Abstract
The ubiquitin/proteasome pathway plays a crucial role in many biological processes. Here we report a novel role for the Arabidopsis 19S proteasome subunit RPT2a in regulating gene activity at the transcriptional level via DNA methylation. Knockout mutation of the RPT2a gene did not alter global protein levels; however, the transcriptional activities of reporter transgenes were severely reduced compared to those in the wild type. This transcriptional gene silencing (TGS) was observed for transgenes under control of either the constitutive CaMV 35S promoter or the cold-inducible RD29A promoter. Bisulfite sequencing analysis revealed that both the transgene and endogenous RD29A promoter regions were hypermethylated at CG and non-CG contexts in the rpt2a mutant. Moreover, the TGS of transgenes driven by the CaMV 35S promoters was released by treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine, but not by application of the inhibitor of histone deacetylase Trichostatin A. Genetic crosses with the DNA methyltransferase met1 single or drm1drm2cmt3 triple mutants also resulted in a release of CaMV 35S transgene TGS in the rpt2a mutant background. Increased methylation was also found at transposon sequences, suggesting that the 19S proteasome containing AtRPT2a negatively regulates TGS at transgenes and at specific endogenous genes through DNA methylation.
Introduction
The 26S proteasome is an ATP-dependent proteinase complex that is responsible for regulated proteolysis of polyubiquitinated proteins in eukaryotic cells and is essential for the development of plants [1], [2]. The 26S proteasome is assembled from two particles: the 20S core particle (20S CP) and the 19S regulatory particle (19S RP). Proteolytic activities reside within the central chamber of the 20S CP, which is a hollow cylinder composed of four stacked rings [3], [4]. The 19S RP binds to one or both ends of the 20S CP and sits directly over the ring pore. The 19S RP recognizes polyubiquitinated proteins and is responsible for their ATP-dependent unfolding and threading through a narrow channel into the 20S CP [5]. The 19S RP is composed of two subcomplexes as follows: a base containing six related AAA-ATPases (designated RPT1–6 for regulatory particle triple-A ATPases) and three non-ATPase subunits (designated RPN1, RPN2, and RPN10, for regulatory particle non-triple-A ATPases), and a lid that contains at least 12 additional RPN subunits (RPN1–3 and −5–13). In plants, most genes encoding 19S RP subunits are duplicated. Such subunit duplication would lead to an increase in not only subunit redundancy but also subunit function. However, the functions of only some of the plant 19S RP subunits are known. RPT2 is essential for the channel opening of the α-ring of the 20S CP in yeast and mammals by its conserved C-terminal motif [6], [7]. The Arabidopsis genome contains two genes, AtRPT2a and AtRPT2b that are paralog RPT2 subunits with a difference of only four amino acids in the protein sequence. We have recently discovered that the rpt2a mutant shows a specific phenotype of enlarged leaves caused by increased cell size correlated with extended endoreduplication, whereas the rpt2b mutant did not show any morphological difference compared with the wild type [8].
DNA methylation is an important epigenetic mark for transcriptional gene silencing including genomic imprinting and repression of transposable elements in plants, vertebrates and some fungi [9], [10]. In general, cytosine methylation is found in both CG and non-CG (CHG and CHH where H is A, T or C) contexts in plants. In the model plant Arabidopsis thaliana, at least three methylation pathways exist and each is associated with a specific methyltransferase. DNA methylation is often considered a stable epigenetic mark, but active demethylation has been observed in both plants and animals and demethylases play an important role in protecting plant genes from potentially deleterious methylation [11]. In Arabidopsis, DNA glycosylases of the DEMETER (DME) family are responsible for removing methylcytosines from DNA. REPRESSOR OF SILENCING1 (ROS1), a DME homolog, is required for demethylating a transgene promoter and some endogenous genes, and for regulating their gene expression [12]. Plants maintain appropriate gene expressions and genomic stability with the coordination of methylases and demethylases.
During subsequent analysis of RPT2a by stable transformation with various expression constructs, many of these constructs showed gene silencing in the rpt2a mutant background. Here, we demonstrate a novel function of RPT2a for the specific regulation of gene silencing that involves DNA methylation in transgenes and some specific endogenous genes.
Results and Discussion
The rpt2a mutant showed transcriptional gene silencing
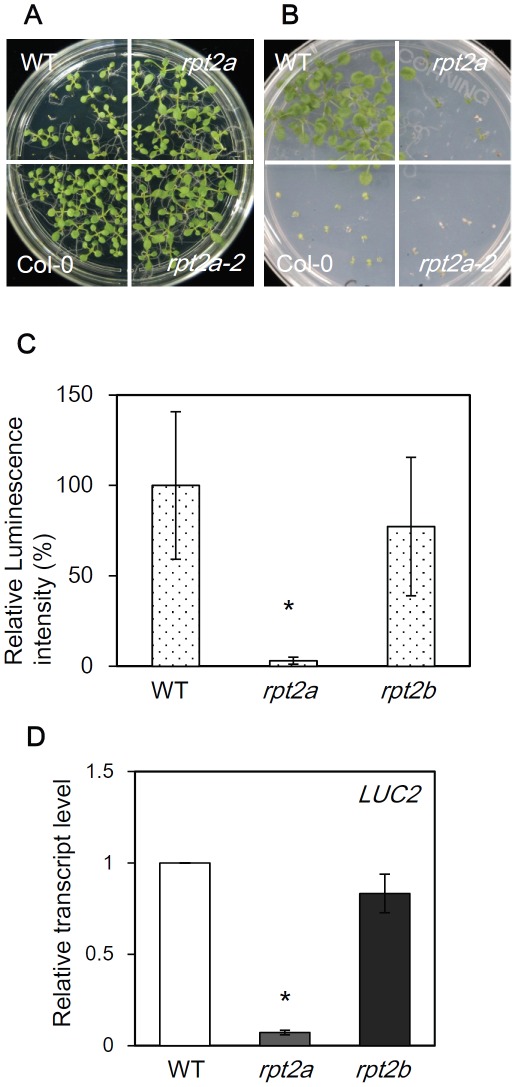
We found that the rpt2a-2 mutant displayed a phenotype of repressed transgene expression during construction of transgenic plants for investigating RPT2a function. As an example of this, introduction of the hygromycin B phosphotransferase gene (HPT) driven by the constitutive CaMV 35S promoter into Col-0 wild-type plants conferred plant survival on MS medium containing hygromycin in Col-0 background. In contrast, the transgene became inactive in rpt2a-2 and transgenic plants showed sensitivity to hygromycin-containing media (Figure 1A and 1B). In order to examine the function of AtRPT2a in transgene silencing quantitatively, luciferase2 (LUC2) overexpressing plants were produced in rpt2a-2 and rpt2b-1 mutant backgrounds by crossing with transgenic wild-type. We confirmed that the transgenic plants expressing LUC2 under the CaMV 35S promoter have a single copy T-DNA inserted in the euchromatin region (Figure S2A, 2C, 2E). Although AtRPT2a and AtRPT2b share an almost identical amino acid sequence, only the rpt2a-2 mutant showed one-tenth the luminescence of the WT, and the rpt2b-1 mutant showed the same level of luminescence as WT (Figure 1C). An identical result was obtained with the rpt2a-1 allele (Figure S1). This result suggests that RPT2a may regulate the expression of transgenes.
Figure 1. The rpt2a mutant shows transcriptional gene silencing.
(A) 35S::HPT in the Col-0 (WT) and rpt2a-2 mutant on MS medium. Col-0 plants without any transgene indicate “Col-0”. (B) 35S::HPT in the WT and rpt2a-2 mutant on MS medium containing 50 µM hygromycin. (C) Relative luminescence intensity of 35S::LUC2 in WT, rpt2a-2 and rpt2b-1 mutants. 35S::LUC2 in WT is set as 100%. *t-test P<0.05, error bar = S.D., n = 20. (D) Quantification of LUC2 gene expression in 35S::LUC2 in WT, rpt2a-2 and rpt2b-1 mutants. 35S::LUC2 in WT is set as 1. Values are the averages of the three experiments, and the level of 18S rRNA was used as an internal control.
To determine whether repression of the transgene in the rpt2a mutant was regulated at the transcriptional or post-translational level, PT-PCR was used to examine the accumulation of LUC2 transcripts. Accumulation of LUC2 transcripts was dramatically decreased in the rpt2a-2 mutant compared to that in the WT (Figure 1D). Consistent with that observed for luciferase activity, LUC2 transcript accumulation in the rpt2b-1 mutant was not different to that in WT (Figure 1D). These results suggest that the luciferase gene was repressed at the transcriptional level in the rpt2a mutant.
The DNA methylation inhibitor released transcriptional gene silencing in the rpt2a mutant
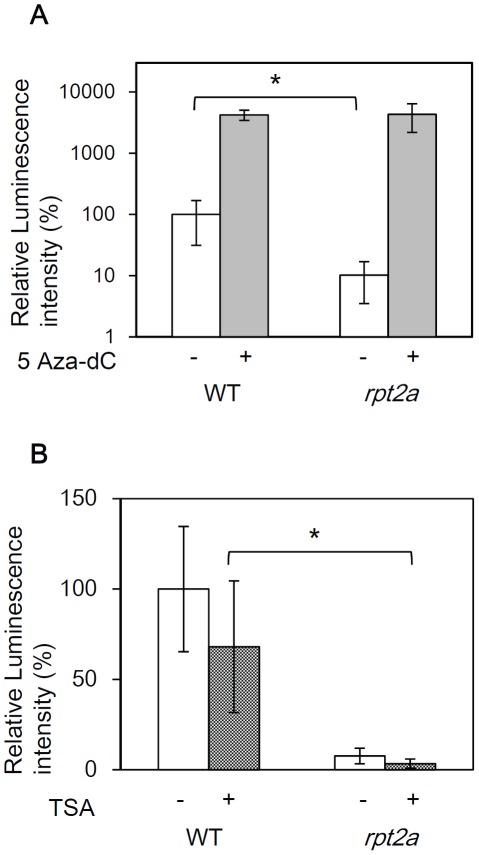
Transcriptional gene silencing (TGS) is often associated with DNA methylation and histone modification. To examine whether such epigenetic changes were involved in the repression observed in the rpt2a mutant, we tested an inhibitor of cytosine methylation, 5-aza-2′-deoxycytidine (5Aza-dC) and an inhibitor of histone deacetylase, TrichostatinA (TSA). Following treatment with 5Aza-dC, gene silencing in the rpt2a-2 mutant was released at the same level as that of 5Aza-dC treated WT (Figure 2A). On the other hand, treatment with TSA did not cause a change in luciferase activity in the rpt2a mutant (Figure 2B). From the experiments using the genes that have been reported to be transcriptionally increased by TSA treatment [13], [14], we confirmed that the TSA treatment is effective (Figure S3). Although these results could not deny the possibility that histone modification, except acetylation, is involved in gene silencing in rpt2a mutant, these results suggest that DNA hypermethylation is correlated with gene silencing in the rpt2a mutant.
Figure 2. 5-aza-2′-deoxycytidine (5Aza-dC) treatment releases gene silencing in the rpt2a mutant.
(A) Relative luminescence intensity of WT and the rpt2a-2 mutant treated with 50 µM 5-aza-2′-deoxycytidine (5Aza-dC). Seedlings grown in MS medium for 2 weeks are transferred to MS liquid medium containing 50 µM 5Aza-dC for one week. 35S::LUC2 in WT without 5Aza-dC treatment is set as 100%. *t-test P<0.05, error bar = S.D., n = 15. (B) Relative luminescence intensity of WT and the rpt2a-2 mutant treated with 0.1 µM TrichostatinA (TSA). Seedlings grown in MS medium for 2 weeks are transferred to MS liquid medium containing 0.1 µM TSA for one week. 35S::LUC2 in WT without TSA treatment is set as 100%. *t-test P<0.05, error bar = S.D., n = 15.
The mutation of DNA methyltransferase released gene silencing in the rpt2a mutant
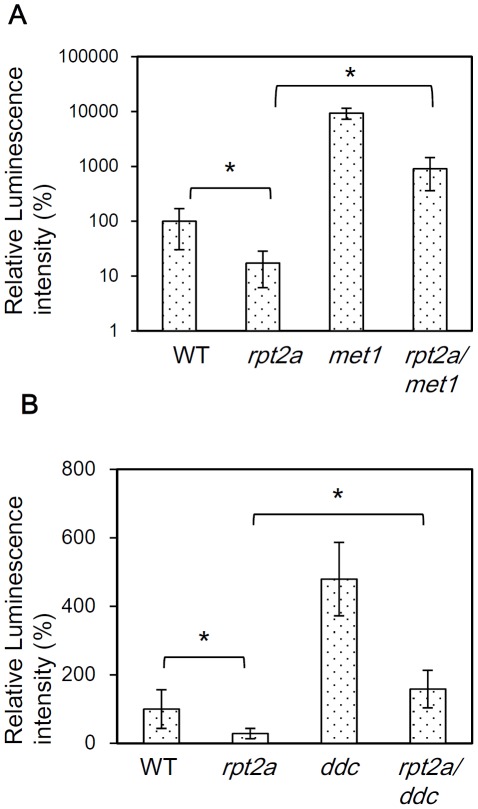
Three classes of DNA methyltransferases: MET1, CMT3 and DRM1/2, regulate DNA methylation in Arabidopsis thaliana. MET1 maintains CG methylation, while DRM1/2 and CMT3 are responsible for methylation at non-CG sites [15]. The above data show that DNA methylation was involved in transgene silencing in the rpt2a mutant (Figure 2A). To further investigate the relationship between gene silencing in the rpt2a mutant and DNA methylation, we next examined luciferase activity in an rpt2a-2 met1-1 double mutant. In addition, DRM1, DRM2 and CMT3 are reported to have a redundant function [16] and thus, we made a rpt2a-2 drm1 drm2 cmt3 quadruple mutant and checked the luciferase activity in this background. LUC activity was found to be much higher in the rpt2a-2 met1-1 double mutant compared to that of the single rpt2a-2 mutant although this was still lower than that observed in the met1 single mutant (Figure 3A). This result suggests a partial release of gene silencing by met1 in the rpt2a mutant.
Figure 3. DNA methyltransferase mutants release gene silencing in the rpt2a mutant.
(A) Relative luminescence intensity of WT, rpt2a-2, met1-1 and rpt2a-2met1-1 double mutants. 35S::LUC2 in WT is set as 100%. *t-test P<0.05, error bar = S.D., n = 10. (B) Relative luminescence intensity of WT, rpt2a-2, drm1 drm2 cmt3 triple mutant (ddc) and rpt2a-2 drm1 drm2 cmt3 quadruple mutant (rpt2a/ddc). 35S::LUC2 in WT is set as 100%. *t-test P<0.05, error bar = S.D., n = 20.
LUC activity in the rpt2a-2 drm1 drm2 cmt3 quadruple mutant was also higher than that in the single rpt2a-2 mutant, whereas that of the drm1 drm2 cmt3 triple mutant was higher than that in the quadruple mutant (Figure 3B). This result suggests a partial release of gene silencing in the rpt2a mutant by the drm1drm2cmt3 triple mutant. These results are consistent with gene silencing in the rpt2a mutant caused by both CG and non-CG hypermethylation. The rpt2a-2 met1 double mutant showed much higher LUC activity than that of the rpt2a-2 drm1 drm2 cmt3 quadruple mutant, due to a greater influence of CG methylation on the as-1 regulatory element within the CaMV 35S promoter [17].
The rpt2a mutation leads to DNA hypermethylation in the promoter of the silenced loci
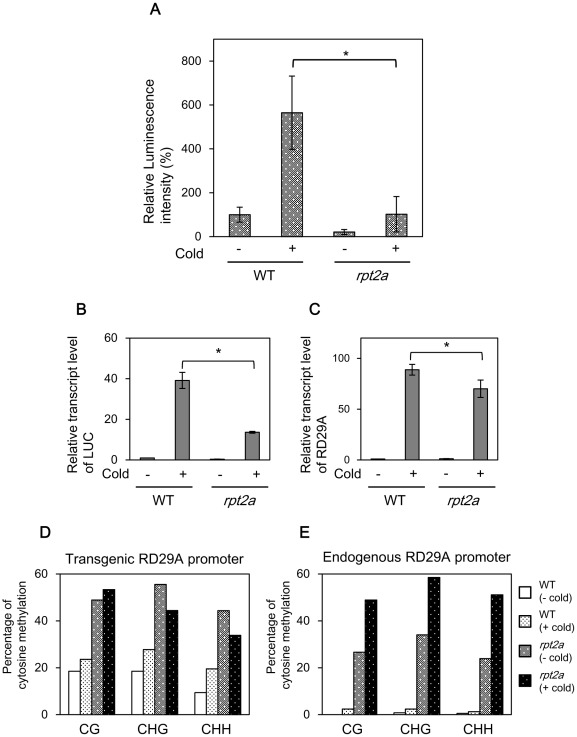
To confirm that the DNA methylation is increased in the rpt2a mutant, we checked the methylation level of the rpt2a mutant by bisulfite sequencing. Methylation levels in the CaMV 35S promoter increased in the rpt2a mutant compared to that in WT. However, the CaMV 35S promoter was also highly methylated in WT, consistent with the results of 5Aza-dC treatment and genetic analysis of DNA methyltransferases, and the differences in DNA methylation level between WT and the rpt2a mutant were obscure. We therefore placed the LUC gene under control of the cold- and drought-responsive RD29A promoter [18]. We confirmed that transgenic plants expressing LUC under the RD29A promoter have a single copy T-DNA inserted in the euchromatin region (Figure S2B, 2D, 2F). Luminescence was induced in WT containing RD29A::LUC when plants were treated with cold stress for 12 hours. On the other hand, luminescence was repressed in rpt2a-2 with and without low temperature treatment (Figure 4A). We confirmed that cold treatment induced transcript accumulation of LUC and endogenous RD29A in WT, whereas these transcripts were repressed in rpt2a-2 upon the cold treatment (Figure 4B, 4C). In contrast, gene expression of other cold-responsive genes COR15A and DREB1 were induced in rpt2a-2 (Figure S4). These results reveal that the rpt2a mutant shows gene silencing of the genes under control of both the RD29A and CaMV 35S promoter, suggesting that TGS in the rpt2a mutant is independent of promoter sequences. DNA methylation levels of exogenous and endogenous RD29A promoters were next investigated. Compared to that in WT, the methylation level of the exogenous RD29A promoter increased and broadened in the rpt2a mutant both before and after cold treatment (Figure 4D, S5). This result may imply that demethylation of the RD29A promoter was abnormal in the rpt2a mutant. The endogenous RD29A promoter contained a very low level of DNA methylation in WT before and after cold treatment. On the other hand, the methylation level of the endogenous RD29A promoter increased in the rpt2a mutant both before and after cold treatment (Figure 4E). The RD29A promoter contains a cis-acting dehydration-responsive element (DRE) involved in the induction by exposure to low temperature [18]. After cold treatment, the methylation level of DRE in endogenous RD29A promoter greatly increased in rpt2a-2 suggesting that DNA methylation of DRE represses the transcription of endogenous RD29A genes in rpt2a-2 (Figure S6). These results are consistent with TGS of transgenes in the rpt2a mutant caused by increased DNA methylation in the promoter region. Interestingly, cold treatment also induced a slight increase of DNA methylation in the Col-0 background (Figure S7). This result may indicate that a rapid increase of transcription induces DNA methylation for marking of activated genes and for monitoring of genome stability.
Figure 4. Gene silencing in the rpt2a mutant is correlated with DNA hypermethylation.
(A) Relative luminescence intensity of RD29A::LUC in WT and rpt2a-2. Plants are untreated or treated with cold (4°C) for 12 hours. (B) Quantification of LUC gene expression in RD29A::LUC in WT and rpt2a-2. Expression levels are relative to that of untreated WT plants. Values are the average of three experiments, and the level of 18S rRNA was used as an internal control. (C) Quantification of RD29A gene expression in RD29A::LUC in WT and rpt2a-2. Expression levels are relative to that of untreated WT plants. Values are the averages of three experiments, and the level of 18S rRNA is used as an internal control. (D) Mean levels of DNA methylation in different cytosine context at the exogenous RD29A promoter in WT and the rpt2a-2 mutant. Frequencies of methylcytosine at CG, CHG and CHH sites are indicated. Twenty clones are sequenced for each sample. (E) Mean levels of DNA methylation in different cytosine contexts at the endogenous RD29A promoter in WT and the rpt2a-2 mutant.
The transposable element is highly methylated in the rpt2a mutant
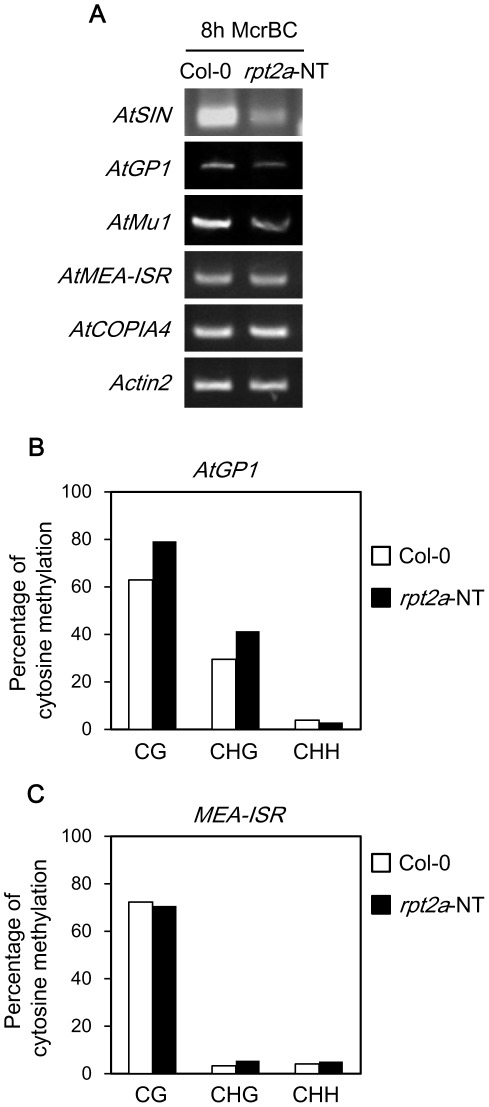
We have shown that the DNA methylation level of the promoter site of transgenes increased in the rpt2a mutant. We next tested the hypothesis that endogenous genes were also hypermethylated in the rpt2a mutant. The Arabidopsis genome contains silenced transposable elements. We tested methylation levels at several transposons by methylation-sensitive PCR with McrBC, which preferentially cuts methylated DNA [19]. Higher levels of methylation result in increased McrBC digestion and consequently reduced amplification products. Compared with Col-0, methylation levels of AtSINE1 and AtGP1 increased in the rpt2a-2 (Figure 5A). On the other hand, the methylation level of AtCOPIA4 and AtMEA-ISR were not different from Col-0. The methylation levels of AtGP1 and AtMEA-ISR were examined further by bisulfite sequencing analysis. We confirmed that the methylation level of AtGP1 increased in rpt2a-2, but that of AtMEA-ISR was not significantly different from Col-0 (Figure 5B, 5C, S8). This result shows that DNA methylations increased in the rpt2a mutant in specific genome loci, but not in the whole genome.
Figure 5. DNA methylation level of transposons is increased in the rpt2a mutant.
(A) McrBC PCR of transposons at Col-0 and rpt2a-2 (no transgene: rpt2a-NT). McrBC-digested genomic DNA is amplified by PCR with primers for the indicated transposons. Input DNA was normalized for each genotype with Actin2. (B) Mean levels of DNA methylation in different cytosine context at the AtGP1 in Col-0 and rpt2a-2 (no transgene). (C) Mean levels of DNA methylation in different cytosine context at the MEA-ISR in Col-0 and rpt2a-2 (no transgene).
We show here that the loss of AtRPT2a function results in TGS and an increase in the DNA methylation of promoter sequences of transgenes. Transposons also showed hypermethylation in the rpt2a mutant. These observations suggest that AtRPT2a, a 19S proteasome subunit protein, is required for the negative regulation of DNA methylation at transgenes and specific genome loci. This is the first report that the proteasome has the potential of regulating DNA methylation.
Genome-wide methylation analysis has shown that about 30% of genes are methylated in Arabidopsis [20] and this DNA methylation status is dynamically regulated by DNA methylation and demethylation reactions [11]. We showed that hypermethylation in rpt2a-2 involved all three methyltransferases. There was no difference in the expression level of these genes for the DNA methyltransferase between Col-0 and rpt2a-2 (Figure S9). PEST motif prediction (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::epestfind) showed that MET1, CMT3 and DRM1/2 all contain PEST motifs suggesting that these DNA methyltransferases can be degraded by the 26S proteasome. Taken together, hypermethylation in the rpt2a mutant is thought to be due to the accumulation of DNA methyltransferase. This hypothesis is supported by a report that, in mammals, Dnmt1 (the homolog of MET1) is degraded by proteasomes upon treatment with a DNA methylation inhibitor [21]. However, increased methylation in rpt2a was observed at specific loci rather than globally, indicating that the accumulation of DNA methyltransferases alone was not responsible for hypermethylation in the rpt2a mutant.
ROS1 and ROS3 are required for demethylation [12], [22]. Mutations in ROS1 cause hypermethylation of the RD29A promoter, leading to silencing of the transgene and its homologous endogenous gene. ROS1 is also required to suppress DNA methylation in a number of other endogenous genomic loci including many transposons [23], [24]. Since we showed that the rpt2a mutant shows a similar phenotype to the ros1 mutant, a hypothesis is presented that AtRPT2a could function with ROS1 and ROS3 in a demethylation pathway. Unfortunately, we have not excluded the relationship between the RPT2a and ROS pathway in this report.
Recent works raise an alternative hypothesis for the function of AtRPT2a. The 19S RP has been shown to be required for methylation of histone H3 lysine 4 (H3K4) in yeast and mammals [25], [26]. In Arabidopsis, genome-wide analysis showed that DNA methylation and H3K4 di- and tri-methylation are mutually exclusive [27]. Although TSA treatment did not release gene silencing in the rpt2a mutant in this report, these observations raise the possibility that an increase of DNA methylation in the rpt2a mutant may be caused by a decrease of H3K4 methylation and expansion of DNA methylation. We could not totally rule out the possibility that the proteasome indirectly controls DNA methylation since the ubiquitin/26S proteasome pathway regulates many biological phenomena. Further studies are required to determine the causes of hypermethylation in the rpt2a mutant.
Epigenetic modification is an important mechanism for adaption of gene expression to development and environmental status [28]. In addition, the Ub/proteasome system plays a crucial role in the response of hormone signaling and environmental stress [2], [29]. We have demonstrated that the RPT2a subunit is required for the regulation of DNA methylation, suggesting that the proteasome participates in epigenetic modification for proper gene expression depending on the environmental status.
Materials and Methods
Plant materials
For germination of Arabidopsis thaliana (ecotype Columbia-0) wild type and mutants, seeds were surface-sterilized and placed on Murashige and Skoog (MS) medium supplemented with 2% sucrose (Germination inducible medium: GIM). After cold treatment for 2 days to synchronize germination, seeds were transferred to 22°C and 50% relative humidity under a 16/8 h light/dark cycle (this time point indicates 0 days after sowing: DAS). The seeds of the met1-1 mutant were provided by Dr. Robert A. Martienssen (Cold Spring Harbor Laboratory). Seeds of the rpt2a-1, rpt2a-2, rpt2b-1, and drm1 drm2 cmt3 triple mutants were obtained from the ABRC (The Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH, USA; stock number: SALK_130019, SALK_005596, SALK_043450, and CS16384 respectively). Sequences bordering the T-DNA insertion were determined using primer pairs listed in Table S1. 35S::HPT plants were obtained by transformation of the wild-type Arabidopsis plants of the Columbia-0 ecotype with T-DNA composed of a HPT gene conferring resistance to hygromycin driven by the 35S promoter of the cauliflower mosaic virus. 35S::LUC2 plants were obtained by transformation of Columbia-0 with a destination vector p7-LUC2. Mutants were crossed to each transgenic line, and F3 progenies homozygous for transgenes and/or the mutations were used for experiments. RD29A::LUC plants were obtained by transformation of Columbia-0 with a destination vector pGWB35 containing genomic fragments of the promoter region of RD29A (824-bp upstream of the ATG). Mutants were crossed to each transgenic line, and F3 progenies homozygous for the transgenes and/or the mutations were used for experiments.
For 5-aza-2′-deoxycytidine (5Aza-dC) treatment and TrichostatinA (TSA) treatment, seedlings grown for one week were transferred to MS liquid medium containing 50 µM 5Aza-dC (Wako) or to MS liquid medium containing 0.1 µM TSA (Wako).
Luciferase activity
Five millimeter diameter leaf sections were floated on 50 µl of Pikkagene cell lysis buffer (TOYO B-Net. CO., LTD) containing 20 µl of 0.1 µM D-Luciferine potassium salt and incubated for 30 min. Samples were measured using a Luminescenceor JNR II (Atto).
Transcript level analysis
Total RNA was extracted by the guanidine thiocyanate method [30]. Total RNA (0.6 µg RNA) was used as a template for first strand cDNA synthesis with ReverTraAce -α-® reverse transcriptase (TOYOBO, Osaka, Japan). First strand cDNA (0.7 µl) was then assayed for gene-specific DNA fragments using the primer pairs listed in Table S1. PCR amplification was performed in the optimum cycles with each gene using the Taq DNA polymerase (New England BioLabs® Japan inc, Tokyo, Japan). Amplified fragments were separated on 1.2% (w/v) agarose gels and visualized by ethidium bromide staining. Real-time PCR was performed with the Power SYBR Green PCR Master Mix (Applied Biosystems) on an Applied Biosystems 7300 Real-Time PCR system (Applied Biosystems). Relative quantitation of gene expression is based on the comparative CT method (User Bulletin No. 2: ABI PRISM 7700 Sequence Detection System, 1997) using 18S rRNA as a reference gene. The following PCR program was used: 2 min at 50°C; 10 min at 95°C; 40 cycles of 15 sec at 95°C, and 1 min at 60°C. Two biological and three technical replicates were performed. The sequences of the primers used are specified in Table S1.
Bisulfite sequencing
For analysis of DNA methylation by bisulfite sequencing, DNA was isolated from the first leaves of 3-week-old plants of WT and mutants using a Nucleon PhytoPure DNA extraction kit (GE healthcare). The protocol of bisulfite treatment in this study is based on the methods of Kanazawa et al., 2007 [31]. DNA was cleaved with the restriction enzyme EcoRI, extracted with phenol/chloroform, and precipitated by ethanol. The cleaved DNA was alkali denatured in 0.3 M NaOH at 37°C for 40 min. Denatured DNA was incubated in a total volume of 600 µM with freshly prepared 5.9 M urea/3.35 M sodium bisulfite/0.5 mM hydroquinone pH 5.0, at 60°C for 36 h under mineral oil. A Quick PCR purification kit (Qiagen) then recovered the DNA. NaOH was added to the DNA solution to a concentration of 0.3 M and then incubated at 37°C for 30 min. Glycogen and ammonium acetate were added to the solution to final concentrations of 0.16 mg/ml and 2.64 M, respectively. DNA was then precipitated with ethanol and dissolved in 10 µl of TE (pH 8.0). Two rounds of PCR were carried out using 1 µl of bisulfite-treated DNAs as a template. Primers for the RD29A promoter and transposons were modified based on the methods of Zheng et al. (2008) and Gao et al. (2010) [22], [32]. To amplify the exogenous RD29A promoter, primers pRD29A nested and pRD29A transR1 were used for the first round PCR, and primers pRD29A converted and pRD29A transR2 were used for the second round PCR. To amplify the endogenous RD29A promoter, primers pRD29A nested and pRD29A endoR1 were used for the first round PCR, and primers pRD29A converted and pRD29A endoR2 were used for the second round PCR. The PCR products were cloned into a pCR2.1 vector (Invitrogen) and 20 clones per one plant were subjected to sequence analysis.
McrBC PCR
McrBC PCR was performed on genomic DNA that was extracted from 3-week-old rosette leaves from 5 plants grown under identical conditions as described above. 250 ng genomic DNA was digested with McrBC for 3 hours and assayed using the PCR primers listed in Table S1.
Supporting Information
Relative luminescence intensity of 35S::LUC2 in WT and rpt2a-1. *t-test P<0.05, error bar = S.D., n = 25.
(TIF)
Analysis of T-DNA insertion site. (A) Southern blot analysis of Col-0, 35S::LUC2 in WT and in rpt2a-2 genomic DNA with the LUC2 as a probe. (B) Southern blot analysis of Col-0, RD29A::LUC in WT and in rpt2a-2 genomic DNA with the LUC as a probe (Methods S1). (C) 35S::LUC2 T-DNA insertion site in At5g58580. (D) RD29A::LUC T-DNA insertion site in At3g11860. (E) Insertion check of 35S::LUC2 by PCR. (F) Insertion check of RD29A::LUC.
(TIF)
RT-PCR analysis of TSA treated plants: ABI3, At3g29650 and 18S rRNA (control).
(TIF)
(A) RT-PCR analysis of cold inducible genes: COR15A, DREB1B and 18S rRNA (control). (B) Quantification of RD29A gene expression in Col-0 and rpt2a-2 (no transgene: rpt2a-NT). Expression levels are relative to that of untreated WT plants. Values are the averages of three experiments, and the level of 18S rRNA is used as an internal control.
(TIF)
(A) Scheme of analyzed region in exogenous RD29A promoter. (B) Bisulfite sequencing of DNA methylation in the exogenous RD29A promoter site (from −346 bp to −51 bp upstream of the promoter). Upper graph shows methylation status in WT and the lower graph shows DNA methylation status in the rpt2a-2 mutant. The height of the vertical lines shows the frequency of methylcytosine. Red, blue and green lines indicate frequencies of methylcytosine at CG, CHG and CHH sites, respectively. Red bars on the x-axis are DRE and DRE/CRT core sequences. Twenty clones are sequenced for each sample.
(TIF)
(A) Scheme of analyzed region in endogenous RD29A promoter. (B) Bisulfite sequencing of DNA methylation in the endogenous RD29A promoter site (from −327 bp to −32 bp upstream of promoter). The upper graph shows methylation status in WT and the lower graph shows DNA methylation status in the rpt2a-2 mutant. The height of the vertical lines shows the frequency of methylcytosines. Red, blue and green lines indicate frequencies of methylcytosine at CG, CHG and CHH sites, respectively. Red bars on the x-axis are DRE and DRE/CRT core sequences. Twenty clones are sequenced for each sample.
(TIF)
(A) Bisulfite sequencing of DNA methylation in the endogenous RD29A promoter site (from −327 bp to −32 bp upstream of the promoter) in Col-0 and rpt2a-2 (no transgene: rpt2a-NT). The upper graph shows methylation status in cold-untreated and treated Col-0, and the lower graph shows cold-untreated and treated rpt2a-2 (no transgene). (B) Mean levels of DNA methylation in different cytosine context at the exogenous and endogenous RD29A promoter in Col-0 and rpt2a-2 (no transgene). Red, blue and green lines indicate frequencies of methylcytosine at CG, CHG and CHH sites, respectively. Twenty clones are sequenced for each sample.
(TIF)
Bisulfite sequencing of DNA methylation in the AtGP1 site in Col-0 and rpt2a-2 (no transgene: rpt2a-NT).
(TIF)
RT-PCR analysis of DNA methyltransferase genes: MET1, CMT3, DRM2 and 18S rRNA (control).
(TIF)
Primers used in this study.
(PDF)
Supplementary methods.
(PDF)
Acknowledgments
We are grateful to Dr. Akira Kanazawa for the bisulfite sequencing analysis. We are also grateful to Dr. Derek B. Goto (Hokkaido University) for critical reading of the manuscript. K.S. acknowledges Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists (2008–2010) and also support by Otsuka Award (2011).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Grants-in-Aid for Scientific Research 2211450100 and 23380198 from the Ministry of Education, Culture, Sports, Science and Technology in Japan (http://www.mext.go.jp/english/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nature reviews. Molecular cell biology. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 3.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 4.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nature reviews. Molecular cell biology. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 6.Köhler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Molecular Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 7.Smith DM, Chang S-C, Park S, Finley D, Cheng Y, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Molecular Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonoda Y, Sako K, Maki Y, Yamazaki N, Yamamoto H, et al. Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. The Plant journal: for cell and molecular biology. 2009;60:68–78. doi: 10.1111/j.1365-313X.2009.03932.x. [DOI] [PubMed] [Google Scholar]
- 9.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 10.Finnegan EJ, Kovac KA. Plant DNA methyltransferases. Plant molecular biology. 2000;43:189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- 11.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annual review of genetics. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant physiology. 2008;146:149–161. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. The Journal of biological chemistry. 2005;280:796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 15.Chan SW-L, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nature reviews. Genetics. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 16.Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Current biology. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 17.Kanazawa A, O'Dell M, Hellens RP. The binding of nuclear factors to the as-1 element in the CaMV 35S promoter is affected by cytosine methylation in vitro. Plant biology (Stuttgart, Germany) 2007;9:435–441. doi: 10.1055/s-2006-924633. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS biology. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, et al. 5-Aza-Deoxycytidine Induces Selective Degradation of DNA Methyltransferase 1 by a Proteasomal Pathway That Requires the KEN Box , Bromo-Adjacent Homology Domain , and Nuclear Localization Signal. Society. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Zheng X, Pontes O, Zhu J, Miki D, Zhang F, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu J-K. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Current biology: CB. 2007;17:54–9. doi: 10.1016/j.cub.2006.10.059. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 24.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, et al. DNA demethylation in the Arabidopsis genome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6752–7. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Molecular cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 26.Koues OI, Dudley RK, Mehta NT, Greer SF. The 19S proteasome positively regulates histone methylation at cytokine inducible genes. Biochimica et biophysica acta. 2009;1789:691–701. doi: 10.1016/j.bbagrm.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome biology. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chinnusamy V, Zhu J-K. Epigenetic regulation of stress responses in plants. Current opinion in plant biology. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurepa J, Wang S, Li Y, Smalle J. Proteasome regulation, plant growth and stress tolerance. Plant signaling & behavior. 2009;4:924–927. doi: 10.4161/psb.4.10.9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 31.Kanazawa A, O'Dell M, Hellens RP. Epigenetic inactivation of chalcone synthase-A transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant & cell physiology. 2007;48:638–647. doi: 10.1093/pcp/pcm028. [DOI] [PubMed] [Google Scholar]
- 32.Gao Z, Liu H-L, Daxinger L, Pontes O, He X, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative luminescence intensity of 35S::LUC2 in WT and rpt2a-1. *t-test P<0.05, error bar = S.D., n = 25.
(TIF)
Analysis of T-DNA insertion site. (A) Southern blot analysis of Col-0, 35S::LUC2 in WT and in rpt2a-2 genomic DNA with the LUC2 as a probe. (B) Southern blot analysis of Col-0, RD29A::LUC in WT and in rpt2a-2 genomic DNA with the LUC as a probe (Methods S1). (C) 35S::LUC2 T-DNA insertion site in At5g58580. (D) RD29A::LUC T-DNA insertion site in At3g11860. (E) Insertion check of 35S::LUC2 by PCR. (F) Insertion check of RD29A::LUC.
(TIF)
RT-PCR analysis of TSA treated plants: ABI3, At3g29650 and 18S rRNA (control).
(TIF)
(A) RT-PCR analysis of cold inducible genes: COR15A, DREB1B and 18S rRNA (control). (B) Quantification of RD29A gene expression in Col-0 and rpt2a-2 (no transgene: rpt2a-NT). Expression levels are relative to that of untreated WT plants. Values are the averages of three experiments, and the level of 18S rRNA is used as an internal control.
(TIF)
(A) Scheme of analyzed region in exogenous RD29A promoter. (B) Bisulfite sequencing of DNA methylation in the exogenous RD29A promoter site (from −346 bp to −51 bp upstream of the promoter). Upper graph shows methylation status in WT and the lower graph shows DNA methylation status in the rpt2a-2 mutant. The height of the vertical lines shows the frequency of methylcytosine. Red, blue and green lines indicate frequencies of methylcytosine at CG, CHG and CHH sites, respectively. Red bars on the x-axis are DRE and DRE/CRT core sequences. Twenty clones are sequenced for each sample.
(TIF)
(A) Scheme of analyzed region in endogenous RD29A promoter. (B) Bisulfite sequencing of DNA methylation in the endogenous RD29A promoter site (from −327 bp to −32 bp upstream of promoter). The upper graph shows methylation status in WT and the lower graph shows DNA methylation status in the rpt2a-2 mutant. The height of the vertical lines shows the frequency of methylcytosines. Red, blue and green lines indicate frequencies of methylcytosine at CG, CHG and CHH sites, respectively. Red bars on the x-axis are DRE and DRE/CRT core sequences. Twenty clones are sequenced for each sample.
(TIF)
(A) Bisulfite sequencing of DNA methylation in the endogenous RD29A promoter site (from −327 bp to −32 bp upstream of the promoter) in Col-0 and rpt2a-2 (no transgene: rpt2a-NT). The upper graph shows methylation status in cold-untreated and treated Col-0, and the lower graph shows cold-untreated and treated rpt2a-2 (no transgene). (B) Mean levels of DNA methylation in different cytosine context at the exogenous and endogenous RD29A promoter in Col-0 and rpt2a-2 (no transgene). Red, blue and green lines indicate frequencies of methylcytosine at CG, CHG and CHH sites, respectively. Twenty clones are sequenced for each sample.
(TIF)
Bisulfite sequencing of DNA methylation in the AtGP1 site in Col-0 and rpt2a-2 (no transgene: rpt2a-NT).
(TIF)
RT-PCR analysis of DNA methyltransferase genes: MET1, CMT3, DRM2 and 18S rRNA (control).
(TIF)
Primers used in this study.
(PDF)
Supplementary methods.
(PDF)