Abstract
Applications in nanomedicine, such as diagnostics and targeted therapeutics, rely on the detection and targeting of membrane biomarkers. In this paper we demonstrate absolute quantitative profiling, spatial mapping, and multiplexing of cancer biomarkers using functionalized quantum dots. We demonstrate highly selective targeting molecular markers for pancreatic cancer with extremely low levels of non-specific binding. We confirm that we have saturated all biomarkers on the cell surface, and, in conjunction with control experiments, extract absolute quantitative values for the biomarker density in terms of the number of molecules per square micrometer on the cell surface. We show that we can obtain quantitative spatial information of biomarker distribution on a single cell, important since tumors cell populations are inherently heterogeneous. We validate our quantitative measurements (number of molecules per square micron) using flow cytometry and demonstrate multiplexed quantitative profiling using color-coded quantum dots.
Keywords: biomarker profiling, cancer biomarkers, quantum dots, core/shell nanoparticles, lipid encapsulation
Background
The detection of cancer biomarkers is important for diagnosis, disease stage forecasting, and clinical management. Since tumor populations are inherently heterogeneous, a key challenge is the quantitative profiling of membrane biomarkers, rather than secreted biomarkers, at the single cell level. The detection of cancer biomarkers is also important for imaging and therapeutics since membrane proteins are commonly selected as targets. Many methods for detection of membrane proteins yield ensemble averages and hence have limited application for analysis of heterogeneous populations or single cells. Fluorescence-based methods allow detection at the single cell level, however, photobleaching presents a major limitation in obtaining quantitative information. Quantum dots overcome the limitations associated with photobleaching, however, realizing quantitative profiling requires stable quantum yield, monodisperse quantum dot - antibody (QD-Ab) conjugates, and well-defined surface chemistry.1 By quantitative profiling we specifically refer to methods that yield absolute values of expression levels (e.g. # μm−2) and not relative values. Here we demonstrate quantitative profiling, spatial mapping, and quantitative multiplexing of molecular biomarkers associated with precursor lesions of pancreatic adenocarcinoma at the single cell level using QD-Ab conjugates.
Pancreatic cancer is the fourth leading cause of cancer death in the US (about 35,000 per year).2 The survival rate amongst pancreatic cancer patients is extremely low, primarily due to the fact that a large fraction (about 80%) of tumors is metastatic at the time of diagnosis.3 The histologic progression from non-invasive precursor lesions, pancreatic intraepithelial neoplasia (PanINs), to invasive and metastatic pancreatic cancer is associated with the sequential accumulation of molecular alterations.
We have selected three biomarkers for pancreatic cancer for quantitative imaging: prostate stem cell antigen (PSCA), claudin-4 (CLDN4), and mesothelin (MSLN). PSCA and MSLN are gylcosylphosphatidyl inositol (GPI)-anchored proteins whereas CLDN4 is one of a large family of tight junction proteins. PSCA is overexpressed in adenocarcinomas and present in the majority of PanIN lesions beginning with early PanIN-1. Claudin-4 overexpression is observed in intermediate PanIN-2 lesions.8–10 Mesothelin overexpression is a late event in the progression model of pancreatic cancer, almost always associated with invasion. All three of these biomarkers are therapeutic targets for pancreatic cancer. Quantitative profiling of these biomarkers was studied in three pancreatic cancer cell lines: Panc-1 (derived from pancreatic ductal adenocarcinoma), MIA PaCa-2 (derived from epithelial pancreatic carcinoma cells), and Capan-1 (derived from a liver metastasis of a grade II pancreatic adenocarcinoma). The immortalized pancreatic ductal cell line HPDE was used for comparison.
QDs exhibit size-dependent absorption and emission properties,13 high fluorescence quantum yields, and with careful functionalization have been widely used for imaging and sensing.14–23 Quantitative QD-Ab targeting requires that each target molecule (e.g. membrane protein) is conjugated with one QD and that non-specific binding is minimized. Although various functionalization schemes have been reported in the literature, here we have developed a method based on encapsulation with a lipid layer optimized for quantitative targeting (Figure 1a). Since quantitative biomarker analysis using QD-conjugates has not previously been reported, we cannot compare our functionalization scheme to other methods, however, through a systematic study of functionalization parameters, we show that: (1) functionalization can be achieved with commercially available reagents, (2) the yield of the functionalization process is high, (3) the QD-conjugates are monodisperse and exhibit good stability in water, and (4) the functionalization method minimizes non-specific binding to cells.
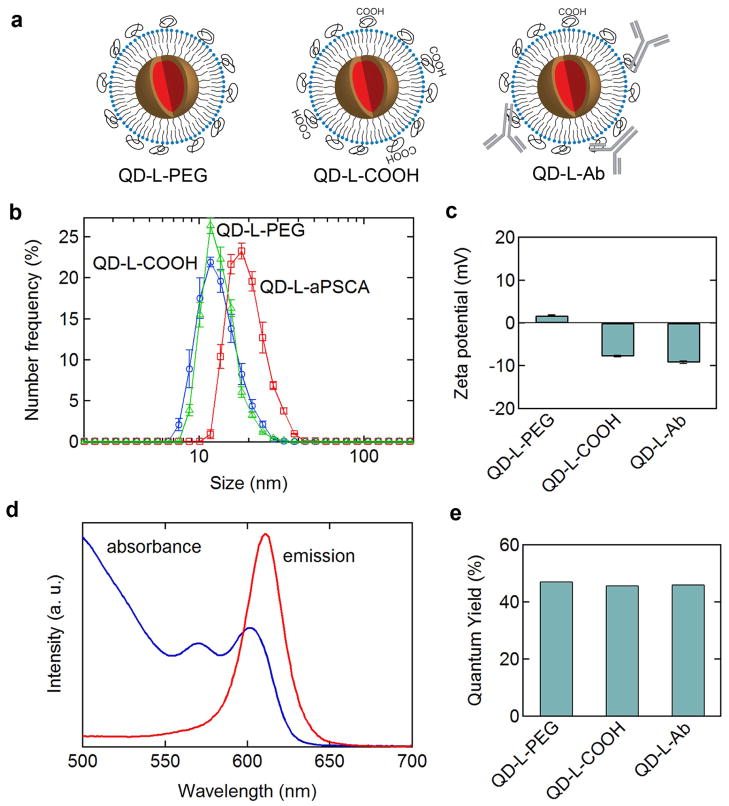
Figure 1.
(a) Schematic illustration of QD conjugates for biomarker targeting: (QD-L-PEG) CdSe/(Cd,Zn)S QDs with 80 mol% MHPC and 20 mol% DPE-Peg 2k. (QD-L-COOH) QDs with 80 mol% MHPC, 15 mol% DPE-PEG2k, and 5 mol% DPE-PEG2k-COOH. (QD-L-Ab) QD-L-COOH covalently conjugated with an average of three targeting antibodies per QD. (b) Particle size distributions for QD conjugates. (c) Zeta potential for QD conjugates. A zeta potential of about −10 mV minimizes aggregation and non-specific binding. (d) Absorbance and emission spectra for QD-L-PEG (Em. 623 nm) in water. (e) Quantum yield for QD conjugates in water.
Methods
Synthesis of QDs
Most experiments were performed using CdSe/(Cd,Zn)S core/shell QDs with an emission wavelength of about 610 nm. For multiplexing experiments we synthesized CdSe/(Cd,Zn)S core/shell QDs with an emission wavelength of 524 nm and CuInSe/ZnS core/shell QDs with an emission wavelength of 707 nm. Details are provided in Supplementary Information.
Water solubilization of QDs
Water soluble QDs were obtained by forming a lipid monolayer composed of MHPC/DPPE-PEG2k (80:20 mol%) or MHPC/DPPE-PEG2k/DPPE-PEG2k-COOH (80:15:5 mole%). Typically 0.25 nmol of QDs, 4 μmol of MHPC, 0.75 μmol of DPPE-PEG2k, and 0.25 μmol of DPPE-PEG2k-COOH were dissolved in 0.3 mL of chloroform. This solution was added to 2 ml of deionized water and heated and maintained at 110 °C for 1 h under vigorous stirring to evaporate chloroform. The resulting solution was sonicated for 1 h, centrifuged, and the supernatant then passed through a syringe filter with a 200 nm PTFE membrane (VWR) to remove any aggregates or unsuspended QDs. Quantum yield measurements were performed on suspensions with about 100 pmol QDs in 4 mL DI water using a Hamamatsu C9920-02 fluorometer. Details are provided in Supplementary Information.
Cell lines
A panel of three human pancreatic cancer cell lines (MIAPaCa-2, Panc-1, and Capan-1) were utilized for these studies. Mia PaCa-2 and Panc-1 were cultured with a growth medium containing DMEM (Dulbecco’s Modified Eagle’s Medium) as the base medium, FBS (fetal bovine serum, 10 %), and P/S (penicillin/streptomycin, 1 %), and Capan-1 was cultured in IMDM (Iscove’s Modified Dulbecco’s Medium) supplemented with 20% FBS and 1% P/S. All three cell lines were incubated at 37 °C and in 5% CO2. The immortalized normal pancreatic cell line HPDE (human pancreatic duct epithelium) was used as a control. HPDE cells were cultured in keratinocyte serum-free (KSF) medium supplemented by bovine pituitary extract and epidermal growth factor (Gibco-BRL, Grand Island, NY).
Antibodies and antibody conjugation
QDs were conjugated with one of three antibodies: anti-Prostate Stem Cell Antigen (aPSCA), anti-claudin-4 (aCLDN4), or anti-mesothelin (aMSLN). The reaction of the primary amines on the antibody with lipid-modified QDs (carboxylic acid-terminated QDs) is catalyzed by 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) resulting in the formation of an amide bond. In a typical reaction, 1 μM QDs was mixed with 2 mM EDC and 5 mM sulfo-NHS in 0.1 M MES (pH 6.0) and incubated for 15 minutes at room temperature with gentle mixing. The remaining unreacted EDC was quenched with the addition of 20 μL of 2-mercaptoethanol (1 M) for 10 minutes. Unreacted reagents and byproducts were removed by centrifugation in 100 kDa MWCO microcentrifuge tubes at 1000 g for 5 minutes. The activated QDs were then resuspended in 1× PBS. The activated QD stock solution was mixed with antibody solution (0.5 – 1 mg mL−1 in PBS) to obtain a 3 – 6 fold molar excess of the antibodies to QDs. The reaction solution was incubated at room temperature for 2 h with gentle mixing. For control experiments QDs were prepared by coating with 80 mol% MHPC and 20 mol% PEGylated lipid DPE-PEG2k (no Ab). To remove excess reagents microfiltration was performed. To ensure that any aggregates are removed, an additional filtration step was carried out using syringe type filters (pore size: 100 nm). The QD suspensions were then characterized using UV-Vis absorption, photoluminescence (PL), dynamic light scattering (DLS), and surface charge (zeta potential).
Imaging
Briefly, about 105 cells (see above for description of cell lines) were pre-seeded in a 12-well culture dish. At 50 – 70% confluency (1 – 2 days), the cell medium was aspirated and the cells washed three times with PBS. Fixing solution (3.7% Formaldehyde) was added to the wells for 20 min and washed three times with PBS. The cells were then incubated with a blocking buffer (10% horse serum or 5% BSA in PBS) for 1 h prior to introducing 500 μL of QD-Ab conjugates to each well and then incubated at RT for 30 min. In all profiling experiments, cells were incubated with 20 pmol QDs corresponding to a dose of about 108 QDs per cell. In experiments to confirm that the membrane biomarkers were saturated with QD-Ab conjugates (Figure 3a), cells were incubated with 0.1 – 20 pmol QDs. Next, the QD-Ab solution was aspirated and the cells washed with PBS three times. The maximum biomarker density (around 500 μm−2) corresponds to about 105 per cell or a maximum QD excess of about 1000 QDs per biomarker.
Figure 3.
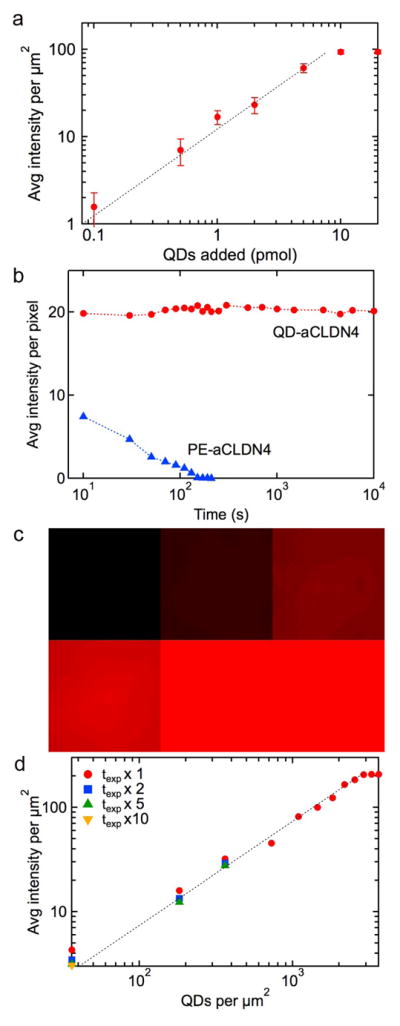
Quantitative analysis of pancreatic cancer biomarkers from fluorescence images with QD-Ab conjugates. (a) Saturation of membrane biomarkers. Average fluorescence intensity for Panc-1 cells incubated with different concentrations of QD-aMSLN. The error bars represent the standard error for measurements over at least 30 cells. The slope at lower concentrations is 1.0 confirming negligible non-specific binding or competitive binding. The plateau at 10 mmol QDs indicates saturation of MSLN at the surface. (b) Stability of fluorescence in QDs and fluorophores. Average fluorescence intensity for Panc-1 cells incubated with QD-aCLDN4 conjugates or PE (phycoerythrin)-aCLDN4 conjugates versus illumination time.(c) Calibration of QD fluorescence. Fluorescence images for different concentrations of QDs confined between two glass slides with fixed area. Top row: 36, 360, 1087 QDs μm−2, bottom row: 1813, 2513, 2900 QDs μm−2. (d) Average fluorescence intensity (normalized for 0.5 s exposure time) versus QD concentration obtained from analysis of images of QD suspensions.
Phase contrast and fluorescence images were taken with a Nikon ECLIPSE TE2000-U microscope equipped with a filter wheel allowing us to mix-and-match excitation and emission filters depending on the QDs (Ex: 350/50, 484/15, 555/25; Em: 457/30, 517/40 (FITC), 605/40 (TRITC), 620/40, or 665/LP). For experiments with QDs (Em. 607 nm), we used Ex: 555/25 and Em: 605/40. Details of emission and filter ranges are provided in Supplementary Information. All images were obtained with a ×20 objective using Nikon Elements software. The focus was set to the top surface of the cell rather than the bottom surface of the cell on the glass slide. Images were recorded using a CoolSNAP HQ2 camera with 2 × 2 binning yielding 696 × 520 pixels, and an output intensity range from 0 – 255. The exposure time was 0.5 s unless otherwise indicated.
Flow cytometry analysis
Cell were centrifuged at 500 × g for 5 mins and washed three times in an isotonic PBS buffer supplemented with 0.5 % BSA to remove contaminating serum components that may be presented in the culture medium. Cells were resuspended in the same buffer to a final concentration of 4 × 106 cells mL−1 and 25 μL of cells (105 cells) transferred to a test tube. 10 μL of PE-conjugated anti-human claudin-4 antibodies (IgG2A) was then added to the test tube and incubated for 30 min. As a control for analysis, cells in a separate tube were treated with a PE-labeled mouse IgG2A isotype control. See Supplementary Information for details.
Image analysis
Immunofluorescence images were acquired and analyzed using Nikon NIS-Elements AR 3.1 software. The software was used to automatically select the cell boundaries and to generate the pixel statistics of the cellular region. The average fluorescence intensity per μm2 within the cellular region was determined quantitatively, which allows us to make quantitative comparisons between different cell lines and different antibodies (i.e. different molecular biomarkers). Control experiments included: (1) PEGylated neutral-charge (zwitterionic) QD-L-PEG (no antibody) incubated with pancreatic cancer cell lines and a normal pancreas epithelial cell line (HPDE), and (2) QD-Ab conjugates incubated with HPDE cells.
Results
Lipid encapsulation
The hydrophobic capping ligands on the QDs after synthesis drive the formation of a lipid monolayer, analogous to the outer leaflet in a bilayer membrane. Due to the high curvature of the QDs, a combination of single and double acyl chain phospholipids was used to form the outer leaflet. To determine the optimum composition, QDs were incubated in solution containing different concentrations of a single alkyl chain phospholipid 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine (MHPC) and a double alkyl chain lipid 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE). The yield of the functionalization process was higher than 60% for compositions in the range from 20 to 50 mol% DPPE (see Supplemental Figure S1a). For ≤ 20 mol% DPPE, the QD-L conjugates are monodisperse with an average hydrodynamic diameter of about 13 nm (see Supplemental Figure S1b), as expected for the addition of a 2 nm lipid to the 8 nm diameter CdSe/(Cd,Zn)S QDs. In contrast, for ≥ 30mol% DPPE, the QDs were polydisperse. The stability in water is also dependent on the lipid composition: QDs with 80 mol % MHPC and 20 mol% DPPE are stable for at least 100 h, significantly longer than other compositions (see Supplemental Figure S1c). Replacing the DPPE with a pegylated version (DPPE-PEG2k), resulted in QD-L-PEG conjugates that were stable for several weeks. Finally, the quantum yield of QD-L conjugates was greater than 40% for QDs with 80 mol% MHPC/20mol% DPPE, and was and significantly higher than other lipid compositions.
Charge and antibody-conjugation
Targeting antibodies were covalently conjugated to the lipid-coated QDs by incorporating a COOH-terminated pegylated lipid (DPPE-PEG2k-COOH). The introduction of charged groups increases stability: QDs that are near-neutral tend to aggregate, resulting in a very low yield after filtration (see Supplemental Figure S1d). Conversely, QDs with significant charge exhibit high levels of non-specific cell surface binding in control experiments. Consequently, there is an optimal range of charge (corresponding to a zeta potential of about −10 mV) to minimize aggregation, maximize yield and stability in water, and minimize non-specific binding. Using zwitterionic lipids, the QDs are almost electrically neutral, with a zeta potential of less than 2 mV (Figure 1c). Introduction of 5 mol% of the COOH-PEG-lipid does not influence the hydrodynamic diameter (Figure 1b) but results in a small negative surface charge, corresponding to a zeta potential of about −7 mV (Figure 1c). The antibodies were covalently conjugated to the QDs through formation of an amide bond between the carboxylic acid of the pegylated lipids and primary amines (lysine or N-terminus) on the antibodies. In control experiments, we separated the antibody fragments not covalently linked to the QDs and determined that at least one antibody per QD was active.
Antibody conjugation resulted in an increase in the average hydrodynamic diameter of the QDs from 13 nm to about 21 nm (Figure 1b) (for a-PSCA) and a small increase in the magnitude of the zeta potential due to the contribution from the antibodies (Figure 1c). The sharp size distribution and absence of aggregates (Figure 1b) is characteristic of successful conjugation and is crucial to minimizing non-specific binding for quantitative profiling. The low concentration of carboxylated PEG-lipids minimizes aggregation during antibody-conjugation and charge-induced non-specific binding. The absorbance/emission spectra (Figure 1d) and the quantum yield (Figure 1e) of the QDs were not influenced by conjugation and the quantum yield remained more than 40%. With careful removal of excess reagents and filtration, the QDs are stable in water for at least several weeks showing no change in optical properties.
Profiling
Figure 2 shows a panel of fluorescence images after incubating Panc-1, MIA PaCa-2, and Capan-1 cells with QD-Ab conjugates. The corresponding phase contrast images are shown in Supplemental Figures S2 – S5. The absence or very low level of fluorescence for HPDE cells (Figure S6) or cells incubated with QDs without antibodies (Figures S3 – S6) indicates that the QD-Ab conjugates exhibit very low non-specific binding. We therefore hypothesize that the fluorescence from the pancreatic cancer cell lines is due to the binding of one QD-Ab conjugate to one target biomarker on the cell surface. This hypothesis is verified in subsequent experiments.
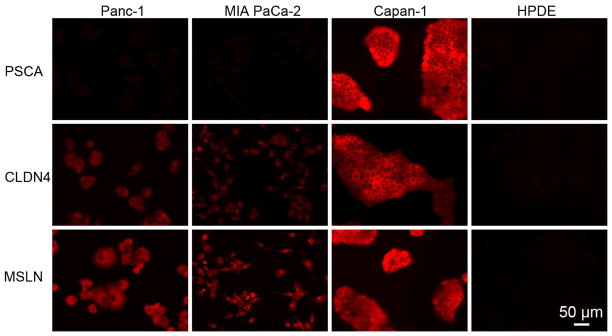
Figure 2.
Profiling of biomarkers for pancreatic cancer. Fluorescence images of pancreatic cancer cells (Panc-1, MIA Paca-2, and Capan-1) and normal pancreatic cells (HPDE) incubated with 20 pmol QD-Ab conjugates (Ab = aPSCA, aCLDN4, and aMSLN).
The fluorescence images from the Panc-1 and MIA PaCa-2 cells are very uniform, in part due to the fact that the cells are relatively isolated. In contrast, the fluorescence from the Capan-1 cells is more pronounced at the cell-cell boundaries. The spatial distribution is discussed in more detail below. Qualitative comparison of the fluorescence images in Figure 2 shows different intensity levels, implying different expression levels. For example, while PSCA shows high expression in Capan-1, MSLN was highly expressed in all three pancreatic cancer cell lines. Similarly, CLDN4 is very highly expressed in Capan-1, moderately expressed in Panc-1, and weekly in expressed MIA PaCA-2. These semi-quantitative observations are in good agreement with results from PCR, Northern blot, and Western blot reported in the literature. We note that these results are only achieved with careful synthesis of the QD-Ab conjugates. Without appropriate functionalization and surface modification, targeting is extremely heterogeneous on the cell surface and control experiments with QDs with no antibody show significant non-specific binding.
To quantitatively determine the expression levels we must (1) confirm that we have saturated all targeted biomarkers on the cell surface and (2) relate the fluorescence intensity to the QD concentration. To confirm that we have saturated all biomarkers on the cell surface, we incubated Panc-1 cells with different concentrations of QD-L-aMSLN conjugates and measured the average fluorescence intensity per cell (Figure 3a). The fluorescence intensity increases linearly with QD concentration up to 10 pmol, at which point the fluorescence intensity remains constant, indicating that all biomarkers are saturated. Prior to saturation, the slope is 1.0 confirming negligible non-specific binding and no competition for binding sites. Finally, we can conclude that for any QD-Ab/cell line combination, all biomarkers are saturated as long as the fluorescence intensity is ≤ 240 μm−2, and this condition is satisfied for all biomarkers and cell lines shown in Figure 2.
Having established that we have saturated the biomarkers on the cell surface, we next relate the fluorescence intensity to the QD concentration. To quantitatively determine biomarker concentrations over a wide range requires that we vary the exposure time when capturing the fluorescence images. To do this we must consider the time dependence of the emission. Figure 3b shows results for experiments where Panc-1 cells were incubated with QD-L-aCLDN4 conjugates or claudin-4 antibody conjugated with the fluorophore phycoerythrin (PE, emission 605 nm), PE-aCLDN4. The emission from QD-L-aCLDN4 is constant for at least 104 s while the emission from the PE-aCLDN4 conjugates decreases exponentially with time due to photobleaching. The stable emission for the QDs shows that we can linearly scale fluorescence intensities from different exposure times. Photobleaching results in an exponential decrease in emission for the PE-aCLDN4 conjugates and highlights the difficulty in using fluorophores for quantitative analysis.1
To relate the fluorescence intensity to QD concentration, a fixed volume of QD suspension was located between two glass slides (Figure 3c). By confining the area of the suspension between the glass slides we can relate the fluorescence intensity to an areal density of QDs (Figure 3d). The average fluorescence intensity per unit area is linearly dependent on the QD concentration and the slope of 1.0 confirms that there are no errors in our procedure.
Having established that we have saturated all biomarkers on the cells and that the fluorescence intensity is proportional to the QD concentration, we can quantitatively analyze the fluorescence images. Figure 4 shows the average biomarker density for PSCA, claudin-4 and mesothelin in the three pancreatic cancer cell lines (see also Supplemental Table S1). The expression levels of these markers are in the range from about 30 μm −2 to 470 μm−2. The expression levels for CLDN4 and MSLN on HPDE cells were less than 15 μm−2 while the expression level for PSCA was about 44 μm−2. From analysis of the background intensity we determined a detection limit of about ±4 μm−2 (SD). The emission from cells incubated with QDs without targeting antibodies (QD-L-PEG, see Supplemental Figures S2 – S5) corresponds to an average level of non-specific binding of 15 μm−2, just above the detection limit.
Figure 4.
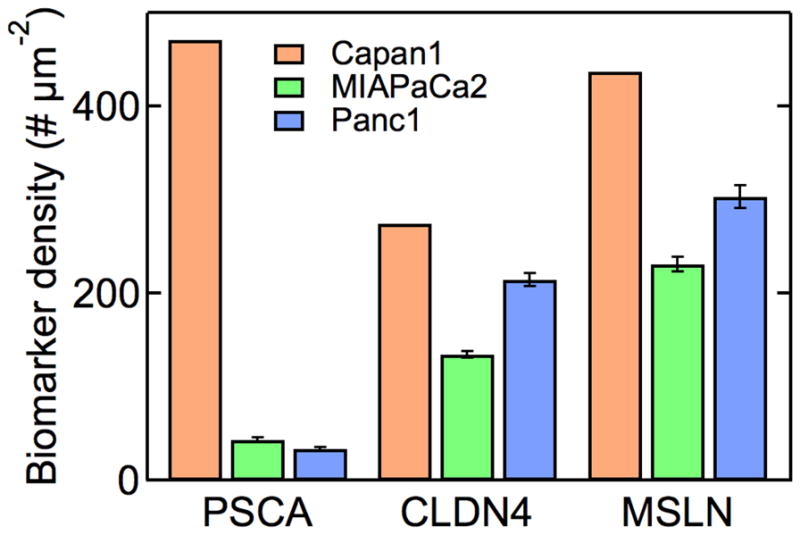
Absolute expression levels for biomarkers for pancreatic cancer. Average biomarker density per μm2 for PSCA, claudin-4 and mesothelin in the three pancreatic cancer cell lines obtained from the average fluorescence intensity per cell and the calibration curve. Data were obtained from at least 300 Capan-1 cells, 100 MIAPaCa-2 cells, and 50 Panc-1 cells. Error bars represent the standard error. Data are tabulated in Table S1.
The expression levels of biomarkers can vary depending on passage and genetic drift. Therefore, to validate the biomarker densities we performed flow cytometer analysis for CLDN4 expression on MIA PaCa-2 cells with phycoerythrin (PE)-conjugated anti-CLDN4, allowing us to make a direct comparison to results from QD-Ab conjugates. From control experiments with beads conjugated with known concentrations of PE and the known ratio of PE to antibodies, the number of PE molecules per cell was converted to antibodies per cell (see Supplemental Figure S6). From flow cytometry analysis we obtain an average CLDN4 density on MIA PaCa-2 cells of 121 ± 0.15 μm−2 (SE, N = 5000 cells), in excellent agreement with the value of 135 ± 3.6 μm−2 obtained from QD-aCLDN4 conjugates (average expression level per cell, N = 100 cells).
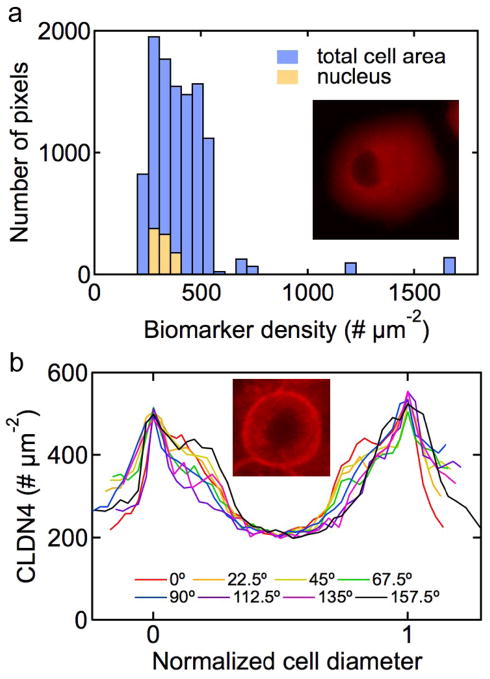
An advantage of biomarker profiling with QD-Ab conjugates, compared to conventional methods such as flow cytometry, is that we can obtain quantitative spatial information at the single cell level. Comparison to the control experiments where cells were incubated with QDs without antibodies (see Supplemental Figures S2 – S5), combined with our validation experiments implies that the fluorescence represents the spatial distribution over the cell surface. From the images in Figure 2, it is evident that the distribution of biomarkers over isolated Panc-1 and MIA PaCa-2 cells is relatively uniform over the cell surface. The nucleus appears somewhat darker since the images were obtained using an inverted microscope. Figure 5a shows the distribution of mesothelin over a Panc-1 cell. The distribution over the single cell is relatively narrow, 304 ± 0.5 μm−2 (SE, N = 10,802 pixels) indicating relatively uniform expression as inferred from the fluorescence image (inset). The intensity over the nucleus is 288 ± 1.4 μm−2 (SE, N = 912 pixels) only slightly lower than the global cell average (see Figure 5a). These results also demonstrate that QD aggregation and non-specific binding can be overcome with careful synthesis and design.
Figure 5.
Spatial distribution of biomarkers. (a) Spatial distribution of mesothelin expression levels over a Panc-1 cell (inset). (b) Quantitative linear profiling of the claudin-4 density across a capan-1 cell (inset). The profiles were along radial lines separated by 22.5° and normalized to the cell diameter.
In contrast to the Panc-1 and MIA PaCa-2 cells, the Capan-1 cells tend to grow in clusters. The distribution of claudin-4 on capan-1 cells is highly non-uniform with significantly higher intensity at the paracellular junctions, consistent with previous immunofluorescence studies8. This paracellular enhancement in cell clusters is expected since claudin-4 is a tight junction protein34. Figure 5b shows quantitative linear profiling of the claudin-4 density along a set of eight radial lines through the center of the cell and separated by an angle of 22.5°. In the paracellular regions, the claudin-4 density is around 500 μm−2, more than double the value in the central region. These results highlight the feasibility of quantitative spatial mapping for isolated cells and monolayer clusters.
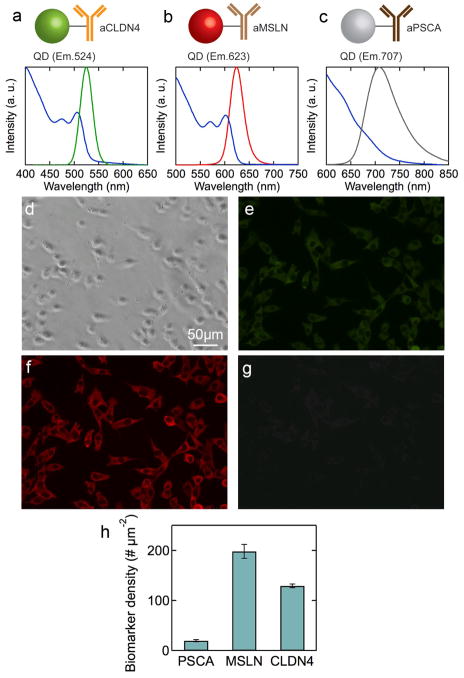
So far we have demonstrated quantitative profiling at the single cell level and spatial profiling. For high throughput profiling of multiple biomarkers, it would be desirable to perform multiplexed imaging. By attaching different antibodies to QDs with different emission wavelength, we prepared color-coded QD-Ab conjugates (see Figure 6) to demonstrate multiplexed targeting in human pancreatic cancer cell lines: QD(Em.524nm)-L-aCLDN4 (green), QD(Em.623nm)-L-aMSLN (red), and QD(Em.707nm)-L-aPSCA (NIR). Figure 8 shows the absorbance and emission spectra for each of the color-coded QD-Ab conjugates. The wavelength of each QD was tuned to minimize the overlap of the emission with those of other QDs, but still to be detectable using different emission filters. Equal amounts of the three different color-coded QDs were simultaneously incubated with MIA PaCa-2 cells and Figure 6 shows the resulting phase contrast image and fluorescence images at the same location taken with different emission filters (see Supplemental Figure S7). Biomarker densities determined from quantitative analysis (see Supplemental Figure S8) of the fluorescence images (Figure 6), are in a good agreement with the results from the individual QD-Ab conjugates (Figure 2) and analysis (Figure 6). Enhanced fluorescence images are shown in Supplemental Figure S9.
Figure 6.
Multiplexed imaging of cancer biomarkers on MIAPaCa-2 cells. Absorbance and emission spectra for (a) QD(Em.524)-L-aCLDN4, (b) QD(Em.623)-L-aMSLN, and (c) QD(Em.707)-L-aPSCA. (d) Phase contrast microscope image for MIAPaCa-2 cells after incubation with the three QD-Ab conjugates. Fluorescence images obtained with (e) FTIC (517/40, green), (f) TRITC (605/40, red), and (g) NIR (665 LP, infra red) filters. (h) Average biomarker density per cell for PSCA, claudin-4 and mesothelin in MIAPaCa-2 cells measured simultaneously. Standard error obtained from 150 cells.
Discussion
We have demonstrated quantitative profiling of biomarkers for pancreatic cancer at the single cell level using QD-Ab conjugates. The key requirements for quantitative profiling of membrane biomarkers using a QD probe are that one QD-Ab conjugate is bound to one target molecule, with no aggregation or non-specific binding. Using our lipid coating strategy for water solubilization and antibody coupling using pegylated lipids, non-specific binding and aggregation are negligible, allowing quantitative profiling of biomarkers for pancreatic cancer.
The expression levels for PSCA, CLDN4, and MSLN in Capan-1, MIA PaCa-2, and Panc-1 cells are in the range from about 30 μm−2 to 470 μm−2. The results are in agreement with results from western blot, northern blot, and PCR where expression levels are scored on a relative scale. The highest expression levels were obtained from PSCA and MSLN in Capan-1 cells, and the lowest expression levels were for PSCA in MIA PaCa-2 and Panc-1 cells. Expression levels were validated using flow cytometry to determine the average expression levels for CLDN4 on MIA PaCa-2 cells. The determination of quantitative expression levels allows direct comparison between cell types at the single cell level. Furthermore, we can provide quantitative spatial information on the distribution of biomarkers.
Despite the complexity of these experiments, measurements performed with QDs that were synthesized and functionalized at different times were reproducible. For example, here we report an average expression level for CLDN4 on Panc-1 of 214 μm−2 (Figure 4). In independent experiments we measured average expression levels of 228 μm−2 and 259 μm−2. Similarly, we measured values for MSLN expression on Panc-1 of 304 μm−2 (Figure 6) and 300 μm−2, and expression levels for PSCA on Panc-1 of 32 μm−2 (Figure 4) and 33 μm−2.
The measured expression levels of 30 μm−2 to 470 μm−2 correspond to average biomarker spacings on the cell membrane of 46 – 190 nm. For a 20 nm diameter QD-Ab conjugate, the maximum expression level that can be measured is 2500 μm−2. As described above, the detection limit reported here was about ±4 μm−2 corresponding to an average spacing of 500 nm. Based on the upper limit due to the size of the QD-Ab conjugates and the detection limit, the dynamic range for measurement is almost three orders of magnitude. An important advantage of QDs for profiling is that photobleaching is negligible (Figure 3b) and hence the intensity is linearly related to exposure time. As a result, longer exposure times can be used when the expression level is low.
We have also demonstrated quantitative multiplexed imaging using color-coded QDs. The expression levels obtained from multiplexed profiling of PSCA, CLDN4, and MSLN in MIA PaCa-2 cells very in excellent agreement with expression levels obtained from single QD-Ab experiments. These results show the feasibility of this technology for staging and forecasting since PSCA, CLDN4, and MSLN are expressed in different stages of progression of pancreatic cancer.
The ability to measure quantitative expression levels of membrane proteins has potential impact in a number of fields. For example, profiling of biomarkers in tissue samples would complement conventional histological staining and morphometric analysis, and may improve staging of disease progression. Similarly, profiling of single cells from blood samples, for example circulating tumor cells, may allow improved diagnosis and clinical management.
Supplementary Material
Quantitative targeting of pancreatic cancer biomarkers is demonstrated using quantum dot - antibody conjugates. Profiling and spatial mapping at the single cell level can be are achieved through minimizing non-specific binding and aggregation.
Acknowledgments
This work was supported by the National Institutes of Health (U54CA151838), NSF (CHE-0905869), and the Sol Goldman Pancreatic Cancer Research Center at Johns Hopkins University.
Abbreviations
- QD
quantum dot
- Ab
antibody
- PSCA
prostate stem cell antigen
- MSLN
mesothelin
- CLDN4
claudin-4
- MHPC
1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine
- DPPE
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lauroyl)
Footnotes
The authors have no conflict of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nature Methods. 2008;5(9):763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society, A. Cancer Facts and Figures 2009. American Cancer Society; Atlanta, GA: 2009. [Google Scholar]
- 3.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes and Development. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 4.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LDN, Goel R, Mathivanan S, Marimuthu A, Kashyap M, Vizza RF, Mayer RJ, DeCaprio JA, Srivastava S, Hanash SM, Hruban RH, Pandey A. A Compendium of Potential Biomarkers of Pancreatic Cancer. Plos Medicine. 2009;6(4):E1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maitra A, Iacobuzio-Donahue C, Argani P, Wilentz RE, Cameron JL, Yeo CJ, Kern SE, Goggins MG, Hruban RH. Expression of mesothelin and prostate stem cell antigen, two novel markers identified by serial analysis of gene expression, in mucinous cystic neoplasms and intraductal papillary mucinous neoplasms of the pancreas. Modern Pathology. 2002;15(1):137A–137A. [Google Scholar]
- 7.Wente MN, Jain A, Kono E, Berberat PO, Giese T, Reber HA, Friess H, Buchler MW, Reiter RE, Hines OJ. Prostate stem cell antigen is a putative target for immunotherapy in pancreatic cancer. Pancreas. 2005;31(2):119–125. doi: 10.1097/01.mpa.0000173459.81193.4d. [DOI] [PubMed] [Google Scholar]
- 8.Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM. Claudin-4: A new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001;121(3):678–684. doi: 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]
- 9.Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer - Support for use as a therapeutic target. American Journal of Clinical Pathology. 2004;121(2):226–230. doi: 10.1309/K144-PHVD-DUPD-D401. [DOI] [PubMed] [Google Scholar]
- 10.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65(21):9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Bharadwaj U, Zhang RX, Zhang S, Mu H, Fisher WE, Brunicardi FC, Chen CY, Yao QZ. Mesothelin is a malignant factor and therapeutic vaccine target for pancreatic cancer. Molecular Cancer Therapeutics. 2008;7(2):286–296. doi: 10.1158/1535-7163.MCT-07-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: Identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clinical Cancer Research. 2001;7(12):3862–3868. [PubMed] [Google Scholar]
- 13.Brus LE. Electron Electron and Electron-Hole Interactions in Small Semiconductor Crystallites - the Size Dependence of the Lowest Excited Electronic State. Journal of Chemical Physics. 1984;80(9):4403–4409. [Google Scholar]
- 14.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nature Materials. 2005;4(6):435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 16.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. In vivo cancer targeting and imaging with semiconductor quantum dots. Nature Biotechnology. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 17.Sapsford KE, Pons T, Medintz IL, Mattoussi H. Biosensing with luminescent semiconductor quantum dots. Sensors. 2006;6(8):925–953. [Google Scholar]
- 18.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Renal clearance of quantum dots. Nat Biotechnol. 2007;25(10):1165–70. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Chen K, Xie R, Xie J, Yan Y, Cheng Z, Peng X, Chen X. In vivo tumor-targeted fluorescence imaging using near-infrared non-cadmium quantum dots. Bioconjug Chem. 2010;21(4):604–9. doi: 10.1021/bc900323v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu AH, Gu WW, Larabell C, Alivisatos AP. Semiconductor nanocrystals for biological imaging. Current Opinion in Neurobiology. 2005;15(5):568–575. doi: 10.1016/j.conb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Noninvasive imaging of quantum dots in mice. Bioconjugate Chemistry. 2004;15(1):79–86. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 22.Smith BR, Cheng Z, De A, Koh AL, Sinclair R, Gambhir SS. Real-time intravital imaging of RGD-quantum dot binding to luminal endothelium in mouse tumor neovasculature. Nano Letters. 2008;8(9):2599–2606. doi: 10.1021/nl080141f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Dvoracek C, Lee KH, Galloway JF, Bhang HE, Pomper MG, Searson PC. CuInSe/ZnS Core/Shell NIR Quantum Dots for Biomedical Imaging. Small. 2011;7(22):3148–52. doi: 10.1002/smll.201101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298(5599):1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Lau SK, Varma VA, Moffitt RA, Caldwell M, Liu T, Young AN, Petros JA, Osunkoya AO, Krogstad T, Leyland-Jones B, Wang MD, Nie SM. Molecular Mapping of Tumor Heterogeneity on Clinical Tissue Specimens with Multiplexed Quantum Dots. Acs Nano. 2010;4(5):2755–2765. doi: 10.1021/nn100213v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howarth M, Liu WH, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nature Methods. 2008;5(5):397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder WJM, Strijkers GJ, van Tilborg GAF, Cormode DP, Fayad ZA, Nicolay K. Nanoparticulate Assemblies of Amphiphiles and Diagnostically Active Materials for Multimodality Imaging. Accounts of Chemical Research. 2009;42(7):904–914. doi: 10.1021/ar800223c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louie A. Multimodality Imaging Probes: Design and Challenges. Chemical Reviews. 2010;110(5):3146–3195. doi: 10.1021/cr9003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cormode DP, Skajaa T, van Schooneveld MM, Koole R, Jarzyna P, Lobatto ME, Calcagno C, Barazza A, Gordon RE, Zanzonico P, Fisher EA, Fayad ZA, Mulder WJM. Nanocrystal Core High-Density Lipoproteins: A Multimodality Contrast Agent Platform. Nano Letters. 2008;8(11):3715–3723. doi: 10.1021/nl801958b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carion O, Mahler B, Pons T, Dubertret B. Synthesis, encapsulation, purification and coupling of single quantum dots in phospholipid micelles for their use in cellular and in vivo imaging. Nature Protocols. 2007;2(10):2383–2390. doi: 10.1038/nprot.2007.351. [DOI] [PubMed] [Google Scholar]
- 31.Koole R, van Schooneveld MM, Hilhorst J, Castermans K, Cormode DP, Strijkers GJ, Donega CD, Vanmaekelbergh D, Griffioen AW, Nicolay K, Fayad ZA, Meijerink A, Mulder WJM. Paramagnetic Lipid-Coated Silica Nanoparticles with a Fluorescent Quantum Dot Core: A New Contrast Agent Platform for Multimodality Imaging. Bioconjugate Chemistry. 2008;19(12):2471–2479. doi: 10.1021/bc800368x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Lee KH, Galloway JF, Searson PC. Synthesis of Cadmium Selenide Quantum Dots from a Non-Coordinating Solvent: Growth Kinetics and Particle Size Distribution. Journal of Physical Chemistry C. 2008;112(46):17849–17854. [Google Scholar]
- 33.Galloway JF, Park J, Lee KH, Wirtz D, Searson PC. Exploiting Nucleation and Growth in the Synthesis and Electrical Passivation of CdSe Quantum Dots. Science of Advanced Materials. 2009;1(1):1–8. [Google Scholar]
- 34.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6(186):1–8. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.