Abstract
Transcriptional profiling highlighted a subset of genes encoding putative multidrug transporters in the pathogen Bacillus cereus that were up-regulated during stress produced by bile salts. One of these multidrug transporters (BC4707) was selected for investigation. Functional characterization of the BC4707 protein in Escherichia coli revealed a role in the energized efflux of xenobiotics. Phenotypic analyses after inactivation of the gene bc4707 in Bacillus cereus ATCC14579 suggested a more specific, but modest role in the efflux of norfloxacin. In addition to this, transcriptional analyses showed that BC4707 is also expressed during growth of B. cereus under non-stressful conditions where it may have a role in the normal physiology of the bacteria. Altogether, the results indicate that bc4707, which is part of the core genome of the B. cereus group of bacteria, encodes a multidrug resistance efflux protein that is likely involved in maintaining intracellular homeostasis during growth of the bacteria.
Introduction
Bacillus cereus sensu lato (the Bacillus cereus group of bacteria) are Gram-positive rod-shaped bacteria that readily form endospores under unfavorable growth conditions. They comprise six phenotypically diverse, but genetically closely related, species - B. cereus sensu stricto, B. anthracis, B. thuringiensis, B. weihenstephaniensis, B mycoides and B. pseudomycoides [1]. Among these bacteria, the last three species are generally considered non-pathogenic. B. thuringiensis is an insect pathogen commercially used as a biopesticide. B. anthracis is notorious for being the agent causing anthrax in humans and animals [2] as well as for its potential role in biological warfare including bioterrorism [3]. B. cereus sensu stricto (from now on referred to as B. cereus) is primarily associated with food borne, self-limiting gastrointestinal infections. It is also an opportunistic pathogen that causes local infections following trauma as well as systemic diseases in predisposed patients [4]–[6]. Although B. cereus is commonly reported as a soil bacterium, the life style is not completely understood. Spores have been isolated from different types of soil [7] as well as faeces of healthy humans [8], [9]. There are reports of B. cereus found in the rhizosphere of plants [10] and guts of invertebrates [11]. The current view is that soil-dwelling spores germinate and grow, either in an animal host or in the rhizosphere of plants, suggesting that the natural lifestyle of B. cereus often involves symbiotic or pathogenic interactions with animals and plants. The diverse environments where B. cereus is found suggest that these bacteria survive and grow in competition with other microorganisms as well as in a range of challenging conditions such as different temperatures, extremes of pH and toxic compounds.
In order to survive challenging extracellular conditions, it is essential to maintain homeostasis in the intracellular environment. This is partly achieved through the action of different types of membrane proteins that transport nutrients and ions into the cell and excrete waste products and toxic compounds from the cell. According to the transportdb database [12], [13] the B. cereus type strain (ATCC14579) has 390 putative membrane transporters, including approximately 100 predicted multidrug transporters. The high number of predicted membrane transporters encountered in B. cereus species may well reflect the diverse environments where they are found.
Multidrug transporters are energy dependent membrane proteins that extrude a wide array of structurally and functionally dissimilar compounds. Multidrug transporters play a significant role in resistance of bacteria to antimicrobial compounds [14]–[16]. It has also been suggested that multidrug transporters have additional/alternative roles in the normal physiology of bacteria, facilitating survival in their ecological niche [17]–[19]. Multidrug transporters are categorized into five superfamilies [20] i. Major Facilitator Superfamily (MFS), ii. Resistance nodulation cell division (RND), iii. Small multidrug resistance (SMR), iv. Multidrug and toxic compound extrusion (MATE) and v. ATP-binding cassette (ABC). The ABC transporters use the energy from ATP hydrolysis directly to force drug expulsion. The other four families of multidrug transporters are secondary metabolite transporters driving drug efflux with either the proton or sodium gradients that are established across the cytoplasmic membrane of cells during respiration (or by ATP hydrolysis in strictly fermentative organisms). Although at present, most of the transporters encoded in B. cereus remain uncharacterized, it was shown that a set of membrane transporters, including several putative multidrug transporters were upregulated in B. cereus ATCC14579 when challenged with sub-lethal concentrations of bile salts [21]. In addition, a partly overlapping set of transporters was induced by sub-lethal concentration of acetic acid [22]. Bacteria would encounter similar stress during passage through the gastrointestinal tract, suggesting that these transporters could be part of a survival system expressed in bacteria in order to inhabit such hostile environments.
In this study, we initiated the biochemical and functional characterization of one putative multidrug transporter (BC4707) shown to be induced by sub-lethal concentrations of both bile salts and acetic acid; by cloning, expressing and characterizing its activities in an E. coli host. In parallel, we deleted the gene (bc4707) and examined its role in the B. cereus ATCC14579 under non-stressful conditions such as exponential growth in LB-medium as well as stressful conditions including bile salts stress, metabolic stress, antibiotics and low pH.
Results
BC4707 is not essential for B. cereus to survive bile salts stress
Induction of bc4707 transcription in B. cereus upon bile salt stress [21], suggested a role in bile salts efflux. To test the role of BC4707 in bile salts protection, a markerless Δbc4707 mutant was constructed in B. cereus ATCC14579 and the two strains were compared under growth in LB medium as well as under bile salts stress. It was revealed that although growth of the bacteria was repressed in the prescence of 50 µg/ml bile salts the Δbc4707 mutant and the wild type grow in a similar fashion (Fig. 1). This result indicated that although bc4707 expression is induced under bile salts stress, the protein is not essential for bile salts protection.
Figure 1. Comparison of the growth of Bacillus cereus ATCC14579 and its isogenic Δbc4707 mutant.
Growth was assayed for the wild type (circles) and the mutant (squares) by monitoring the absorbance at 600 nm in LB medium (solid lines; closed symbols) and during 50 µg/ml bile salts stress initiated at OD600 of 0.5 (dashed lines; open symbols). Shown are averages and standard deviations of two independent experiments.
Transcriptional profiling suggests that BC4707 is not involved in protection of B. cereus against bile salts stress
Considering that bc4707 expression was induced by bile salts stress whereas BC4707 was non-essential for growth in the presence of sub-lethal concentrations of bile salts, we hypothesized that a compensatory mechanism was induced in the Δbc4707 mutant under such conditions. In order to test for compensatory mechanisms, a series of microarray analyses were performed comparing the transcriptional profiles of the wild type B. cereus and the Δbc4707 mutant under bile salts stress. Bacteria were grown to an OD600 of 0.5. At this point bile salt was added to a concentration of 50 µg/ml and the cultures were incubated for 15 additional minutes. RNA was prepared for each strain at OD600 of 0.5 (WTOD0.5, mutantOD0.5) as well as after 15 min bile salts stress (WTBS15, mutantBS15). To determine the bile salt induced stress response in LB medium, the mRNA content of the bile salt stressed cells was compared to the mRNA content of the pre-stressed cells by microarray analysis (i.e. WTBS15 was compared to WTOD0.5 and mutantBS15 was compared to mutantOD0.5. Arrayexpress accession numbers E-MEXP-3535 and E-MEXP-3536 respectively). These microarray analyses demonstrated a substantial stress response in both the wild type and the Δbc4707 mutant (Table S1), with significant differential regulation of approximately 10% of the genome, including at least 10 efflux proteins (counting bc4707 in the wt strain, see also Kristoffersen et al. [21]). The transcriptional profiles of the wild type and mutant after 15 min bile salts stress were similar (Table S1) with few and negligible differences between the two data sets. To identify potential compensatory mechanisms that allow the Δbc4707 mutant to grow similar to the wild type under bile salt stress, the mRNA content of the Δbc4707 strain after bile salts stress for 15 min (mutantBS15) was compared to the corresponding mRNA of the wild type (WTBS15). This microarray analysis (Arrayexpress accession number E-MEXP-3529) confirmed that there are few differences between the two strains in the bile salts induced stress response (Table 1). Four genes were differentially expressed more than 1.5-fold. Three of these genes were up-regulated in the Δbc4707 mutant: bc2989 annotated as a hypothetical protein; bc4111, which, from sequence similarity, is described as a bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrase II protein, involved in riboflavin synthesis; and bc4904, annotated as a hydrolase of the alpha/beta fold family. Furthermore, the cold shock protein CspD (bc4859), was down regulated in theΔbc4707 mutant compared to the wild type. This indicates that the absence of BC4707 does not induce a major compensatory response to bile salts stress at the mRNA level.
Table 1. Transcriptional profile of B. cereus ATCC14579 Δbc4707 (AH1598) compared to wild type (AH1709) grown to logarithmic stage (OD600∼0.5) in rich medium followed by bile salt stress for 15 min.
| Gene | Predicted protein function | Δbc4707/WT |
| (fold change) | ||
| bc2989 | Hypothetical protein | 1.68 |
| bc4111 | Bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrase II protein | 1.51 |
| bc4707 | drug resistance transporter, EmrB/QacA family | 0.32 |
| bc4859 | Cold shock protein CspD | 0.61 |
| bc4904 | Hydrolase, alpha/beta fold family | 1.96 |
BC4707 is not involved in the acid stress defense of B. cereus
According to Mols et al. bc4707 is slightly up-regulated under sub-lethal acetic acid stress [22], indicating a potential role for BC4707 in maintaining the intracellular pH of B. cereus in an acidic environment. To test this, the experiments described by Mols et al. were reproduced. Bacteria were grown in a neutrally buffered BHI medium to mid-logarithmic phase, at which point the medium was acidified and the OD was measured for an additional 4.5 hours. No difference was observed between the wild type and Δbc4707 mutant (data not shown), indicating a non-essential role for BC4707 under acetic acid shock.
Inactivation of the bc4707 gene renders B. cereus more susceptible to norfloxacin
To elucidate the role of BC4707 in B. cereus drug tolerance, the markerless knock out mutant was compared to the wild type in susceptibility assays (Table 2). The Δbc4707 mutant and wild type strains only differed in the sensitivity to one (norfloxacin) of 12 toxic compounds tested. The two-fold difference in susceptibility to norfloxacin was consistent between the wild type and Δbc4707 mutant. Growth curves of the wild type and mutant supplemented with a sub-lethal concentration of norfloxacin, demonstrated a moderate, but consistent reduction in growth of the Δbc4707 mutant compared to the wild type (Fig. 2). Taken together, BC4707 is involved in norfloxacin tolerance of B. cereus ATCC14579.
Table 2. Susceptibilities of B. cereus ATCC14579 (wild type) compared to its isogenic Δbc4707 mutant.
| MIC (µg/ml) | |||
| B. cereus ATCC14579 | |||
| Compound | Wild type | Δbc4707 | § |
| Chloramphenicol | 2.5 | 2.5 | 1 |
| Ciprofloxacin | 0.32 | 0.32 | 1 |
| Bile salts | 125 | 125 | 1 |
| Deoxycholate | 250 | 250 | 1 |
| SDS | 100 | 100 | 1 |
| Crystal violet | 0.1 | 0.1 | 1 |
| Kanamycin | 15 | 15 | 1 |
| Nalidixic acid | 1.875 | 1.875 | 1 |
| Erythromycin | 0.2 | 0.2 | 1 |
| Ethidium bromide | 40 | 40 | 1 |
| Norfloxacin | 1.25 | 0.625 | 2 |
| Tetracycline | 2.5 | 2.5 | 1 |
represents the fold difference in MIC between the wild type B. cereus ATCC14579 and its isogenic Δbc4707 mutant.
Figure 2. Growth of B. cereus ATCC14579 and Δbc4707 mutant under stress with sublethal concentration of norfloxacin.
Over night cultures were diluted 1∶100 and grown to mid-logarithmic phase. These cultures were diluted to OD600 of 0.02 in 25 ml LB and norfloxacin was added. The cultures were incubated for 240 min at 30°C and 220 rpm shaking. The growth curves show averages and standard deviations of two independent experiments.
Microarray analysis suggests a physiological role for BC4707 in exponential growth of B. cereus
Although bioinformatics analysis has predicted BC4707 to be a multidrug transporter, susceptibility testing of the Δbc4707 mutant only identified norfloxacin as a potential substrate (Table 2). A few multidrug transporters have been shown to have physiological functions in addition to multidrug efflux [17], [18], [23]–[25]. The large number of putative multidrug transporters in B. cereus suggests alternative or additional functions to multidrug efflux for some of these transporters, possibly under conditions that do not impose exogenous stress on the bacteria. We therefore measured the transcription level of bc4707 under logarithmic growth in rich medium. Quantitative RT-PCR analysis at different time points of B. cereus ATCC14579 growing in LB medium indicated that although the absolute expression level of bc4707 is relatively low (data not shown) it is increasingly expressed during logarithmic phase up to the onset of the transition preceding stationary phase in B. cereus (Fig. 3). The increasing expression of bc4707 during logarithmic phase supported the hypothesis of a potential role for BC4707 under exogenously imposed non-stressful growth conditions. Growth of the Δbc4707 mutant in LB-medium, however, was identical to the growth of the wild type strain (Fig. 1), suggesting a non-essential role for BC4707 in rich medium. Potentially, overlapping functions of constitutively expressed efflux proteins may conceal the function of the moderately expressed BC4707. Alternatively, a compensatory mechanism may be induced that masks the necessity for the BC4707 efflux protein. Microarray analysis was therefore carried out in mid logarithmic phase at OD600 0.5 of cells growing in LB medium (WTOD0.5, mutantOD0.5), comparing the wild type and the BC4707 deletion mutant (Arrayexpress accession number E-MEXP-3537). The results (Fig. 4 and Table 3) revealed that although the Δbc4707 mutant and the wild type displayed the same growth rate, 27 genes were differentially regulated, at least 1.5 fold at OD600 0.5. Thirteen genes were up-regulated, whereas 14 genes were down-regulated, thus indicating a potential role for BC4707 in the normal physiology of B. cereus ATCC14579 under exogenously non-stressful conditions, most likely by relieving the cell of toxic metabolites.
Figure 3. Growth of B. cereus ATCC14579 and expression analysis of bc4707.
A. Bacteria were grown in LB-medium. Circles indicate OD600 measurements. Bacteria were harvested for RNA extraction (open circles). B. Gene expression analysis of bc4707 by qRT-PCR. Averages and standard deviations from three experiments are shown. The relative expression at different OD600 values is compared to the expression level at OD600 of 0.05. 16S RNA was used as a control to normalize the data.
Figure 4. COG distribution of differentially regulated genes in exponentially growing Δbc4707 mutant and wild type.
The proportion of genes belonging to a COG class is shown as percentage of the total number of differentially regulated genes. Open bars indicate downregulated genes in the mutant compared to the wild type , solid bars represent upregulated genes. i. the proportion of regulated genes that are poorly characterized, ii. the proportion of genes with a predicted function in cellular processes and signaling, iii. the proportion of genes involved in information storage and processing, iv. the proportion of regulated genes involved in metabolism and transport.
Table 3. Transcriptional profile of B. cereus ATCC14579 Δbc4707 (AH1598) compared to wild type (AH1709) under logarithmic growth (OD600∼0.5) in rich medium.
| Gene | Predicted protein function | Δbc4707/WT |
| (fold change) | ||
| bc0579 | malate sodium symporter | 2.06 |
| bc0580 | malate dehydrogenase | 1.61 |
| bc0659 | ribose operon repressor | 1.54 |
| bc0660 | ribokinase | 1.52 |
| bc0662 | ribose ABC transporter, ATP-binding protein | 1.52 |
| bc0872 | cystine-binding protein | 0.41 |
| bc0873 | cystine permease protein | 0.39 |
| bc0874 | cystine ATP-binding protein | 0.44 |
| bc0909 | oligopeptide ABC transporter, permease protein | 0.63 |
| bc0942 | hypothetical protein | 0.65 |
| bc1034 | glycerol uptake facilitator protein | 2.12 |
| bc1035 | glycerol kinase | 1.53 |
| bc1251 | dihydrolipoamide acetyltransferase | 1.59 |
| bc1252 | 2-oxoglutarate dehydrogenase, E1 component | 1.57 |
| bc1739 | proton/sodium-glutamate symporter | 1.65 |
| bc2045 | hypothetical protein | 1.68 |
| bc2300 | oxalate∶formate antiporter, putative | 1.57 |
| bc3718 | PTS system, fructose-specific IIABC component | 0.60 |
| bc3719 | 1-phosphofructokinase | 0.60 |
| bc3720 | transcriptional regulator, DeoR family | 0.41 |
| bc3760 | 6-phospho-beta-glucosidase | 0.59 |
| bc4242 | proton/sodium-glutamate symporter | 0.47 |
| bc4366 | cystathionine beta-lyase | 0.52 |
| bc4367 | cystein synthase A | 0.50 |
| bc4368 | 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase | 0.56 |
| bc4369 | Dimethyladenosine transferase | 0.47 |
| bc4396 | molybdopterin biosynthesis protein | 0.64 |
| bc4707 | drug resistance transporter, EmrB/QacA family | 0.38 |
| bc4789 | S-ribosylhomocysteinase | 0.65 |
| bc5239 | enterotoxin/cell wall binding protein | 1.80 |
Bold text indicates that the gene is differentially regulated more than or equal to two times (i.e. the values are ≥2 or ≤0.5).
Phenotype analyses did not identify a condition where BC4707 is essential
To screen for potential substrates or conditions where BC4707 is essential, or has a major role, Phenotype MicroArray™ experiments were conducted. In these experiments, the metabolic activity was compared between the Δbc4707 mutant and the wild type strain in 672 different conditions, including peptide nitrogen sources, carbon sources, and pH. There were no consistent differences in metabolic activity between the wild type and mutant under any of the conditions (PM plates 1, 2, 6–8 and 10) testing for carbon source, peptide nitrogen source or pH preferences (data not shown).
To complement the phenotype microarray, growth tests were conducted in minimal medium using different compounds as sole carbon and/or nitrogen sources. The growth experiments were initially carried out in liquid cultures, but the OD600 measurements failed to give a reading, since bacteria exclusively grew as aggregates on the glass at the air-liquid interface. To circumvent this problem, subsequent experiments were carried out on GGGS-agar plates. In agreement with the phenotype microarrays, no consistent differences were found between the Δbc4707 mutant and the wild type in growth behaviour (data not shown). There were indications of a small difference in motility behavior on GGGS plates, where the mutant appeared to be more motile than the wild type. However, a consistent difference between the wild type and mutant could not be confirmed on swarming or swimming plates (data not shown).
Expression of the bc4707 gene in Escherichia coli
Although bc4707 expression is induced under bile salts stress [21] as well as under acetic acid shock [22], the protein is not essential for growth under such conditions (Fig. 1, Table 1 and data not shown). Sequence similarity has classified BC4707 as a multidrug transporter of the Major Facilitator Superfamily [12]. Deletion of bc4707 from B. cereus ATCC14579 only affected growth of the mutant in one of 12 tested toxic compounds. It is likely that redundancy among efflux proteins conceal an eventual multidrug transport ability of BC4707. A common and sensitive method to demonstrate multidrug transport capacity in a membrane protein is by testing for increased drug tolerance after heterologues expression in an E. coli strain that is hypersensitive to multiple drugs. For this purpose, the gene (bc4707) was cloned into the E. coli expression vector pTTQ18 as described previously [26]. The resulting plasmid potentiates IPTG inducible heterologous expression of a BC4707-RGSHis6 tag fusion protein in E. coli.
Induction of protein expression with IPTG yielded a relatively high level of BC4707-RGSHis6 in the membranes of the expression host BL21 (DE3), as demonstrated by Coomassie Blue staining as well as Western blotting using an antibody raised against the RGSHis6 tag (Fig. 5 a and b). The observed molecular weight of the BC4707-RGSHis6 protein after SDS-PAGE (Fig. 5) was substantially lower (35 kDa) than that (52.5 kDa) predicted from the amino acid sequence, a common feature for membrane proteins [27], [28]. The protein was solubilized from the inner membrane of the E. coli host using the detergent dodecyl–beta-D-maltoside (DDM) and purified to approximately 90% homogeneity by immobilized metal affinity chromatography (IMAC) as described in Materials and Methods (Fig. 5 c). The impurities detected by Coomassie staining in the purified protein fraction, likely represent histidine-containing host proteins that co-purified with the target protein [29]. Two protein bands in addition to the target one of 35 kDa repeatedly showed up on Western blots (Fig. 5 d). The band of higher molecular weight than the target, probably represented the completely unfolded form of BC4707-RGSHis6 [27], whereas the faint band of lower molecular weight is likely a degradation product of the target protein. Edman degradation confirmed that the N-terminal amino acid sequence of the protein that was detected as the major band on the Coomassie stained gel was the B. cereus BC4707 protein (data not shown).
Figure 5. Expression and purification of BC4707-RGSHis6.
Protein expression was induced for 3 hours by addition of IPTG (1 mM) to exponentially growing bacteria. Cells were harvested and mixed membrane preparations were made from each culture. The proteins in the mixed membrane fractions prepared by the water-lysis technique [47] were separated by SDS-PAGE and detected by: (A) Coomassie Blue stain; or (B) Western blotting with an antibody to the RGSHis6-tag (+ and − represent mixed membrane preparations of induced and uninduced cells, respectively). Purification of BC4707-RGSHis6 was done by immobilized metal affinity chromatography (IMAC). Staining with Coomassie Blue (C) shows the enrichment and purity of BC4707-RGSHis6 at different stages of purification. The gel was loaded as follows: 1) molecular weight marker 2) inner membranes solubilized in dodecyl-beta-D-maltoside detergent; 3) supernatant of the solubilized inner membranes after ultracentrifugation; 4) pellet of the solubilized inner membrane fraction after ultracentrifugation; 5) flow-through after IMAC-binding; 6) elution fraction (5 µg protein). Western blot (D) of the final elution fraction (7.5 µg protein). The sizes of the bands in the molecular weight markers are indicated.
The integrity of the purified protein was further demonstrated by circular dichroism (CD) measurements. Negative bands at 208 nm and 222 nm and a positive band at 193 nm indicated predominantly α-helical secondary structure (Fig. 6), which is consistent with the predicted topology of BC4707 of 14 α-helices (data not shown). These experiments demonstrated that BC4707 was expressed as a full sized RGSHis6 fusion protein in E. coli and that the protein localized to the membrane verifying the integrity and localization of the protein upon heterologues expression.
Figure 6. SRCD analysis of purified BC4707-RGSHis6 protein.
CD spectral analysis of BC4707-RGSHis6 was performed over 260-170 nm at 1 nm intervals with an integration time of 1 s. The spectrum shown is buffer subtracted and is an average of 10 scans. The dashed line represents the HT voltage at the photomultiplier.
BC4707 is a multidrug transport protein
E. coli DH5α ΔacrAB contains a deletion in acrA encoding a membrane fusion protein and acrB encoding a multidrug efflux system protein causing inactivation of the RND-transporter complex, which results in an E. coli mutant that is hypersensitive to many toxic compounds compared to the wild type [30]–[33]. This permits sensitive functional characterization of multidrug transporters following their expression from a plasmid [34], [35]. The presence of pBC4707 in E. coli DH5α ΔacrAB resulted in improved tolerance to 3 of 12 tested xenobiotics compared to the strain carrying the empty vector, pTTQ18 (Table 4). Increased expression of BC4707-RGSHis6 by IPTG induction resulted in even higher tolerance to the fluoroquinolones ciprofloxacin and norfloxacin, whereas no further tolerance to kanamycin was observed (Table 4). It is possible that the increased tolerance to kanamycin in the pBC4707 carrying strain is due to leaky expression of BC4707-RGSHis6 prior to the addition of kanamycin, which potentiates drug expulsion. The reason there was no further increase in tolerance after IPTG induction could be that since kanamycin and IPTG was added simultaneously inhibition of protein translation by kanamycin could have prevented further increase of BC4707-RGSHis6 in the membrane.
Table 4. Susceptibilities of E. coli DH5α ΔacrAB expressing bc4707 (pBC4707) compared to its vector control (pTTQ18) under uninduced (LB (0 mM IPTG)) and induced (LB (0.05 mM IPTG)) conditions.
| MIC (µg/ml) | MIC (µg/ml) | |||||
| LB (0 mM IPTG) | LB (0.05 mM IPTG) | |||||
| E. coli DH5α ΔacrAB | E. coli DH5α ΔacrAB | |||||
| Compound | pTTQ18 | pBC4707 | § | pTTQ18 | pBC4707 | § |
| Chloramphenicol | 0.3125 | 0.3125 | 1 | 0.3125 | 0.3125 | 1 |
| Ciprofloxacin | 0.01 | 0.04 | 4 | 0.01 | 0.16 | 16 |
| Bile salts | 3200 | 3200 | 1 | 3200 | 3200 | 1 |
| Deoxycholate | 1600 | 1600 | 1 | 1600 | 1600 | 1 |
| SDS | 50 | 50 | 1 | 50 | 50 | 1 |
| Crystal violet | 0.25 | 0.25 | 1 | 0.25 | 0.25 | 1 |
| Kanamycin | 2.5 | 10 | 4 | 2.5 | 10 | 4 |
| Nalidixic acid | 30 | 30 | 1 | 30 | 30 | 1 |
| Erythromycin | 3.13 | 3.13 | 1 | 3.13 | 3.13 | 1 |
| Ethidium bromide | 2.5 | 2.5 | 1 | 2.5 | 2.5 | 1 |
| Norfloxacin | 0.08 | 0.32 | 4 | 0.08 | 1.28 | 16 |
| Tetracycline | 1.56 | 1.56 | 1 | 1.56 | 1.56 | 1 |
represents the fold difference in MIC between the E. coli DH5α ΔacrAB (pBC4707) strain compared to the vector control (DH5α ΔacrAB (pTTQ18)).
Transport by BC4707 utilizes metabolic energy
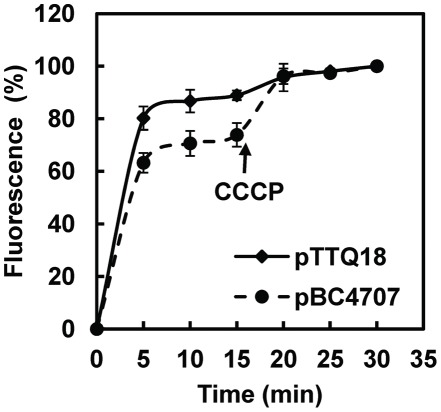
The classification of BC4707 as an MFS protein implies that it is a secondary active transporter for efflux driven by the cation gradient, probably H+ built up during cell respiration [36]. To test if the function of BC4707 is dependent on an intact proton gradient a norfloxacin accumulation assay was performed in E. coli (Fig. 7). Cells expressing BC4707-RGSHis6 had lower fluorescence intensity compared to cells carrying the empty plasmid pTTQ18, indicating that the presence of BC4707-RGSHis6 prevents accumulation of norfloxacin in the cells, most likely via active efflux of the drug (Fig. 7). Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) is a protonophore that destroys the proton gradient [37]–[39]. Addition of CCCP to the assay buffer resulted in increased accumulation of norfloxacin in cells, and the increase in accumulation is relatively more pronounced in the cells expressing BC4707-RGSHis6 (Fig. 7), indicating that efflux mediated by BC4707-RGSHis6 is dependent on an intact proton gradient. Taken together these data demonstrate that BC4707, as predicted by in silico analysis, is a multidrug efflux protein driven directly or indirectly by the proton motive force.
Figure 7. Accumulation of Norfloxacin in E. coli DH5α ΔacrAB (pBC4707) and DH5α ΔacrAB (pTTQ18).
The fluorescence of intracellularly accumulated norfloxacin was measured at 280 nm excitation and 445 nm emission in the supernatant of bacterial extracts. Bacteria expressing BC4707-RGSHis6 are indicated by circles whereas bacteria carrying the empty vector are shown as diamonds. The assay was started by the addition of 100 µM norfloxacin at time 0 min. At 15 min, the protonophore CCCP (100 µM) was added to disrupt the proton motive force. Shown are averages and standard deviations from two independent experiments.
Transcription analyses demonstrate that the expression level of bc4707 is not altered in response to sub-lethal concentrations of selected drugs
The susceptibility assay of E. coli DH5α ΔacrAB indicate that BC4707 has the capability to transport xenobiotics such as fluoroquinolones and possibly kanamycin, whereas the susceptibility assay in B. cereus ATCC14579 showed involvement of BC4707 in norfloxacin tolerance. We therefore tested the transcriptional level of BC4707 in B. cereus in the presence and absence of the drugs. B. cereus ATCC14579 was grown in LB medium to logarithmic phase (OD600 of 0.5), norfloxacin, ciprofloxacin or kanamycin were added and the culture was incubated for 15 minutes followed by RNA extraction. Quantitative RT-PCR-analysis (using 16S RNA as reference) indicated that bc4707 transcription was not significantly affected by stress with these antibiotics compared to an unstressed control (Fig. 8). Similar results were obtained using GatB RNA [40] as reference (data not shown).
Figure 8. Relative expression of bc4707 in stressed cells compared to unstressed cells.
The expression level of bc4707 was measured by qRTPCR and was normalized against the expression of16S RNA. The values shown are ratios of the normalized expression levels of cells stressed with antibiotics for 15 min compared to unstressed cells. Shown are averages and standard deviations of at least two independent experiemnts each done with two technical replicates.
Discussion
Previous results mapping the stress response induced by bile salt in B. cereus [21] indicated a role for BC4707 in bile salts tolerance. By contrast, phenotype analyses comparing the response of B. cereus ATCC14579 and its isogenic Δbc4707 mutant to bile salt-induced stress showed that BC4707 is not vital for B. cereus under these conditions (Fig. 1 and Table 2). This suggests that the cells either compensate for the absence of BC4707 under bile salt stress or it is a minor contributor to the bile salt protection under the conditions tested. Microarray analyses comparing the bile salts stress responses of the wild type and Δbc4707 mutant did not reveal major compensatory alterations in gene expression (Table 1). The general conclusion is therefore that BC4707 does not have a significant role in the defense of B. cereus against bile salts stress.
Similarly, it has been shown that bc4707 expression is induced under acetic acid shock [22] but direct comparison of the Δbc4707 mutant and the wild type failed to demonstrate a major role for BC4707 under this condition (data not shown). Taken together, these experiments show that although transcriptional data suggested participation of BC4707 in protecting B. cereus ATCC14579 against bile salt stress and acetic acid stress, which are two conditions that enteropathogenic bacteria theoretically would encounter, it is not essential under these conditions.
Functional characterization of BC4707 in E. coli DH5α ΔacrAB confirmed the in silico prediction that it is a secondary metabolite multidrug transporter (Fig. 7, Table 4). Yet, the deletion of bc4707 in B. cereus failed to confirm a major role in multidrug transport (Table 2). The Δbc4707 strain had a somewhat increased susceptibility to only one (norfloxacin) of 12 tested xenobiotics compared to the wild type. It is likely that the reason for this discrepancy is due to the vast number of multidrug transporters in B. cereus; these may offer elasticity to the antibiotic stress response and conceal the contribution of the moderately expressed BC4707 under the tested conditions. Moreover, transcriptional data showed that BC4707 expression is not induced by norfloxacin, ciprofloxacin or kanamycin stress, indicating that BC4707 only protects against these antibiotics when it is already present in the membrane. Therefore it is possible that the involvement of BC4707 in multidrug transport is small under specific conditions, but may be physiologically relevant to the bacteria under other conditions. The effect of bc4707 inactivation on norfloxacin tolerance in addition to the increased tolerance of E. coli DH5α ΔacrAB expressing BC4707-RGSHis6 suggests that BC4707 is involved in protecting the cells against the action of at least some fluoroquinolone compounds. The potential involvement of membrane transport proteins in fluoroquinolone resistance by members of the Bacillus cereus group of bacteria is becoming increasingly evident. It was recently reported that during ciprofloxacin stress of B. anthracis, the gene GBAA0834, a TetR type transcriptional regulator was a hotspot for mutations leading to ciprofloxacin resistance. As a consequence of mutations leading to disruption of GBAA0834, several adjacent multidrug transporters were up regulated and were probably responsible for the increased ciprofloxacin tolerance [41]. These findings are of concern, since fluoroquinolones are the first line of antimicrobial compounds used in the treatment of B. anthracis infections [42], [43].
Although norfloxacin transport is an important function for BC4707 from a clinical perspective, it is likely not the original native function. A BLAST search showed that chromosomally encoded orthologues of BC4707 with high amino acid identity (>91%) exist in all 88 members of the B. cereus group of bacteria that have been sequenced to date (data not shown), indicating this gene as being part of the core genome with an ancient and conserved function. Fluoroquinolone compounds, on the other hand are synthetic drugs, first introduced in the early 1960s. Multidrug transporters have been shown to have additional fundamental physiological functions in addition to the multidrug transport capability [17]–[19], [23]–[25]. Furthermore, it has been speculated that environmental stress from conditions in the natural habitat of bacteria may be responsible for the conservation of multidrug transporters and the spread of multidrug resistant bacteria [44]. It is likely that the fluoroquinolone transport capability of BC4707 is an additional effect to the original native function. Based on this it was hypothesized that the reason BC4707 is conserved in all members of the B. cereus group is due to an indigenous function that is important in the natural environment of the bacteria.
Attempts using phenotype experiments comparing the wild type and Δbc4707 mutant under a range of stressful conditions, including metabolic stress, different salt conditions and pH, failed to elucidate an essential indigenous role of BC4707 in the normal physiology of B. cereus. The microarray data from comparison of the wild type and Δbc4707 mutant during exponential growth in rich medium showed differential regulation of a set of genes involved in metabolism of cysteine, as well as alternative carbon sources (Table 3). We hypothesized that the differential regulation of these genes may reflect a compensatory mechanism to counteract accumulation of toxic metabolites. It is possible that this compensation is enough to sustain normal growth of the mutant in the absence of BC4707. Alternatively, the difficulty in identifying an alternative role for BC4707 in addition to multidrug transport is due to redundancy among the encoded multidrug transporters at the conditions tested. It is likely that inactivation of additional efflux proteins is required in order to clarify the role of BC4707 in the normal physiology of B. cereus as well as in the potential multidrug transport.
Materials and Methods
Bacterial strains and plasmids used in this study are listed in table 5. If not otherwise stated, culture conditions were LB medium at 30°C and 220 rpm.
Table 5. Strains and plasmids used in this study.
| Strains or plasmids | Relevant genotype or description | Source or reference |
| Bacillus cereus strains | ||
| ATCC14579 | Wild type | Laboratory collection |
| AH1598 | ATCC14579 Δbc4707 (pBClin15−) | This study |
| AH1709 | ATCC14579 (pBclin15−) | This study |
| Escherichia coli strains | ||
| BL21 (DE3) | F− ompT hsdS B(rB − mB −) gal dcm (DE3) | Novagen |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ (lacZYA-argF)U169 (φ80lacZΔM15) | Laboratory collection |
| AH1805 | DH5α ΔacrAB | This Study |
| Plasmids | ||
| pTTQ18 | E. coli expression plasmid, Ampr | [26] |
| pBC4707 | pTTQ18[PlacIQ/bc4707] | This study |
| pBKJ236 | E. coli-Bacillus shuttle vector, Eryr, Ori(Ts) | [46] |
| pBKJ236_bc4707 | pBKJ236[Δbc4707] | This study |
| pBKJ223 | [Pamy/IsceI], Tetr | [46] |
Construction of mutants
The E. coli DH5α ΔacrAB mutant was constructed according to the λ-Red recombinase based method described by Datsenko and Wanner [45]. Briefly, approximately 300 ng of processed PCR product was introduced by electroporation into E. coli DH5α containing pKD46, which expresses λ-Red recombinase. The PCR product was amplified from pKD3 and consists of a chloramphenicol cassette flanked by FRT-sites and approximately 40 bp homologous to the 5′ end of acrA as well as the 3′ end of acrB. Primers used in the study are listed in Table 6. Recovered colonies carrying the chloramphenicol cassette in place of acrAB were purified twice on LB agar containing chloramphenicol (20 µg/ml). A markerless mutant was constructed by introducing the pCP20 plasmid encoding a FLP-recombinase into the chloramphenicol resistant mutant followed by screening for antibiotic sensitive mutants. Recovered colonies were purified twice on LB agar. The mutation was confirmed by sequencing.
Table 6. Primers used in this study.
| Purpose and name | Sequence |
| Construction of KO-mutants | |
| B. cereus ATCC14579 | |
| AV19_upBC4707_Xba_F_II | CTATCTAGAATTTCGGCCTATCTGGACCT |
| AV20_upBC4707_SalI_R_II | ATAGTCGACCATTTGTTTTCCCCTTACCC |
| AV9_downBC4707_SalI_F_new | ATGTCGACTAAAAAGGATGACCATTTTGGTCATCCT |
| AV10_downBC4707_PstI_R_new | ATACTGCAGCCATTTGGGCCATCACCATGTAA |
| E. coli DH5α | |
| acrAB-KO-Start | ATGAACAAAAACAGAGGGTTTACGCCTCTGGCGGTCGTTCGTGTAGGCTGGAGCTGCTTC |
| acrAB-KO-Stop | TGTGCTCGATATCTTCATTCTTGCGGCTAAAGCGGCGGCGCATATGAATATCCTCCTTAGT |
| Sequencing of pBKJ236Δ bc 4707 | |
| pBKJ236_MCS_1 | ACACTTTATGCTTCCGGCTC |
| pBKJ236_MCS_2 | CAGTCTATCCCCTGGCGAA |
| Sequencing of KO-mutants | |
| Bacillus cereus Δbc4707 | |
| AV55_forwKO4707_1CO | GTGACCGCTCATTACAAGGTC |
| AV59_revKO4707_2CO | ATGTTGTAACGCCGAGTAAAGG |
| E. coli Dh5α ΔacrAB | |
| acrAB-KO-control-for | CATCGAGGATGTGTTGGC |
| acrAB-KO-control-rev | TTTGGGTGAGTATTCATCCAT |
| Construction of pBC4707 | |
| Bc4707-Forward_EcoRI | CCGGAATTCGCATATGAGAAAAAAAGTCATGATGTCTTTAATGTT |
| Bc4707-Reverse_PstI | AAACTGCAGCTTGTTGCTCACTTGTTTGTGTAGCTGGCTTTAATA |
| Sequencing of pBC4707 | |
| pTTQ18-MCS-start | TTGCGCCGACATCATAACG |
| pTTQ18-MCS-stop | CCTCTTCGCTATTACGCCA |
A markerless knock-out mutant in B. cereus ATCC14579 was constructed essentially as described by Janes & Stibitz [46]. Briefly, the fused up- and downstream regions flanking bc4707 were amplified by PCR and introduced into the suicide shuttle vector pBKJ236. The vector contains the recognition site for the homing endonuclease I-SceI. After transformation, pBKJ236 was incorporated into the chromosome of B. cereus via homologous recombination. Selection of recombinants were done on LB plates containing erythromycin (5 µg/ml) and was facilitated by the temperature sensitive origin of replication in pBKJ236 which does not permit replication of the plasmid at 37°C. Following transformation of a second plasmid (pBKJ223) carrying the gene encoding the homing endonuclease I-SceI, double strand breaks were introduced in the chromosome at the site of the inserted plasmid. The double strand breaks force homologous recombination events to repair the DNA damage which results in either wild type or mutant genotypes. Recombinants that had undergone the second recombination step were identified as erythromycin sensitive colonies. Screening for markerless mutants was done by PCR amplification using primers annealing up- and downstream of bc4707. The mutation was confirmed by sequencing.
Construction of the E. coli expression vector pBC4707
The gene bc4707 was amplified by PCR from genomic DNA extracted from the B. cereus type strain ATCC14579 using the primers Bc4707-forward_EcoRI and Bc4707-reverse_PstI (Table 6). The PCR product was cloned into the plasmid pTTQ18 by restriction digestion using EcoRI and PstI and ligation using T4-DNA ligase (New England Biolabs). Ampicillin resistant colonies were clean streaked twice and the vector insert was verified by sequencing.
Expression of BC4707-RGSHis6 and preparation of membrane fractions
Optimization of protein expression in E. coli BL21 (DE3) (pBC4707) was done in 100 ml cultures at 37°C, 150 rpm. IPTG (0–1 mM) was added to the cultures in the mid-logarithmic growth phase (OD600 of 0.4–0.6). Protein expression was induced for 3 h. The bacterial cells were disrupted by the water-lysis method [47] and the membrane fraction was prepared as described previously [47]. For large scale production of BC4707-RGSHis6, E. coli BL21 (DE3) (pBC4707) was grown in a 30 liter culture in a fermenter at 37°C. Protein expression was induced for 3 h following addition of 1 mM IPTG at OD600 0.4–0.6. Bacterial cell disruption was achieved by explosive decompression in a Constant Cell Disruption System (WWW.constantsystems.com). Inner membranes were prepared as previously described [47].
Solubilization and purification of the BC4707-RGSHis6 membrane protein
E. coli inner membranes containing overexpressed BC4707-RGSHis6 were added to solubilization buffer [20 mM Tris pH 8.0, 30 mM imidazole pH 8.0, 300 mM NaCl, 20% glycerol, 1% dodecyl-beta-D-maltoside (DDM)] to give a final protein concentration of 3 mg/ml. The suspension was incubated at 4°C for a minimum of 1 hour with gentle mixing. Insoluble proteins were removed by ultracentrifugation at 100,000× g for 1 hour at 4°C. The solubilized protein was added to the Ni-NTA resin (Qiagen™, Germany) and incubated for a minimum of 1.5 hours at 4°C with gentle mixing. The unbound protein was allowed to run off and the protein-bound resin was washed with 10× resin volumes of wash buffer (20 mM Tris pH 8.0, 30 mM imidazole, 10% glycerol, 150 mM NaCl, 0.05% DDM) and eluted using imidazole elution buffer (20 mM Tris pH 8.0, 200 mM imidazole, 5% glycerol, 150 mM NaCl, 0.05% DDM). The eluted protein was concentrated using a Vivaspin 20 (100 kDa MWCO) concentrator (Sartorius Ltd., Epsom, UK) and desalted using a Econo-Pac® 10DG desalting gel column (BioRad, California, USA) to remove any extraneous imidazole remaining from the elution step. The concentrated protein was analyzed immediately or stored at −80°C.
Preparation and analysis of samples by Synchrotron Radiation Circular Dichroism spectroscopy
Purified BC4707-RGSHis6 protein was resuspended to a final concentration of 0.5 mg/ml in CD buffer (10 mM potassium phosphate, pH 7.6; 0.05% DDM) and the sample placed into a 0.02 cm pathlength circular cell. CD measurements were performed on Beamline B23 (Diamond Light Source, Didcot, UK) at 18°C. Data points were recorded at over 260-170 nm at 1 nm intervals with an integration time of 1 s. A spectrum of the CD buffer was taken and subtracted as background.
RNA isolation
Bacterial cells were harvested by diluting the cultures with an equal volume of ice-cold methanol followed by centrifugation at 4,000 g for 5 min at 4°C. Cells were disrupted using a Precellys®24 tissue homogenizer. RNA isolation was performed using the RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's protocol, including the optional on column DNAse treatment. After elution, the RNA was treated with Turbo DNAse (Applied Biosystems, USA) according to the manufacturer's manual followed by a second round of purification using the RNeasy Mini Kit. The concentration of RNA was determined by UV-spectrometry and the quality was controlled by agarose gel electrophoresis as well as UV-spectrometry.
Quantitative RT-PCR
Complementary strand DNA was synthesized from 1 µg of RNA using the Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA was diluted 1∶5 (1∶2500 for the reference gene 16S). Three microliter of diluted cDNA was mixed with primers (0.5 µM) as well as qPCR&GO™ LC480 green master mix (MPBiomedicals, USA) to a final volume of 15 µl. Primers were designed to result in an amplicon of approximately 100 bp (Table 6). Quantitative PCR was performed with a Roche Lightcycler 480 (Roche Diagnostics GmbH, Germany). The cycling conditions were 95°C for 1.5 min followed by 45 cycles at 95°C for 10 seconds, 58°C for 10 seconds and 72°C for 10 seconds. The crossing point (Ct) values were determined by the 2nd derivative maximum of two technical replicates per biological replicate. Results were calculated by the ΔΔCt approximation. Expression ratios are averages of at least 2 biological replicates including 2 technical replicates using 16S as the reference gene for normalization. In addition, GatB RNA was used as reference to confirm the results.
Microarray analysis
The microarray slides, containing 70-mer oligonucleotides representing all the genomic ORFs of B. cereus (ATCC14579), were printed at the Microarray core facility of the Norwegian University of Science and Technology (NTNU) in Trondheim. The microarray experiments were conducted as previously described by Gohar et al. [48]. Briefly, the microarray slides were pre-incubated at 42°C for 45 minutes in pre-hybridization buffer (5×SSC, 0.1% SDS, 0.1% BSA); washed in MQ water followed by isopropanol and finally spun dry. Complementary strand DNA synthesis, labeling and purification was carried out with the FairPlay III™ microarray labeling kit (Stratagene, USA), using 500 ng random hexamers (Applied Biosystems, USA), 20 µg of RNA, and Cy3 or Cy5 dyes (GE Healthcare Bio-Sciences AB, Sweden). After purification, the samples were concentrated with a Microcon column (Millipore, USA). Labeled DNA was mixed with hybridization solution (30% formamide, 5×SSC, 0.1% SDS and 0.1 mg/ml salmon sperm DNA) denatured at 95°C for 2 minutes, and incubated at 42°C before hybridization. Hybridization was carried out over night at 42°C in a hybridization chamber (Monterey Industries, CA, USA) humidified with 5×SSC. After hybridization, the slides were washed at 42°C in a buffer containing 0.5×SSC and 0.01% SDS followed by 0.06×SSC, and finally in isopropanol before they were spun dry. The slides were scanned with an Axon 4000B scanner using the GenePix Pro 6.0 software (Molecular Devices, CA, USA). Microarray data was analyzed with a custom R-script in R 2.10.1 [49]. The GenePix files were imported to R, and the Limma package [50] was used for filtering, normalization, and further analysis. Microarray experiments were conducted with two independent experiments analyzed in two dye swapped technical replicates each. The Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession numbers E-MEXP-3529, E-MEXP-3535, E-MEXP-3536 and E-MEXP-3537.
Susceptibility assays
Bacteria were grown in LB medium at 30°C, 220 rpm overnight, diluted 1∶100 into a pre-culture and incubated at the same conditions until mid-logarithmic phase (OD600∼0.5). The pre-culture was diluted to OD600 of 0.02 and distributed in 100 µl aliquots in the wells of 96 wells plates (Becton Dickinson Labware, Franklin Lakes, USA). Compounds were added to the wells at appropriate concentrations in 2× dilution series from 100× concentrated stock solutions. For susceptibility assays using E. coli DH5α ΔacrAB carrying pTTQ18 or pBC4707, ampicillin (100 µg/ml) was included in the medium. These assays were carried out in protein expression uninduced (0 mM IPTG) as well as induced (0.05 mM IPTG) conditions. The plates were incubated at 30°C, 220 rpm for 20 h and then analyzed by ocular inspection as well as by a Victor™×multilabel plate reader (Perkin-Elmer) at 595 nm. Minimum inhibitory concentrations were determined as the concentration of xenobiotics at which the OD after 20 h incubation was less than double the starting OD. In case visual inspection showed aggregated cells at the bottom of the wells in otherwise transparent media, the OD measurements were considered unreliable whereas the MIC value was determined as the concentration where the media turned from turbid to transparent. In most cases, the two methods to determine the MIC correlated.
Growth in GGGS medium
Bacterial growth under nutrient poor conditions was tested using a minimal medium derived from GGGS medium [51]. The composition of the medium was K2HPO4 (3.0 mM), KH2PO4 (3.5 mM), MgSO4 (0.8 mM), MnCl2 (0.04 mM), NaCl (0.2 mM), CaCl2 (0.2 mM), ZnCl2 (0.05 mM), and FeCl3 (0.04 mM). In addition to this, 2 mM nitrogen source as well as 20 mM carbon source was added. Combinations of 7 different nitrogen sources and 6 different carbon sources were tested. The carbon sources investigated were: glucose, lactate, maltose, sorbitol, galactose, pyruvate, and xylose. The nitrogen sources investigated were: L-serine, L-glutamate, L-arginine, L-proline, urea and ammonium chloride. Bacteria were grown overnight in LB medium at 30°C and 220 rpm shaking. The culture was diluted 1∶100 in fresh LB medium and cultured under the same conditions until mid-logarithmic phase. The bacteria were washed once in GGGS medium and resuspended in fresh GGGS medium including nitrogen and carbon sources to an OD600 of 0.01. Initially, attempts were made to establish growth curves for the mutant and wild type. The cultures were incubated in 100 ml Erlenmeyer flasks filled to a fourth of the volume, at 220 rpm at 30°C for up to 140 h. The absorbance was measured at regular intervals. In a subsequent experiment the bacterial suspensions (OD600 0.01 in GGGS medium) were spotted onto 1.5% agar plates containing GGGS medium supplemented with nitrogen and carbon sources. These plates were incubated at 30°C for 140 h. Bacterial growth was scored depending on the density and appearance of the colonies.
Motility assays
Swimming motility was observed on 0.3% LB agar plates whereas swarming motility was observed on 1.0% LB agar plates supplemented with 0.5% glucose. The plates were inoculated with 5 µl of culture (OD600∼5) that had been concentrated from a logarithmically growing culture (OD600∼1). Plates were incubated at 37°C. The diameter of the zone created by the swimming bacteria and the radius of the zone created by the swarming bacteria respectively were measured at regular intervals for 70 h. Similar experiments were conducted using 0.3% and 1.0% GGGS agar plates supplemented with different combinations of carbon and nitrogen sources as described above.
Phenotype microarray
Bacteria were inoculated onto tryptic soy agar (TSA), incubated at 28°C over night and clean streaked onto TSA for an additional over night incubation. Single colonies, picked using sterile cotton swabs, were suspended in inoculation fluid (IF-0, Biolog, Hayward, CA, USA). The transmittances of the bacterial suspensions were adjusted to 85% on a Biolog turbidometer. Cell suspension, inoculation fluids, tetrazolium dye, and suitable additives were mixed according to the manufacturer's protocol optimized for Bacillus subtilis. The mixture was inoculated into the Phenotype Microarray plates (100 µl per well), which were incubated at 28°C in an Omnilog reader. Quantitative color changes were recorded every 15 min by a charge-coupled camera device for a period of 48 h. The kinetic responses were analyzed with the Omnilog-PM software. The data was analyzed visually as well as by comparing the calculated areas under the kinetic response curves.
Norfloxacin accumulation assay
Bacterial cells for the norfloxacin accumulation assay were prepared by growing E. coli DH5α ΔacrAB (pBC4707) as well as E. coli DH5α ΔacrAB (pTTQ18) at 37°C in LB broth containing ampicillin (100 µg/ml) over night. The cultures were diluted to an OD600 of 0.05 in fresh LB broth including ampicillin (100 µg/ml). These cultures were grown to OD600∼0.5, at which point IPTG was added to a concentration of 0.1 mM. Expression of proteins was induced for 3 hours. Following this, the cells were spun down, washed twice in Tris-Cl (pH 7.0) including 100 mM NaCl and resuspended in the same buffer. The OD600 was adjusted to 1 and glucose was added to a final concentration of 20 mM. The assay was started by the addition of 100 µM norfloxacin. Samples (1 ml) were taken every five minutes for 30 min starting before (time 0 min) the addition of norfloxacin. CCCP (100 µM) was added at time 15 min. Samples were centrifuged at 13000 rpm for 30 seconds, the supernatant was discarded. The bacterial pellet was washed once in Tris-Cl (pH 7.0) including 100 mM NaCl and resuspended in 0.1 M glycin (pH 3.0). The Norfloxacin was extracted over night at 37°C. The bacterial debris was removed by centrifugation and the fluorescence of the supernatant was measured at 280 nm excitation and 445 nm emission on a Perkin-Elmer LS-55 fluorescence spectrophotometer.
Acid stress
Acid stress experiments were conducted basically as described by Mols et al. [22]. Pre-cultures of the B. cereus wild type as well as the Δ4707 isogenic mutant were inoculated from overnight cultures. The pre-cultures were grown until mid-logarithmic phase (OD600 was between 0.5–1.0) at which point they were diluted 1∶100 in brain heart infusion medium (BHI) buffered to pH 7.1 with 100 mM or 10 mM sodium phosphate. The buffered cultures were grown to an OD600 of 0.5. At this point the medium was acidified to pH 5.5 using 0.238% (v/v) 12 M hydrochloric acid, 0.69% (v/v) lactic acid, 0.205% (v/v) 12 M hydrochloric acid with 0.074% (v/v) acetic acid or 0.571% (v/v) acetic acid, respectively. The OD600 of the cultures were recorded for an additional 4.5 h.
Supporting Information
Bile salt induced transcriptional profiles of B. cereus ATCC14579 and its isogenic Δbc4707 mutant. Bacteria were grown to OD600 0.5 (tOD 0.5) at which point bile salts (50 µg/ml) were added followed by additionally 15 min incubation (tBS 15). Cells were harvested and total RNA was prepared from each strain before (tOD 0.5) and after (tBS 15) bile salts stress. Microarray analysis was conducted comparing the gene expression at time tBS 15 with time tOD 0.5. Shown are genes with significantly (P<0.05) different relative expression (tBS 15/tOD 0.5>1.5 or tBS 15/tOD 0.5<0.67) in at least one strain after bile salts stress for 15 min. * indicates relative expression values that were not statistically significant (i.e. p>0.05).
(DOC)
Acknowledgments
We thank Drs Siligardi and Hussain for assistance with determinations of the SRCD spectra at the DIAMOND Light Source.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Work in the Kolstø laboratory is supported by the Norwegian Research Council (Functional Genomics FUGE II grant 183421; http://www.forskningsradet.no/servlet/Satellite?c=Page&cid=1226993493114&p=1226993493114&pagename=fuge%2FHovedsidemal). Work in the PJFH laboratory is supported by the European Drug Initiative on Channels and Transporters (EDICT) consortium grant number 201924 (http://www.edict-project.eu/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kolsto AB, Tourasse NJ, Okstad OA. What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol. 2009;63:451–476. doi: 10.1146/annurev.micro.091208.073255. [DOI] [PubMed] [Google Scholar]
- 2.Mock M, Fouet A. Anthrax. Annu Rev Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 3.Jernigan JA, Stephens DS, Ashford DA, Omenaca C, Topiel MS, et al. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis. 2001;7:933–944. doi: 10.3201/eid0706.010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottone EJ. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev. 2010;23:382–398. doi: 10.1128/CMR.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drobniewski FA. Bacillus cereus and related species. Clin Microbiol Rev. 1993;6:324–338. doi: 10.1128/cmr.6.4.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotiranta A, Lounatmaa K, Haapasalo M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000;2:189–198. doi: 10.1016/s1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 7.von Stetten F, Mayr R, Scherer S. Climatic influence on mesophilic Bacillus cereus and psychrotolerant Bacillus weihenstephanensis populations in tropical, temperate and alpine soil. Environ Microbiol. 1999;1:503–515. doi: 10.1046/j.1462-2920.1999.00070.x. [DOI] [PubMed] [Google Scholar]
- 8.Turnbull PC, Kramer JM. Intestinal carriage of Bacillus cereus: faecal isolation studies in three population groups. J Hyg (Lond) 1985;95:629–638. doi: 10.1017/s0022172400060733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh AC. Prevalence of Bacillus cereus in the faeces of healthy adults. J Hyg (Lond) 1978;80:233–236. doi: 10.1017/s0022172400053572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7:1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 11.Jensen GB, Hansen BM, Eilenberg J, Mahillon J. The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol. 2003;5:631–640. doi: 10.1046/j.1462-2920.2003.00461.x. [DOI] [PubMed] [Google Scholar]
- 12.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic acids res. 2007;35:D274–279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren Q, Paulsen IT. Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J mol microbiol biotechnol. 2007;12:165–179. doi: 10.1159/000099639. [DOI] [PubMed] [Google Scholar]
- 14.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 15.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 16.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 17.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 18.Krulwich TA, Lewinson O, Padan E, Bibi E. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat Rev Microbiol. 2005;3:566–572. doi: 10.1038/nrmicro1181. [DOI] [PubMed] [Google Scholar]
- 19.Neyfakh AA. Natural functions of bacterial multidrug transporters. Trends Microbiol. 1997;5:309–313. doi: 10.1016/S0966-842X(97)01064-0. [DOI] [PubMed] [Google Scholar]
- 20.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12:205–213. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 21.Kristoffersen SM, Ravnum S, Tourasse NJ, Okstad OA, Kolsto AB, et al. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579 [corrected]. J Bacteriol. 2007;189:5302–5313. doi: 10.1128/JB.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mols M, van Kranenburg R, Tempelaars MH, van Schaik W, Moezelaar R, et al. Comparative analysis of transcriptional and physiological responses of Bacillus cereus to organic and inorganic acid shocks. Int J Food Microbiol. 2010;137:13–21. doi: 10.1016/j.ijfoodmicro.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Chan YY, Bian HS, Tan TM, Mattmann ME, Geske GD, et al. Control of quorum sensing by a Burkholderia pseudomallei multidrug efflux pump. J Bacteriol. 2007;189:4320–4324. doi: 10.1128/JB.00003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Xiao M, Horiyama T, Zhang Y, Li X, et al. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J Biol Chem. 286:26576–6584. doi: 10.1074/jbc.M111.243261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramon-Garcia S, Martin C, Thompson CJ, Ainsa JA. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob Agents Chemother. 2009;53:3675–3682. doi: 10.1128/AAC.00550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szakonyi G, Leng D, Ma P, Bettaney KE, Saidijam M, et al. A genomic strategy for cloning, expressing and purifying efflux proteins of the major facilitator superfamily. J Antimicrob Chemother. 2007;59:1265–1270. doi: 10.1093/jac/dkm036. [DOI] [PubMed] [Google Scholar]
- 27.Findlay HE, Rutherford NG, Henderson PJ, Booth PJ. Unfolding free energy of a two-domain transmembrane sugar transport protein. Proc Natl Acad Sci U S A. 2010;107:18451–18456. doi: 10.1073/pnas.1005729107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward A, Hoyle C, Palmer S, O'Reilly J, Griffith J, et al. Prokaryote multidrug efflux proteins of the major facilitator superfamily: amplified expression, purification and characterisation. J Mol Microbiol Biotechnol. 2001;3:193–200. [PubMed] [Google Scholar]
- 29.Structural Genomics C, China Structural Genomics C, Northeast Structural Genomics C, Graslund S, Nordlund P, et al. Protein production and purification. Nature Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother. 2001;45:1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura H. Gene-Controlled Resistance to Acriflavine and Other Basic Dyes in Escherichia coli. J Bacteriol. 1965;90:8–14. doi: 10.1128/jb.90.1.8-14.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura H. Genetic determination of resistance to acriflavine, phenethyl alcohol, and sodium dodecyl sulfate in Escherichia coli. J Bacteriol. 1968;96:987–996. doi: 10.1128/jb.96.4.987-996.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Morita Y, Huda MN, Kuroda T, Mizushima T, et al. VmrA, a member of a novel class of Na(+)-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J Bacteriol. 2002;184:572–576. doi: 10.1128/JB.184.2.572-576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldsby RA, Heytler PG. Uncoupling of Oxidative Phosphorylation by Carbonyl Cyanide Phenylhydrazones. Ii. Effects of Carbonyl Cyanide M-Chlorophenylhydrazone on Mitochondrial Respiration. Biochemistry. 1963;2:1142–1147. doi: 10.1021/bi00905a041. [DOI] [PubMed] [Google Scholar]
- 38.Heytler PG. uncoupling of oxidative phosphorylation by carbonyl cyanide phenylhydrazones. I. Some characteristics of m-Cl-CCP action on mitochondria and chloroplasts. Biochemistry. 1963;2:357–361. doi: 10.1021/bi00902a031. [DOI] [PubMed] [Google Scholar]
- 39.Cavari BZ, Avi-Dor Y. Effect of carbonyl cyanide m-chlorophenylhydrazone on respiration and respiration-dependent phosphorylation in Escherichia coli. Biochem J. 1967;103:601–608. doi: 10.1042/bj1030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiter L, Kolsto AB, Piehler AP. Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J Microbiol Methods. 2011;86:210–217. doi: 10.1016/j.mimet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Serizawa M, Sekizuka T, Okutani A, Banno S, Sata T, et al. Genomewide screening for novel genetic variations associated with ciprofloxacin resistance in Bacillus anthracis. Antimicrob Agents Chemother. 2010;54:2787–2792. doi: 10.1128/AAC.01405-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inglesby TV, O'Toole T, Henderson DA, Bartlett JG, Ascher MS, et al. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA. 2002;287:2236–2252. doi: 10.1001/jama.287.17.2236. [DOI] [PubMed] [Google Scholar]
- 43.Swartz MN. Recognition and management of anthrax–an update. N Engl J Med. 2001;345:1621–1626. doi: 10.1056/NEJMra012892. [DOI] [PubMed] [Google Scholar]
- 44.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janes BK, Stibitz S. Routine markerless gene replacement in Bacillus anthracis. Infect Immun. 2006;74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward A, Sanderson NM, O'Reilly J, Rutherford NG, Poolman B, et al. The amplified expression, identification, purification, assay and properties of histidine-tagged bacterial membrane transport proteins. In: Baldwin S, editor. Membrane Transport - a Practical Approach. Oxford: Blackwell's Press; 2000. pp. 141–166. [Google Scholar]
- 48.Gohar M, Faegri K, Perchat S, Ravnum S, Okstad OA, et al. The PlcR virulence regulon of Bacillus cereus. PLoS One. 2008;3:e2793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth GK. Gentleman VC R, Dudoit S, Irizarry R, Huber W, editors. Limma: linear models for microarray data. 2005. pp. 397–420. Bioinformatics and Computational Biology Solutions using R and Bioconductor: Springer.
- 51.Mols M, de Been M, Zwietering MH, Moezelaar R, Abee T. Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics. Environ Microbiol. 2007;9:2933–2944. doi: 10.1111/j.1462-2920.2007.01404.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bile salt induced transcriptional profiles of B. cereus ATCC14579 and its isogenic Δbc4707 mutant. Bacteria were grown to OD600 0.5 (tOD 0.5) at which point bile salts (50 µg/ml) were added followed by additionally 15 min incubation (tBS 15). Cells were harvested and total RNA was prepared from each strain before (tOD 0.5) and after (tBS 15) bile salts stress. Microarray analysis was conducted comparing the gene expression at time tBS 15 with time tOD 0.5. Shown are genes with significantly (P<0.05) different relative expression (tBS 15/tOD 0.5>1.5 or tBS 15/tOD 0.5<0.67) in at least one strain after bile salts stress for 15 min. * indicates relative expression values that were not statistically significant (i.e. p>0.05).
(DOC)