Plants are embattled in a war with rasping, sucking, and chewing insects, deadly viruses, debilitating bacteria, and castrating fungi. This war costs billions of dollars in crop losses each year, making the study of plant-pathogen and plant-herbivore interactions one of the most significant branches of applied biology (1). But the study of how plants and their enemies interact also has inspired major advances in fundamental research regarding species interactions, particularly concerning the interplay of evolution and ecology. Especially influential has been the idea that herbivorous insects have driven the evolution of plants, and in turn, plant adaptations to insect attack have stimulated a diversification of insects (2). This evolutionary dance between insects and plants is a widely cited example of what generally is referred to as “coevolution”—that is, reciprocal adaptive genetic changes within populations of interacting species that act as selective agents for one another. Coevolution fascinates biologists because it suggests a view of nature in which close associations between species have shaped their life histories and ecologies in a way that fundamentally alters how they interact. If coevolution is a widespread and dominant process, then one of humankind’s more insidious impacts on the world is likely to be the perturbation of coevolved systems.
Original ideas about coevolution were inspired by studies of plant-insect and plant-pathogen interactions, and those who study the impact of diseases or herbivores on plants inevitably are indoctrinated with the notion of coevolution. Ironically, even though coevolution is conceptually compelling, we lack definitive empirical studies that show how it works. Alternative hypotheses for the course of coevolution include: (i) escalating arms races in which plants relentlessly add to their chemical arsenals, whereas herbivores follow suit with new mechanisms for overriding those defenses, (ii) cyclical selection in which highly defended plants are favored in times when virulent pathogens or herbivores exert a severe toll, but which gradually decline in prevalence because of costs associated with resistance traits when the plants are not under attack, and (iii) a stasis that entails little evolutionary change in either plants or their enemies because a paucity of genetic variation or the presence of specific constraints limit the opportunities for evolution.
Apart from classical work detailing gene-for-gene interactions between virulent pathogens and resistant plant varieties (3), evidence in support of different modes of coevolution is lacking from natural populations. However, in an illuminating culmination of almost 20 years of work, Berenbaum and Zangerl (4) have pieced together one of the most compelling examples of coevolution for plant-herbivore systems. The plant is wild parsnip, Pastinaca sativa, an introduced European weed that now occurs throughout much of eastern North America in disturbed habitats. The herbivore is the parsnip webworm, Depressaria pastinacella, which is the dominant (and in fact only) herbivore associated with wild parsnip in most of North America. Previous work has documented that parsnip is defended against webworms by furanocoumarins, with the production levels for individual furanocoumarin compounds possessing heritabilities ranging from 0.54 to 0.62. But the webworms are not passive targets for the plant defenses—they are able to metabolize these plant toxins at rates with heritabilities that range from 0.33 to 0.45. Of course, simply discovering that furanocoumarin production is heritable in parsnip, and that furnaocoumarin metabolism is heritable in webworms does not in itself reveal anything about the nature of coevolution. It merely provides evidence that coevolution is plausible. Berenbaum and Zangerl added two additional critical pieces of information:
(i) Both parsnips and webworms can be grouped into one of four phenotypic clusters, where each cluster corresponds to a particular mix of furanocoumarins in the case of plants (bergapten, xanthotoxin, isopimpinellin, and sphondin) and a particular mix of metabolic abilities on the part of the webworms (i.e., ability to metabolize the four furanocoumarin compounds).
(ii) When one samples associations of plants and their herbivores from populations along a latitudinal gradient, there is a remarkable matching of the frequencies of insect clusters and plant clusters. For example, if a plant population has a high percentage of a cluster characterized by high bergatpin production, the associated herbivore population would have a matching high percentage of the cluster corresponding to high bergaptin metabolism.
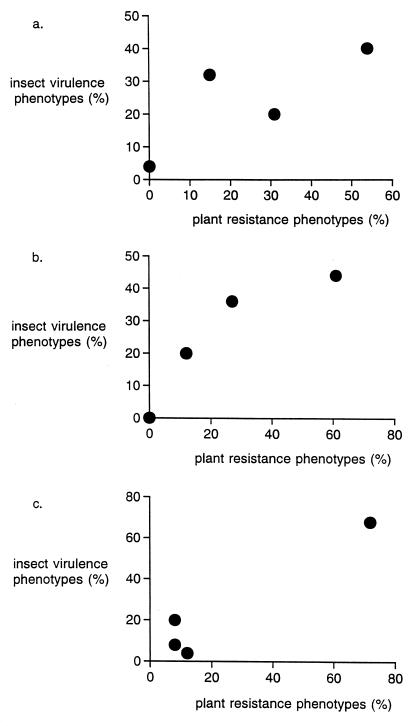
The correspondence between plant and herbivore populations in their pertinent chemical profiles was detected by sampling 26 seeds from plants in four different populations and assaying for the detoxification abilities of 25 webworm larvae associated with each parsnip sample. While Fig. 1 clearly indicates a “matching” between plant and herbivore populations in terms of their furanocoumarin production versus detoxification clusters, it does not reveal how close to “perfect” this matching actually is. If we view the frequency of each cluster type among plants as a template to be matched, we can ask how many webworm larvae would need to switch their phenotype cluster to make a perfect match? When we analyze the data underlying Fig. 1 in this way, we find that the matching is almost too good to be true: in one population only five of 25 larvae would need to switch phenotypes to make a perfect match, in the second population four of 25 larvae would need to be a different phenotype, and in the third population an amazingly low three of 25 larvae is all that would need to be in different clusters for a perfect match. Given the vagaries of sampling, these low number of “switches” required for a perfect match are extraordinary.
Figure 1.
Correlation between plant and insect phenotype frequency distributions for insect metabolism of furanocoumarins (horizontal axes) versus plant production of furanocoumarin compounds (vertical axes). Data obtained correspond to figure 4 of ref. 4: (a) Charleston, IL samples, (b) Winoma, MN samples, and (c) Peotone, IL samples.
But perhaps the most intriguing detail involves the one set of populations of parsnips and webworms that did not match at all (not shown in Fig. 1, but from samples from Urbana, IL). At the Urbana site there was no matching whatsoever between the frequencies of plant phenotypes and herbivore phenotypes; in particular, more than half of the webworm larvae would have had to switch to different phenotype clusters within the Urbana sample to obtain a perfect match. What was different about this “disobedient” Urbana association? In the three population associations depicted in Fig. 1, the insects were collected in very close proximity to the plants from which the seeds were collected. In contrast, for the mismatched Urbana data, the seeds were collected from only a tiny portion (2.5%) of a much larger tract of land from which the insects were gathered. If this lack of spatial congruence in sampling explains the mismatch, then the coevolution implicated in Fig. 1 must be an extremely localized phenomenon, which would have profound consequences. Clearly, the next step is to go back and sample the “exceptional” Urbana association in a way that better matches the insects to their plants, and to see whether a match such as those displayed in Fig. 1 subsequently is detected when sampling is spatially congruent.
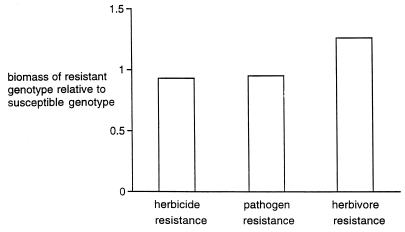
Apart from the pattern evident in Fig. 1, the other important lesson from the webworm/parsnip interaction is the hint of cycling selection as a key to the coevolutionary interaction. The different populations displayed in Fig. 1 each display different frequencies of furanocoumarin clusters, with the suggestion of a geographically varying cycling of selection, each population slightly out of phase with one another. Alternative explanations for the polymorphisms in plant defenses and insect counter-adaptations do not seem to fit the facts. For example, there are no clear environmental gradients underlying the geographic variation in phenotype frequencies, and chance founder effects seem an unlikely explanation because all phenotypes tend to be present at all sites (with none being lost by genetic drift). But while the data in Fig. 1 are consistent with a hypothesis of cyclic selection, there is no direct evidence to support this model of coevolution. One ingredient of the cyclic selection hypothesis that is absolutely essential is the presence of a “cost” associated with resistance or “defensive” traits. If there were no cost to resistance, then through time all plants would come to possess beneficial resistance traits (and we would not see the tremendous diversity of defensive levels evident in Fig. 1). The notion of costs and constraints is critical whenever a mechanistic model of coevolution is proposed, and indeed it is impossible to predict the course of coevolution without a clear hypothesis about these costs and constraints. While costs of resistance traits in plants are widely assumed, data regarding their frequency and strength are not so compelling. When Bergelson and Purrington (5) reviewed experiments aimed at detecting the costs of resistance traits, they reported costs that were surprisingly modest to nonexistent (Fig. 2). Clearly costs in these coevolutionary arms races are not a simple matter, leading Bergelson and Purrington (6) to conduct experiments that suggest the costs associated with resistance are substantially modified by environmental stress.
Figure 2.
Magnitude of costs of resistance measured as the biomass of near-isogenic lines of plants with resistance trait divided by biomass of same lines lacking the resistance trait; these measurements are made in the absence of the stress agent ameliorated by the resistance trait. No cost is represented by a value equal or higher than 1.0, whereas the presence of a cost is indicated by a value less than 1.0, with the magnitude of the cost proportional to how much below 1.0 the value is. Data are extracted from Bergelson and Purrington’s table 3 in ref. 5.
Studies of natural populations of plants and their enemies intimate a picture of coevolution in which, in lieu of escalating arms races, there may be a standoff—a sort of trench warfare with advances and retreats—all mediated by the complex costs associated with either resistance traits or virulence traits. Whether or not this idea can be firmly substantiated remains to be seen and will require both research at the molecular level and research that pinpoints the mechanisms underlying the costs suffered by resistant plants in the absence of its pathogen or herbivore. As mentioned above, if coevolution is a potent force, then novel associations between species should differ fundamentally in their character because of an absence of an evolutionary history. Because humans are increasingly creating novel associations between species, it is important that we understand whether the absence of an opportunity for coevolution exacerbates the numerous environmental risks associated with exotic species and biological invasions.
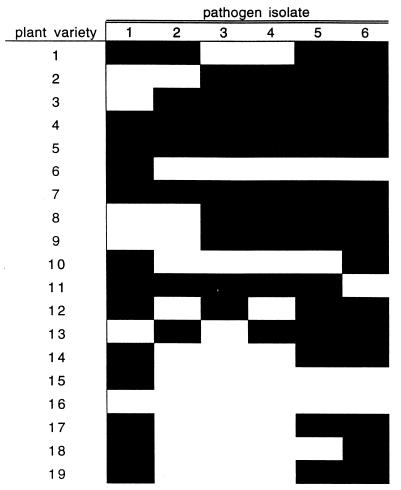
The matching of herbivores to their food plants evident in Fig. 1 foreshadows a well-known problem in agriculture and plant breeding. Pathogen and herbivore populations contain so much genetic variation that new crop varieties bred for resistance attributes, eventually end up selecting for “matched” enemy populations that have counter-adaptations capable of overriding the plant defenses. A major challenge for sustainable agriculture is the design of strategies for thwarting pathogen and herbivore evolution. In agriculture there is no coevolution in the traditional sense, because the genetic composition of crop populations is determined by humans (through breeding programs and patterns of seed distribution). However, the key questions arising from studies of natural plant-insect or plant-pathogen systems are also key questions in agriculture. For example, plant breeders seek resistance traits that do not cause reductions in crop yield. More ingeniously, plant breeders seek resistance traits for which the herbivore or pathogen countermeasures are likely to extract major costs from these enemies, which would make the evolution of virulence a slower and less certain process (7). Similarly, the population structure of crop pests is important to know, because this structure dictates the spatial scale at which we can expect herbivores and pathogens to adapt to different varieties (8). The amount of diversity in pathogen populations within a very small area can be staggering, with discouraging implications for the development of durable resistance in crops. For example, collecting from only two nearby nurseries in the Philippines, six distinct lineages of the fungal pathogen rice blast were identified (7). When 19 different rice varieties were exposed to these fungal lineages only three of the varieties were resistant to all six lineages (Fig. 3). One can easily imagine that if the rice blast was collected from a few more sites, additional disease lineages would have been uncovered, which would have been virulent to those three apparently resistant rice varieties. This genetic variation is the problem faced by crop breeders: a tremendous diversity in pathogens and herbivores so that virulence traits overriding just about any plant resistance factor are already present somewhere, and such negligible costs for these virulence traits that they do not readily disappear from pathogen or herbivore populations in the absence of selection favoring virulence (9).
Figure 3.
Resistance spectra for 19 rice cultivar lines tested against six rice blast lineages isolated from two nearby nurseries in the Philippines. Black represents resistant and white represents susceptible, so that only solid black rows are resistant across the board to all six blast lineages. The figure is derived from data presented in table 16.4 of ref. 7.
Regardless of whether coevolution involves an escalating arms race or the advances and retreats of trench warfare in the form of cyclical selection, the details of population structure and costs for resistance or virulence traits can be expected to govern its outcome. These same details will determine the feasibility of different plant breeding and genetic engineering technologies as routes toward sustainable agriculture (9). The tight phenotypic matching between parsnip webworm populations and populations of their food plants that Berenbaum and Zangerl (4) have discovered indicates that the enemies of plants may be able to fine-tune themselves so rapidly and effectively to plant defenses that agriculture just may have to accept some substantial level of crop losses as unavoidable.
Acknowledgments
I thank May Berenbaum for providing her raw data so that Fig. 1 could be drawn and Joy Bergelson for loaning me usage of her phrase “trench warfare.”
Footnotes
The companion to this Commentary is published on page 13743 in issue 23 of volume 95.
References
- 1.Pimentel D. Bull Entomol Soc Am. 1976;22:20–26. [Google Scholar]
- 2.Ehrlich P, Raven P. Evolution. 1964;18:586–608. [Google Scholar]
- 3.Flor H H. Phytopathology. 1955;45:680–685. [Google Scholar]
- 4.Berenbaum M R, Zangerl A R. Proc Natl Acad Sci USA. 1998;95:13743–13748. doi: 10.1073/pnas.95.23.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergelson J, Purrington C. Am Nat. 1996;148:536–558. [Google Scholar]
- 6.Bergelson, J. & Purrington, C. (1999) Am. Nat., in press. [DOI] [PubMed]
- 7.Ziegler R, Tohme J, Nelson R, Levy M, Correa-Victoria F. In: Rice Blast Disease. Ziegler R, Teng P, Leong A, editors. Wallingford, U.K.: Commonwealth Agricultural Bureau International; 1994. pp. 267–292. [Google Scholar]
- 8.Leung H, Nelson R, Leach J. Adv Plant Pathol. 1993;10:157–205. [Google Scholar]
- 9.Mundt C. Phytopathology. 1990;80:221–223. [Google Scholar]