-54/55/56 had no effect on the mean defecation cycle time of mir-240/786 worms (data not shown).
mir-35 family
The mir-35 family comprises eight miRNAs, mir-35 through mir-42. These family members are redundantly required for embryonic development and mutants lacking mir-35 through mir-41 exhibit temperature sensitive embryonic lethality [13]. We found that loss of mir-54/55/56 did not suppress the embryonic lethal phenotype of mir-35/41 mutants, but rather significantly enhances this phenotype (Figure 3G).
mir-1
mir-1 is necessary for normal neuromuscular function [40]. mir-1 mutants display a resistance to levamisole-induced paralysis due to an increase in levels of its targets, UNC-29 and UNC-63 [40]. We found that loss of mir-52 weakly suppressed the levamisole resistance phenotype of mir-1 worms (Figure 3H). We found that after 140 minutes on 200 µM levamisole mir-52; mir-1 worms were less resistant to levamisole compared to mir-1 (Figure 3H). We also found that mir-52 worms appeared to be slightly more sensitive to levamisole compared to wild type worms. Loss of mir-54/55/56 had no effect on levamisole sensitivity of mir-1 or wild-type worms (data not shown).
Loss of mir-51 family members does not broadly enhance miRNA biogenesis or activity
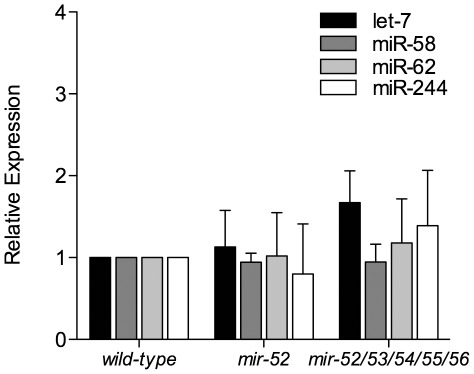
To account for the observation that the loss of mir-52 suppressed multiple miRNA-dependent phenotypes, we proposed that mir-52 may act to broadly regulate miRNA biogenesis or activity. To examine if the mir-51 family regulates the miRNA pathway, we measured mature miRNA levels for a set of miRNAs that display various expression and biogenesis characteristics. We analyzed levels of the let-7 miRNA, a developmentally-regulated miRNA that functions in the developmental timing pathway in the hypodermis [5], miR-58, a highly abundant miRNA [26], miR-62, a miRtron that displays Drosha independent biogenesis [41], and miR-244, a miRNA that is expressed at lower levels primarily in hypodermal seam cells [29]. We found that the levels of these miRNAs are unchanged in mir-52 mutants as well as in mir-52/53/54/55/56 mutants (Figure 4). mir-52/53/54/55/56 mutant worms display impenetrant embryonic lethality, slow growth, and mating defects [13], [25] indicating that mir-51 family targets are sufficiently misregulated to result in severe, penetrant mutant phenotypes. However, no change in miRNA levels were detected for the four miRNAs analyzed. These results indicate that the observed suppression of developmental timing defects is not likely due to an increase in overall miRNA levels and that mir-51 family miRNAs likely do not function broadly to regulate miRNA biogenesis.
Figure 4. Levels of mature let-7, miR-58, miR-62, and miR-244 are unchanged in the absence of mir-51 family members.
Levels of mature miRNAs in wild type, mir-52, and mir-52/53/54/55/56 mutant worms were measured and normalized to the average of two control RNAs, U18 and sn2343. The graph represents the level of mature miRNAs relative to wild type. Error bars represent the standard deviation (SD) between biological replicates. No differences in mature miRNA expression was observed (student′s t-test, p>0.24).
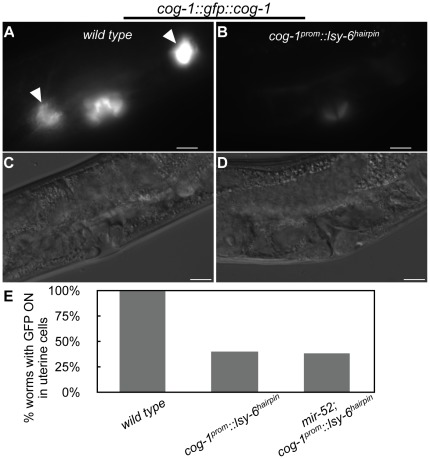
In order to determine if loss of mir-52 can act to enhance miRNA activity, we analyzed the activity of ectopically expressed lsy-6 in the repression of a cog-1::gfp reporter [14]. Ectopic expression of lsy-6 under the control of the cog-1 promoter allowed us to examine the activity of the lsy-6 miRNA in cells where it is normally not found, including uterine and vulva cells [14]. We found that in 60% of worms examined, ectopic expression of the lsy-6 miRNA resulted in the down-regulation of cog-1::gfp in uterine cells (Figure 5). We found that loss of mir-52 had no effect on the activity of ectopic lsy-6 repression of cog-1 (Figure 5). These data indicate that lsy-6 activity is not enhanced in the absence of mir-52, thereby suggesting that the mir-51 family does not function broadly to regulate the activity of miRNAs.
Figure 5. Loss of mir-52 does not enhance the ability of ectopically-expressed lsy-6 to regulate its target, cog-1.
(A–E) Effect of mir-52 on lsy-6 mediated regulation of cog-1::gfp::cog-1 expression. Representative fluorescent image of cog-1::gfp::cog-1 transgene expression in (A) wild type worms and (B) worms with cog-1::lsy-6 transgene with corresponding DIC images (C and D, respectively). White triangles point to uterine cells. Bars represent 10 µm. (E) Percentage of worms of given genotype with cog-1::gfp expression in either uterine cell, n≥20 (range 20–68). Worms were scored in mid-to-late L4 stage.
Discussion
The goal of this study was to define the mechanism whereby loss of mir-51 family members can suppress the developmental timing defects of alg-1 mutant worms. Our genetic evidence indicates that mir-51 family members act early in the developmental timing pathway to regulate L2 versus L3 cell fate decisions. We observed that loss of the mir-51 family member, mir-52, strongly suppressed the L2 stage reiteration phenotype of mir-48/84/241 mutants and lin-46 mutants. No significant suppression was observed with later acting genes in the developmental timing pathway, such as let-7 and puf-9. Similarly, we observed genetic enhancement of precocious phenotypes due to mutations that result in omissions of early larval stage programs, like lin-14 and hbl-1, but not mutations that result in omission of later larval stage programs, like lin-41. This suggests that the developmental timing pathway is the most sensitive to the loss of mir-52 in the L2 stage.
In many species, including humans and flies, mir-100, let-7, and lin-4 family members are located in a genomic cluster [42]-[44]. In flies, these three miRNAs are polycistronic and function together to regulate adult behaviors [42]. Although this clustered organization in the genome is not observed in worms, evidence herein supports a functional relationship between the let-7 and mir-51 family of miRNAs in the regulation of the developmental timing pathway.
Although mir-51 family members interact with developmental timing genes, such as let-7 family members and lin-46, mir-51 family members are atypical developmental timing genes. First, unlike other developmental timing miRNAs, such as lin-4 and let-7, mir-51 family members do not display stage-specific expression but rather display nearly ubiquitous expression throughout development. In addition, loss of nearly all mir-51 family members, which results in multiple defects including slow growth and embryonic lethality, did not result in developmental timing defects [13,25; Table 1 and Table 2]. We therefore propose that the mir-51 family miRNAs are not themselves regulators of developmental timing decisions but that they likely act downstream in the execution phase of developmental programs.
Surprisingly, we found that mir-51 family members displayed genetic interactions with multiple miRNA genes. These miRNAs function in diverse developmental and physiological processes, which include developmental timing, vulva fate specification, neuronal fate specification, defecation, and neuromuscular function. Loss of the mir-51 family member, mir-52, partially suppressed the vulva cell fate defects of let-60gf mutants, the ASEL cell fate defects of lsy-6 mutants, the defecation defects of mir-240/786 mutants, and the levamisole resistance of mir-1 mutants. These activities for the mir-51 family may reflect the regulation of a single target that functions broadly in many pathways or the regulation of multiple targets that each function in distinct pathways. Our analysis of candidate targets failed to conclusively identify downstream mir-51 family targets (data not shown).
One hypothesis to account for the observed suppression of multiple miRNA-regulated pathways is that the loss of an abundant miRNA such as miR-52 frees up miRNA-induced silencing complex (miRISC) so that it is available for binding by other miRNAs in a cell. Consistent with this model, the miR-51 family is both abundantly [26]–[28] and broadly expressed in tissues in which we observed a genetic interaction, including the hypodermis, the ASEL neuron, the vulva, the intestine, and muscle [25], [29], [45], [46]. In this model, excess miRNAs would compete for a limited pool of miRISC in wild-type worms in order to effectively repress their targets. In genetic backgrounds in which the activity of miRISC factors are reduced, such as in alg-1 mutants, miRISC becomes limiting as evidenced by an increased amount of stem-loop miRNA precursors in both worms [18] and in human cells [47]. In human cells, overexpression of Argonaute-encoding genes results in an elevation of ectopically-expressed mature miRNAs [47]. However, it was not determined if endogenous miRNAs were elevated following Argonaute overexpression. In wild-type worms, precursor miRNAs are often detected in relatively low abundance [18], [48], [49]. These low levels of miRNA precursors that are detected may indicate either the competition for limited miRISC or the normal, steady state level of miRNA precursor in the biogenesis pathway. Although our genetic data are consistent with this limiting miRISC model, our molecular and transgene expression data are inconsistent with this model. First, it is expected that overall levels of all mature miRNAs would be elevated in mutants that lack the abundant miR-52 due to increased loading into miRISC. However, no such elevation in the mature miRNA levels was observed for the four miRNAs that were analyzed in the absence of mir-52 or mir-52/53/54/55/56 activities. In addition, loss of mir-52 did not result in an enhancement of ectopic lsy-6 activity in uterine cells as would be predicted by the limiting miRISC model. Future research will be directed at testing this model in order to determine if critical miRISC factors, such as Argonaute proteins, are limiting. Interestingly, in human cells and early Xenopus embryos, Argonaute protein levels are low and can be readily saturated by exogenous siRNAs [50], [51].
We found that the interactions with multiple miRNA-dependent pathways likely does not reflect the regulation of the miRNA pathway by the mir-51 family. As described above, neither increased miRNA biogenesis nor increased miRNA activity was observed in mir-51 family mutant backgrounds. Thus, mir-51 family members likely do not function to regulate the core pathway required for all miRNA biogenesis or activity. However, it remains possible that mir-51 family members may function to modulate miRNA activity in specific cells or for only a subset of miRNAs not included in our analysis.
We propose that the broad activity of mir-51 family members reflects the repression of a target or multiple target mRNAs that act in distinct genetic pathways, possibly acting to fine tune, or buffer target protein levels. This broad activity described for the mir-51 family is unlike that of previously described miRNAs in C. elegans. A direct target or targets of the mir-51 family in the diverse development pathways described herein remain unknown. Of the 293 conserved targets predicted by Targetscan [52], [53], only 6 contain more than one binding site for the mir-51 family and none of these 6 have more than two binding sites for the mir-51 family. Because the mir-51 family is broadly and abundantly expressed throughout development in C. elegans [25]–[28], multiple binding sites for the mir-51 family within the 3′UTR of a gene would be expected to cause robust repression of that gene throughout development. Multiple sites or sites with high binding affinity may therefore be selected against during evolution. We speculate that the function of mir-51 family members may be to weakly repress or fine-tune the protein levels for a large set of diverse downstream targets.
Materials and Methods
Nematode Methods
C. elegans were maintained using standard conditions. Strains used in this study are listed in Table S1. Worms were kept at 20°C unless otherwise indicated. All strains were constructed using standard genetic techniques [54]. PCR was used to confirm the genotype of strains that contained miRNA deletion alleles [17]. Fluorescence and differential interference contrast (DIC) microscopy were performed with a Nikon Eclipse 80i compound microscope equipped with a Photometrics CoolSNAP HQ2 monochrome digital camera and RS Image software (Roper Scientific) or with NIS Elements software (Nikon).
Defecation assay
Worms were transferred to NGM plates at room temperature and allowed to equilibrate on plates for at least 5 minutes prior to scoring the time between consecutive pBoc contractions. 10 consecutive contractions was scored per worm.
Levamisole sensitivity assay
Worms were transferred to NGM plates supplemented with 200 µM levamisole at room temperature, as done elsewhere [40]. Paralysis was assessed every 20 minutes over the course of 140 minutes by prodding worms with a wire. 20 worms were scored per strain in three independent assays.
GFP reporter transgenes
The syIs63 transgene was used to monitor cog-1 repression in the presence of ectopic lsy-6 expression driven from the otEx1450 transgene array expressing cog-1prom::lsy-6hairpin as described in Johnston and Hobert (2003) [14]. This array was chromosomally integrated to generate otIs193 (kindly provided by L. Cochella and O. Hobert).
RNA Preparation
A mix of 1000 late L4 and L4 molt worms were collected for wild type, mir-52, and mir-52/53/54/55/56 mutant worms. Total RNA was prepared using Trizol (Invitrogen) followed by isopropanol precipitation. RNA samples were DNase treated (DNA-free Kit, Ambion).
qRT- PCR
10 ng of total RNA was used to analyze the levels of mature miRNAs with Applied Biosystems Taqman miRNA assays following the manufacturers protocol. Data was analyzed using 2−ΔΔCt analysis [55] with the mean of U18 and sn2343 as reference. Two technical replicates were performed using two independently isolated total RNA samples for each strain. Each qPCR reaction was performed in triplicate. Student's t-tests were used to statistically compare the fold change of miRNA expression relative to wild type. No significant difference was identified between strains, p>0.24. RNA isolated from N2 worms was used to determine the % efficiency for each PCR assay, which was found to be >95% in each case.
Supporting Information
Strains used in this study.
(PDF)
Acknowledgments
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank Luisa Cochella and Oliver Hobert for providing a strain carrying the otIs193 chromosomally-integrated transgene array.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a National Institutes of Health grant, R15 GM084451 awarded to ALA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, et al. Regulation by let-7 and lin-4 miRNAs Results in Target mRNA Degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Mukherji S, Ebert MS, Zheng GXY, Tsang JS, Sharp PA, et al. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbott AL. Uncovering New Functions for MicroRNAs in Caenorhabditis elegans. Curr Biol. 2011;21:R668–R671. doi: 10.1016/j.cub.2011.07.027. doi: 10.1016/j.cub.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, et al. The let-7 MicroRNA Family Members mir-48, mir-84, and mir-241 Function Together to Regulate Developmental Timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston RJ, Hobert O. Nature 426: 845–849. doi:doi; 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 15.de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20:2159–2168. doi: 10.1016/j.cub.2010.11.015. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato M, Paranjape T, Müller RU, Ullrich R, Nallur S, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 19.Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- 20.Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren H, Zhang H. Developmental Biology; 2010. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. doi: 10.1016/j.ydbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw WR, Armisen J, Lehrbach NJ, Miska EA. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics. 2010;185:897–905. doi: 10.1534/genetics.110.117515. doi: 10.1534/genetics.110.117515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato M, de Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 29.Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Research. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepper AS-R, McCane JE, Kemper K, Yeung DA, Lee RC, et al. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development. 2004;131:2049–2059. doi: 10.1242/dev.01098. doi: 10.1242/dev.01098. [DOI] [PubMed] [Google Scholar]
- 31.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Developmental Biology. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 34.Lin S-Y, Johnson SM, Abraham M, Vella MC, Pasquinelli A, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 35.Ambros V, Horvitz HR. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1987;1:398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- 36.Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Developmental Biology. 1999;205:240–253. doi: 10.1006/dbio.1998.9096. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- 37.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Eisenmann DM, Kim SK. Mechanism of activation of the Caenorhabditis elegans ras homologue let-60 by a novel, temperature-sensitive, gain-of-function mutation. Genetics. 1997;146:553–565. doi: 10.1093/genetics/146.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas JH. Genetic analysis of defecation in Caenorhabditis elegans. Genetics. 1990;124:855–872. doi: 10.1093/genetics/124.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, et al. The MicroRNA miR-1 Regulates a MEF-2-Dependent Retrograde Signal at Neuromuscular Junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Developmental Biology. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 44.Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- 45.Didiano D, Cochella L, Tursun B, Hobert O. Neuron-type specific regulation of a 3′UTR through redundant and combinatorially acting cis-regulatory elements. RNA. 2010;16:349–363. doi: 10.1261/rna.1931510. doi: 10.1261/rna.1931510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Emmons SW. Regulation of the Caenorhabditis elegans posterior Hox gene egl-5 by microRNA and the polycomb-like gene sop-2. Dev Dyn. 2009;238:595–603. doi: 10.1002/dvdy.21876. doi: 10.1002/dvdy.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 48.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 49.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 50.Lund E, Sheets MD, Imboden SB, Dahlberg JE. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 2011;25:1121–1131. doi: 10.1101/gad.2038811. doi: 10.1101/gad.2038811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 54.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains used in this study.
(PDF)