Abstract
EZH2/H3K27me3 and polycomb group complex (PcG) play a major role in regulating global gene expression including tumor suppressor genes. EZH2 is linked to cell cycle regulated EZH2 phosphorylation by CDK1, a mitotic kinase which increases in arrested mitosis compared to S phase. CDK1 phosphorylation of EZH2 accelerates the degradation of pEZH2. Phospho-EZH2 is subjected to ubiquitination. The half-like of pEZH2 is shorter when compared to total EZH2. In the present study, pEZH2 was found concentrated together with ubiquitin in the Mallory-Denk bodies (MDB) that were formed in hepatocytes in the livers of drug primed mice refed DDC and humans with alcoholic hepatitis or hepatocellular carcinoma. The cells that formed MDBs in the mice livers studied were associated with a growth advantage and a high proliferative index. However, the livers from patients with alcoholic hepatitis showed evidence of cell cycle arrest where PCNA, cyclin D1 and p27 positive nuclei were numerous but Ki-67 positive nuclei were scarce. It is concluded that MDB formation is linked to the cell cycle and global gene expression (i.e. loss of gene silencing) through its association with the regulation of the polycomb group PRC2/EZH2/H3K27me3 complex.
Keywords: EZH2 (enhancer of Zest 2), H3K27me3 (trimethylated H3K27), MDB (Mallory-Denk bodies), Alcoholic hepatitis (AH), Hepatocellular carcinoma (HCC)
INTRODUCTION
Liver cells proliferate, over express FAT10 and form Mallory-Denk bodies (MDB), when DDC primed mice are refed DDC for 7 days. This response is prevented by feeding S-adenosyl methionine (SAMe) or betaine with DDC (Li et al., 2008; Bardag-Gorce et al., 2007; Oliva et al., 2009; Oliva et al., 2008). Betaine and SAMe provide methyl groups for methylation of DNA by the H3K27 me3/EZH2 polycomb group complex (PcG) which prevents this liver cell phenotype from forming (Chen et al., 2010). DDC feeding in this model ultimately leads to liver tumor formation long after DDC is withdrawn (Oliva et al., 2008; Nan et al., 2006). The significance of the change in liver cell phenotype to over express FAT10 and form MDBs is that these changed hepatocytes have a decrease in the 26s proteasome formation, which is due to a shift from the 26s proteasome to form the immunoproteasome. This leads to loss of the function of the 26s proteasome and consequentially the formation of MDB aggresomes (Oliva et al., 2010; Bardag-Gorce et al., 2010). MDBs also form in human livers in chronic liver diseases as well as in hepatocellular carcinomas (HCC). This raises the intriguing question as to whether changes in EZH2/H3K27me3 are involved in the pathogenesis of MDBs that form in man (French et al., 2011).
In preliminary studies it was shown that pEZH2 co-localized with ubiquitin in the MDBs formed in the livers of mice refed DDC (French et al., 2011). Ubiquitin binds to keratins localized in the MDB aggresome (Ohta et al., 1988). MDB forming hepatocytes lack adequate functioning 26S proteasomes (Bardag-Gorce et al., 2010) which could explain the failure of these liver cells to degrade pEZH2. The degradation of EZH2 by the 26s proteasome is accelerated when EZH2 is phosphorylated by CDK1 (Wu and Zhang et al., 2011). Phosphorylation of EZH2 suppresses methylation of H3K27 (Wei et al., 2011).
Methylation of DNA is reduced in the MDB forming hepatocytes since 51 methylcytosine is reduced in the hepatocytic nuclei in MDB forming cells (Li et al., 2008; Oliva et al., 2008). H3K27me3 is reduced in the nuclear fraction isolated from the livers of DDC primed mice refed DDC (Bardag-Gorce et al, 2010). This leads to the loss of methylated DNA, the reduction in global gene silencing and the increase in the expression of a large number of genes (Li et al., 2008; Oliva et al., 2008).
EZH2 (enhancer of zeste 2) is a member of the polycomb group complex (PeG) of proteins together with H3K27me3. EZH2 has the ability to trimethylate lysine 27 of histone 3 (Wu and Zhang, 2011). EZH2 plays an important role in epigenetic silencing, the cell cycle and stem cell biology where cell differentiation or self renewal is determined by regulating nanog expression in the link with promoters of pluripotent and differentiated cells and cancer progenitor cell formation (Kheradmand et al., 2009; Wei et al., 2011; Zeng et al., 2011; Chen, 2010; Villasante et al., 2011; Chen et al., 2010; Shen et al., 2008).
EZH2 plays a role in cell proliferation giving cells a proliferative advantage (Braken et al., 2003). EZH2 is also often over expressed in several different types of cancers (Simon and Lange, 2008; Varambally et al., 2002). RNAi mediated knockdown of EZH2 compromises cell proliferation (Braken et al., 2003), Varambally et al., 2002). EZH2 expression is high in proliferating cells so that the over expression of EZH2 contributes to oncogenesis by inappropriately silencing tumor suppressor genes (Wu and Zhang, 2010).
In the DDC refeeding mouse model, in which the cells proliferate and form MDBs, FAT10 a marker for hepatocellular carcinoma (HCC) is also over expressed (Lee et al., 2003) and later liver tumors form (Oliva et al., 2008). These cells have a growth advantage over normal cells and proliferate to replace the normal hepatocytes (Oliva et al., 2008; Nan et al., 2006; Oliva et al., 2009). Thus, the phenotypically changed hepatocytes that over express FAT10 and form MDBs are hepatic progenitor cells. The question addressed here is, what role EZH2 plays in the formation of MDBs and FAT10 over expression.
METHODS
In order to determine the relationship between MDB formation, FAT10 over expression, H3K27me3 methylation, and EZH2 phosphorylation to liver cell proliferation in chronic liver disease and HCC, mice and human livers were studied using Western blot, RT PCR and immunohistochemistry.
Archived mouse liver tissue stored at −80C°, nuclear fractions isolated from mouse livers and mouse zinc/formalin-fixed liver tissues embedded in paraffin were utilized. Human liver resections surgically removed from HCCs in patients with HCV and/or HBV and stored in paraffin were also studied using immunohistochemical stains. Biopsies of livers from patients who had alcoholic hepatitis were similarly studied by immunohistochemistry. In all cases the presence of liver cells forming MDBs was the focus of the examination.
Mouse Liver
Mice were fed 0.1% DDC (diethyl 1.4-dehydro-1, 4, 6-trimethyl-3,5-pyridine-dicarboxylate according to the DDC priming protocol (Yuan et al., 1996) where DDC was fed mixed in a solid, high protein, nutritionally adequate diet, for 10 weeks followed by one month of DDC withdrawal. This was followed by 7 days of DDC refeeding.
Three mice were used in each of four groups as follows: 1) control, 2) DDC fed for 10 weeks, withdrawn 1 month, refed DDC 7 days, 3) refed DDC+betaine, 4) refed DDC+S-adenosyl methionine (SAMe). SAMe was fed daily for 7 days (4 g/kg body weight daily by gavage). (Bardag-Gorce et al., 2010). Betaine was fed in the drinking water (2%) for 7 days. RTPCR was performed to measure EZH2 mRNA.
Mouse livers were used in the immunofluorescent studies. All mice were treated in a humane manner as approved by the Animal Care Committee at Harbor-UCLA LA Biomed Research and Education Institute according to the Guidelines of the National Academy of Science. Seven human archived AH liver biopsies and 4 human archived HCC tissues forming MDBs were utilized in the immunofluorescent studies.
Nuclei Isolation
The isolation was carried out according to the method of Umlauf et al., 2004. Liver tissue, frozen in isopentane, which was immersed in liquid nitrogen and stored at −80°C, was homogenized in a Dounce homogenizer with 10 strokes in 1 ml of buffer-I. The homogenates were centrifuged for 10 min at 6000 g. The Pellets were then re suspended in buffer-II, placed on ice for 10 min and then centrifuged 20 min at 9000 g on a sucrose cushion (buffer III). All buffers used contained protease inhibitors: 10 mM benzamidine, 0.7 µg/ml leupeptin, 50 µg/ml soy bean trypsin inhibitor, 0.2 µg/ml aprotinin, 2 µg/ml antipain, 0.7 µg/ml pepstatin, 0.5 mM PMSF (Calbiochem, La Jolla, CA), sodium butyrate 5 mM and DTT 1 Mm. Protein concentrations were measured using the Bradford method (Bradford, 1976), and bovine serum albumin was the protein standard.
Western Blot Analysis
Proteins (50 µg) from livers which were fast frozen in liquid nitrogen and stored at −80°C, and nuclear extracts were separated by SDS-PAGE gels and transferred to a PVDF membrane (Bio-Rad, Hercules, CA) for 1 h in 25 mM Tris-HC1 (ph 8.3), 192 mM glycine and 20% methanol. The membranes were stained using primary antibodies to antigens. Appropriate species polyclonal and monoclonal HRP-conjugated antibodies were used as the secondary antibodies. The membranes were subjected to chemiluminescence detection using luminal, according to the manufacturer’s instructions (Amersham Pharmacia Biotech, NJ). The antibodies used were to EZH2 (Cell Signaling, Danvers MA) and pEZH2 (Epitomics, Burlingame CA.).
Quantitative real-time RT-PCR assay
Total liver RNAs were extracted with Trizol Plus RNA Purification Kit (Invitrogen, Carlsbad, CA) as described previously (Li et al., 2008). The sequence of PCR primers for EZH2 were: NM-001146689.1
Forward
GGTGAAGAGTTGTTTTTTGATTACAGA
Reverse
TCTCGTTCGATGCCCACATA
Immunohistochemistry
Liver tissue was fixed in 10% buffered zinc formalin. These sections were double stained using a rabbit polyclonal antibody to ubiquitin (Dako, Carpenteria CA) or a mouse antibody to ubiquitin (Millipore, Billerica MA) for one stain. A second stain was done with antibodies to H3K27me3, (Abcam, Cambridge MA), EZH2, (Cell Signaling, Danvers MA) or pEZH2 (Bethyl, Montgomery AL). Texas-red (Jackson, Westgrove PA) and FITC (Jackson, Westgrove PA) conjugated second antibodies were used. DAPI (Invitrogen, Eugene OR) was the nuclear stain. H3K27me3 was measured morphometrically in the nuclei by comparing the staining intensity of the nuclei in MDB forming hepatocytes with neighboring normal hepatocytes. The results were displayed as a graph attached to the immunofluorescent photograph transferred from the monitor to the memory stick electronically as a screen hunter. A Nikon 400 fluorescent microscope was used and morphometric monitoring was done using Nikon software. In addition immunoperoxidase staining was done using mouse anti Ki-67 nuclear antigen (Zymed, SanFrancisco CA) mouse anti PCNA (Signet, Dedham MA), rabbit monoclonal anti cyclin D1 (Biocare, Concord CA), anti p21 and anti p27 (Dako, Carpenteria Ca).
Statistical analysis
P values were determined by ANOVA and Student Newman-Keuls for multiple group comparisons (Sigma-Stat software, San Francisco, CA).
RESULTS
Mouse livers
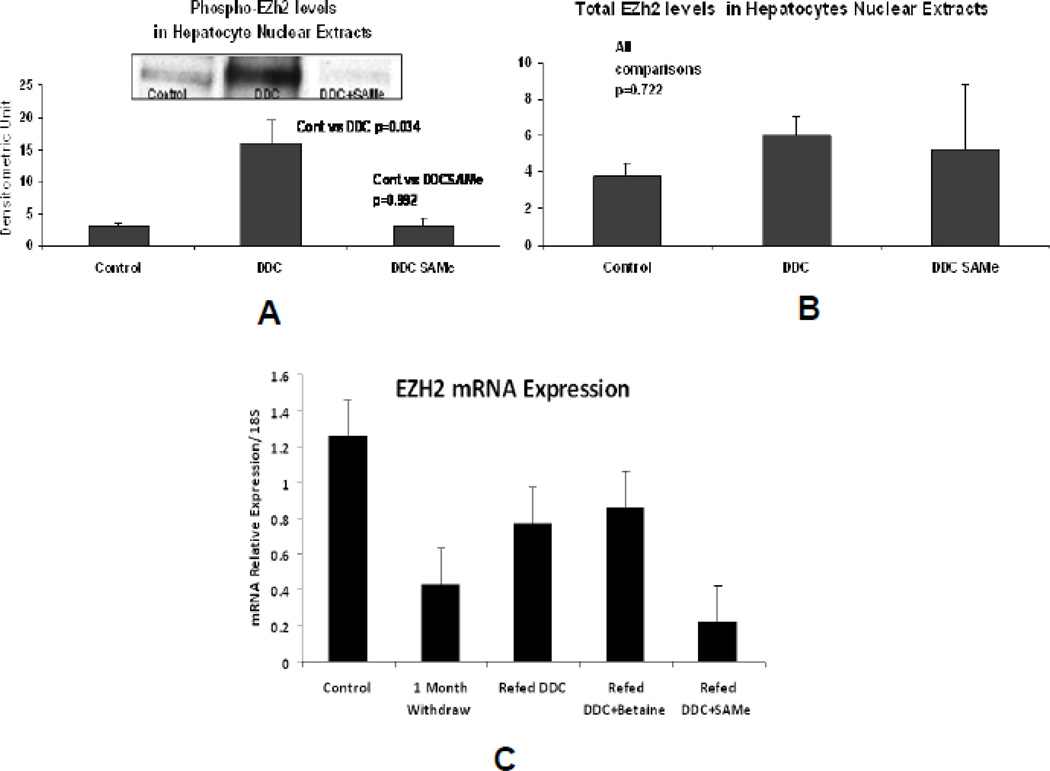
Western blots of livers from mice refed DDC compared with normal controls showed that the level of EZH2 was not changed (Fig.1a). The pEZH2 band was increased in the DDC refed mice livers and this increase was prevented when SAMe was fed with DDC (Fig 1b).
Fig 1.
A) Western blot using the antibody to pEZH2 showed an increase in proteins in the n mean±SEM). B) Western blots using the antibody to EZH2 showed there was no significant difference between the 3 groups of mice used in A (n=3). C) qPCR of the nuclear levels of EZH2 mRNA showed no significant difference in the levels between the controls and the DDC refed groups of mice except when SAMe was fed the DDC refed mice (n=3, mean±S.E.M., p<0.05).
The RTPCR results showed that EZH2 in the DDC withdrawal group of the DDC refed group and the DDC refed + betaine group tended to have lower values compared to the normal control group (Fig 1c). Only the DDC refed + SAMe group had significantly lower values.
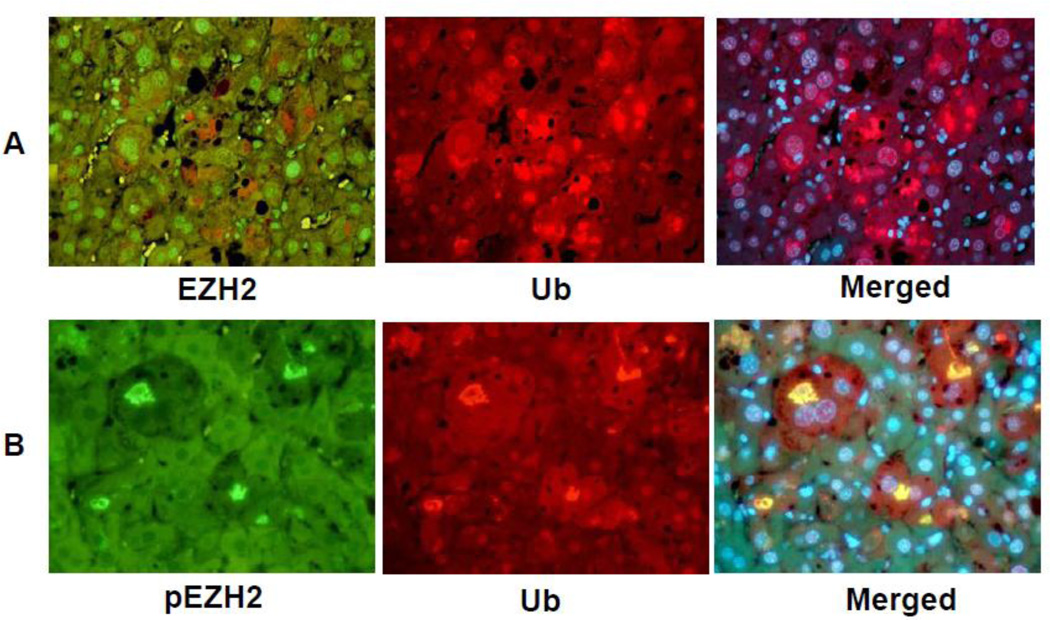
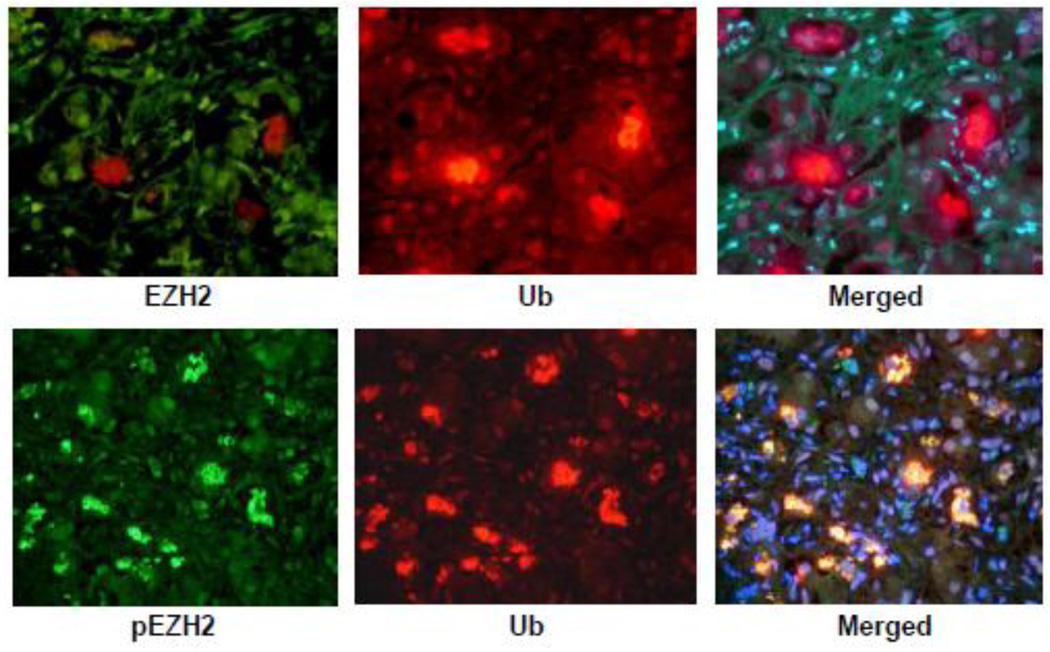
The immunofluorescent (IF) results of the livers from DDC fed mice showed no change in the liver cells forming MDBs compared to normal hepatocytes when double stained for ubiquitin (Ub) and EZH2 (Fig 2A). However, pEZH2 stained strongly positive in the MDBs within the liver cells as indicated by co localization of Ub positive stain (red) and the pEZH2 positive stain (green) and merged in the tricolor stain (yellow) (Fig 2B).
Fig 2.
Liver cells which formed MDBs stained positive for ubiquitin (A and B, red) and pEZH2 (A, green) but not EZH2 (B, green). pEZH2 and ubiquitin were co localized in MDBs (B, yellow). (A×479, B×572.)
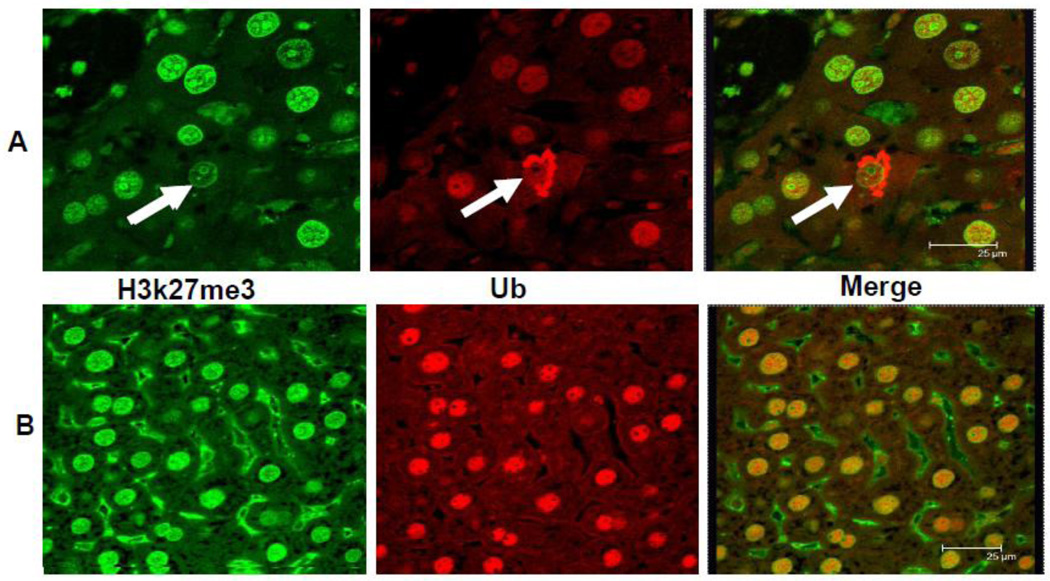
The immunofluorescent antibody stain for H3K27me3 was positive in the nuclei in control and DDC refed mice livers (Fig 3). Screen Hunters were obtained from the morphometric screen to visualize and compare the intensity of nuclear staining for H3K27me3 in the nuclei of MDB forming cells compared to the nuclei of neighboring normal hepatocytes (Fig 4). Note that the intensity of nuclear staining was positive and equal in all the livers cells in the control. Next, note that for comparison in the mouse refed DDC where MDBs (yellow stained) had been formed by hepatocytes, the nuclei had variable decreased staining intensity compared to normal hepatocytic nuclei (see white line which defines the traced nuclear area for the green line intensity). The blue line on the photomicrograph indicates where the intensity was measured where it runs over the nuclei compared here. This shows that the nucleus in the MDB forming cells stains with less intensity in contrast to the nucleus in the normal liver cell, which shows an increase in intensity. Fig 5 shows mitosis in a liver cell that formed a MDB, indicating that these MDB forming cells were proliferating. Fig. 6 shows that nuclei stained with the PCNA antibody were increased in DDC refed mouse livers indicating that there is increased liver cell proliferation in the DDC refed mice.
Fig 3.
The Mallory-Denk body (MDB) hepatocytic balloon cell is shown in the upper panel. The stain for H3K27me3 in the nucleus of the balloon cell is less intense than the adjacent normal hepatocytic nucleus (arrow) and the nuclei in the control liver in the lower panel. The balloon cell MDB stains positive for ubiquitin (upper panel). The merged photo of the balloon cell further documents the reduced nuclear staining for H3/K27me3 (Confocal microscope).
Fig 4.
The MDB forming hepatocytic balloon cell nucleus in the liver of a DDC refed mouse shows the loss of staining intensity (green) for H3K27me3 (B) when compared to the neighboring normal hepatocytes. The normal control (A) shows equal intensity staining of the nuclei for H3K27me3, (B) (arrow) MDBs stain yellow because H3K27me3 (green) is double stained with ubiquitin (red). The reduction in nuclear levels of H3K27me3 induced by DDC refeeding has been confirmed by Western blot (Bardag-Gorce et al., 2010). (Screen Hunter) (A×543, B×654)
Fig 5.
Liver from a DDC refed mouse double stained with an antibody to ubiquitin and DAPI (blue) shows that the nucleus of a ballooned hepatocyte that had formed an MDB was in mitoses (arrow) (x 1962).
Fig 6.
Sections of liver from mice refed DDC and control stained with an antibody to PCNA showing rare nuclear staining positive in the control (A) and numerous positive staining in the DDC refed mouse liver (C). Similarly liver from the mouse refed DDC stained for cyclin D1 (D) shows numerous positive stained nuclei. The control mouse liver shows only a few positive stained nuclei. Magnifications are; (A×606), (B×606), (C×606), (D×910).
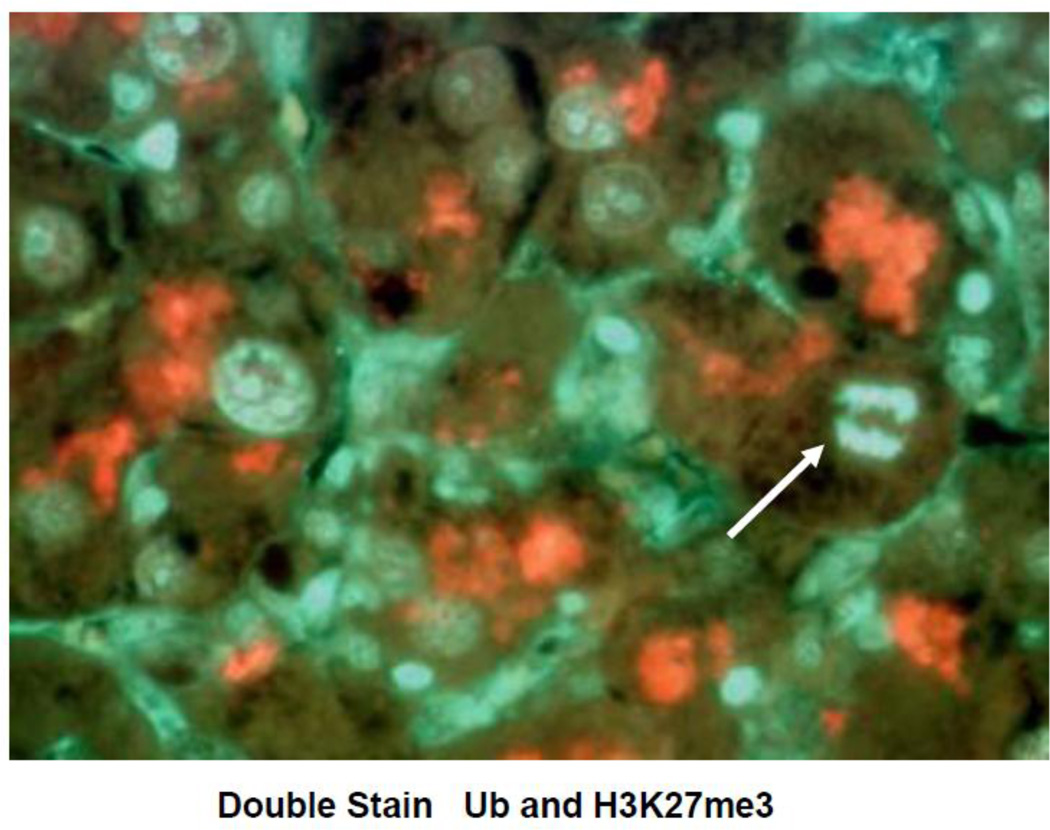
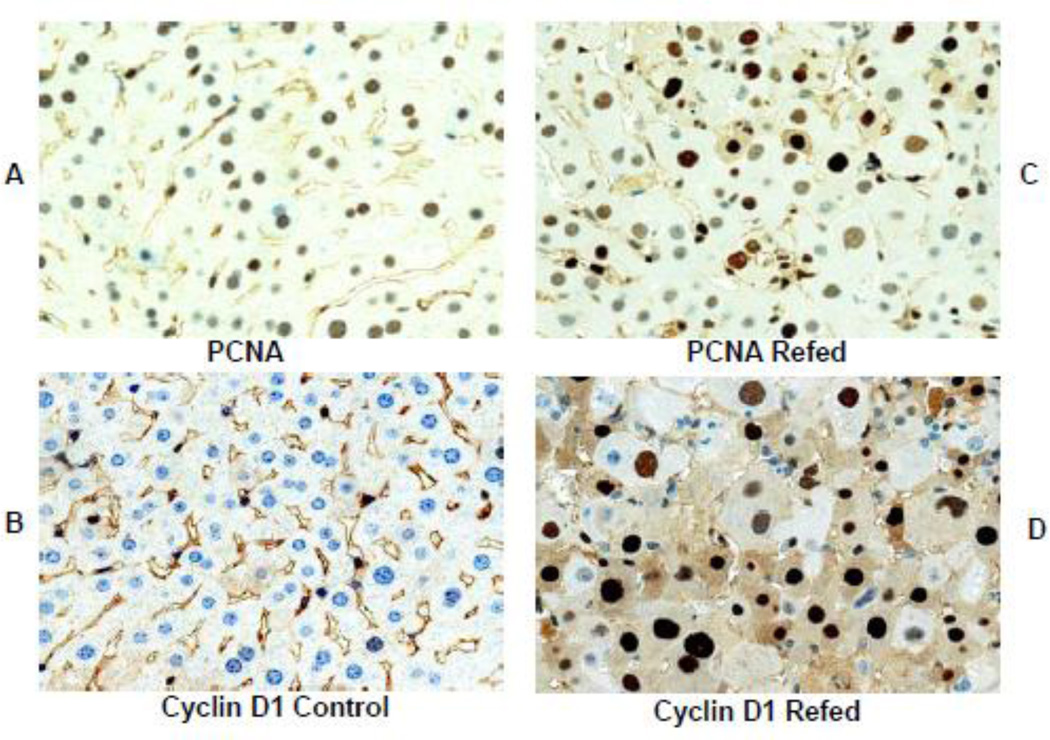
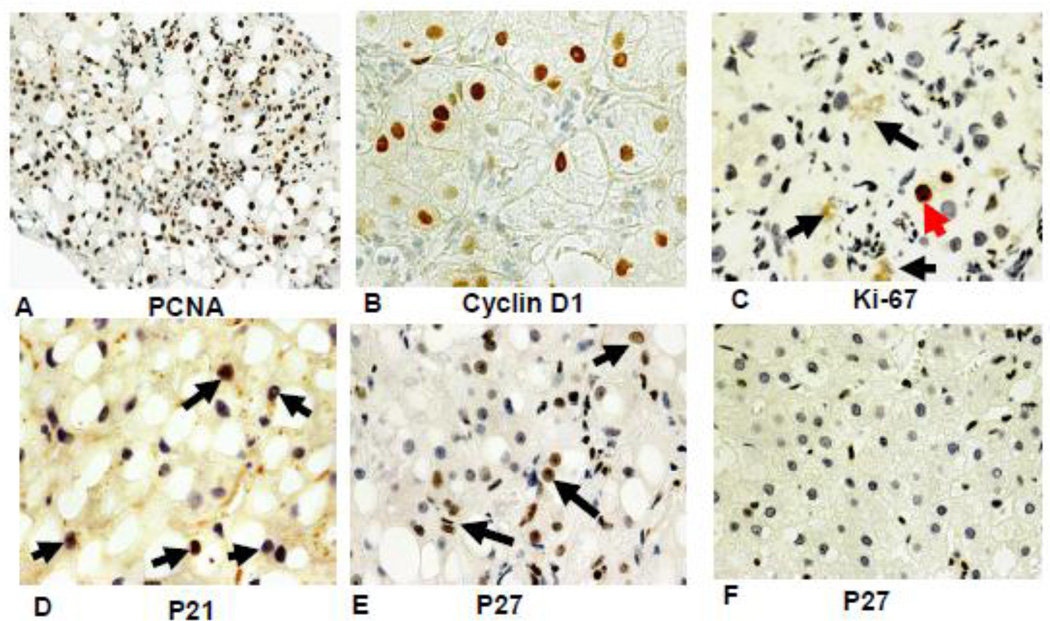
Human liver Human livers in 7 alcoholic hepatitis (AH) cases and 4 hepatocellular carcinoma cases (HCC) where MDBs were formed were studied in order to compare them with the changes in mouse livers forming MDBs. Immunostains for EZH2 and pEZH2 were done. In AH where MDBs formed the antibodies to UB and pEZH2 co localized in MDBs, as was seen in the mice liver MDBs. (Fig 7). The immunostain for PCNA and Cyclin D1 indicated that most of the liver cells stained positive in 6 out of the 7 biopsies of AH (Fig 8A&B). However, the stain for Ki67 was positive in only a few hepatocytes (8C). The stain for p21 (8D) and p27 (8E) were positive in numerous hepatocytic nuclei in the liver biopsies from patients with alcoholic hepatitis. Since p21 and p27 delay the cell cycle and cause cell cycle arrest, this finding could explain the discrepancy between the PCNA and Ki-67 results. Double stains for Ub and H3K27me3 of the AH livers showed reduced H3K27me3 in the nuclei of the MDB forming liver cells compared to adjacent normal liver cells (Fig 9). Electron microscopy of the MDB forming hepatocytes showed euchromatin in the nuclei (Fig 10).
Fig 7.
A liver biopsy from a patient who had alcoholic hepatitis was stained with an antibody to EZH2 (upper panel) and an antibody to pEZH2 (lower panel). Note that the nuclei stained for EZH2 were all stained with the same intensity (left photo). The MDBs were stained green for pEZH2, red for ubiquitin and yellow when the tricolor filter was used indicating that pEZH2 was increased in MDBs. pEZH2 was also increased in the Western blot in mice refed DDC (Fig 1) and the liver section stained with the pEZH2 antibody (Fig 2). (Upper and Lower Panel×436).
Fig 8.
The liver biopsy slides from patients with alcoholic hepatitis were stained with antibodies to PCNA (A), Cyclin D1 (B), Ki-67 (C), p21 (D) and p27 (E). Only a few scattered nuclei were positive for Ki67 (arrow). No nuclei stained positive in a liver from an alcoholic hepatitis patient. (F). Magnification (A×218), (B×654), (C×654), (D×684), (E×436), F×436).
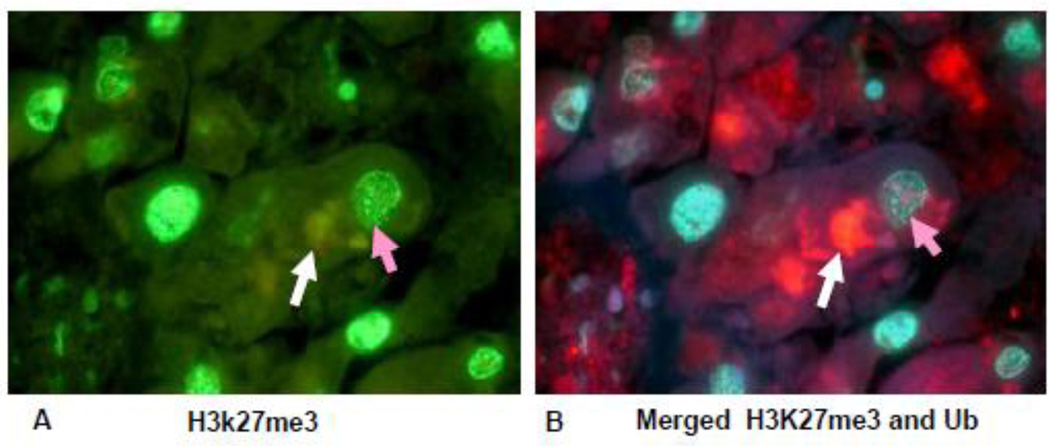
Fig 9.
Liver section from a patient who died with alcoholic hepatitis, stained for H3K27me3 and ubiquitin. Note that the liver cell containing an MDB (white arrow, red) B) stained with less intensity for H3K27me3 (orange arrow) when compared to the nucleus of a neighboring hepatocyte. (A×1040), (B×1040).
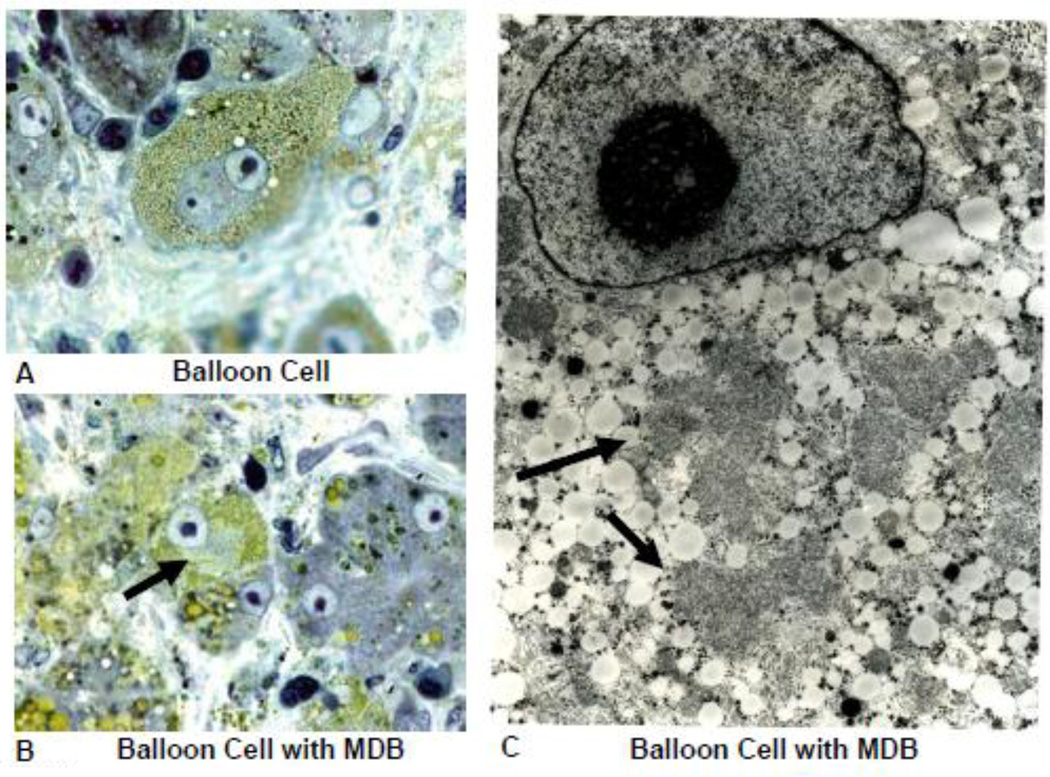
Fig 10.
Electron microscopic photograph of balloon cells from a patient with alcoholic hepatitis. A) Balloon cell with micro vesicular fat stained by osmium and a nucleus which contains a large nucleolus. B) Ballooning cell that formed an MDB (arrow). C) EM of balloon cell with micro vesicular fat in the cytoplasm together with an MDB (arrows). Note the nuclear euchromatin and prominent nucleolus, (A×1451), (B×1040), (C×6710).
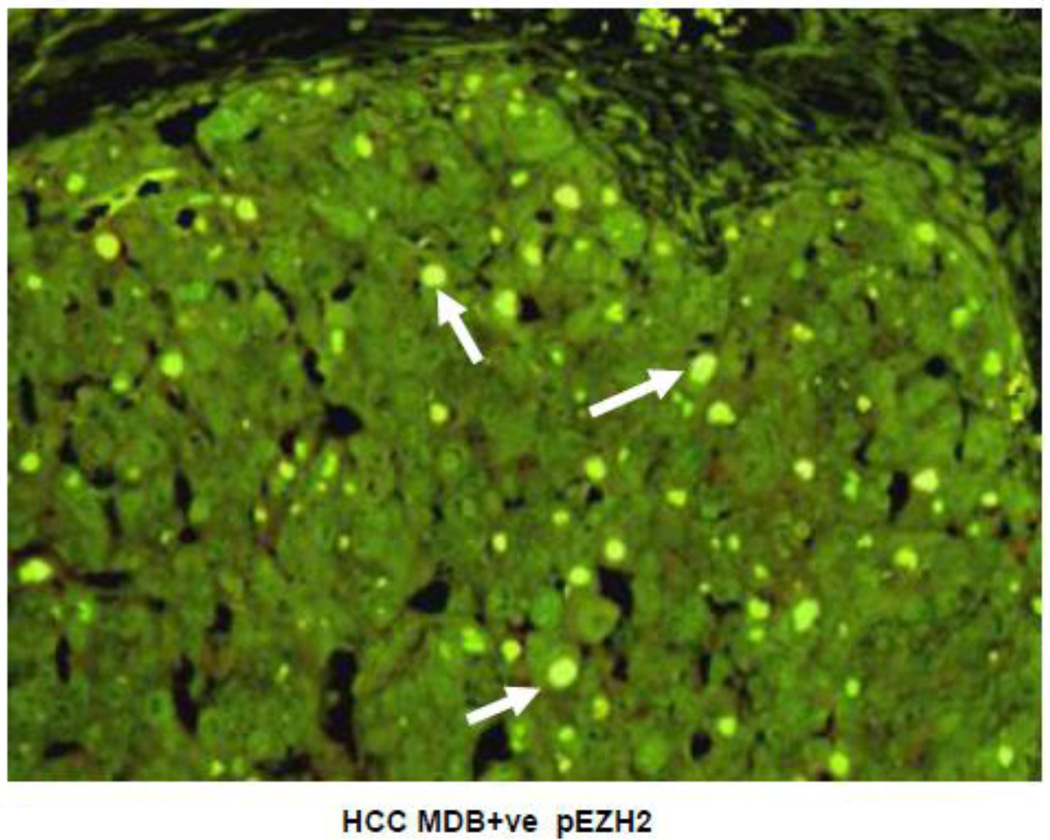
The double stain of hepatocellular cells showed co-localization of UB and pEZH2 in the MDBs (Fig 11). The MDB forming HCC cells showed uniform PCNA positivity of the tumor cell nuclei in 3 out of 4 HCCs studied where MDBs were formed. This was compared to tumor areas where no MDBs were formed (Fig 11). When the tumors forming MDBs were stained for pEZH2, the MDBs stained positive for pEZH2 (12) in the same way as the MDBs in the drug fed mice and human ALD did. Tumor cells forming MDBs that were stained for H3K27me3 had nuclei which stained less intensely when compared with tumor cells in adjacent tumor areas where no MDBs were formed (Fig 13).
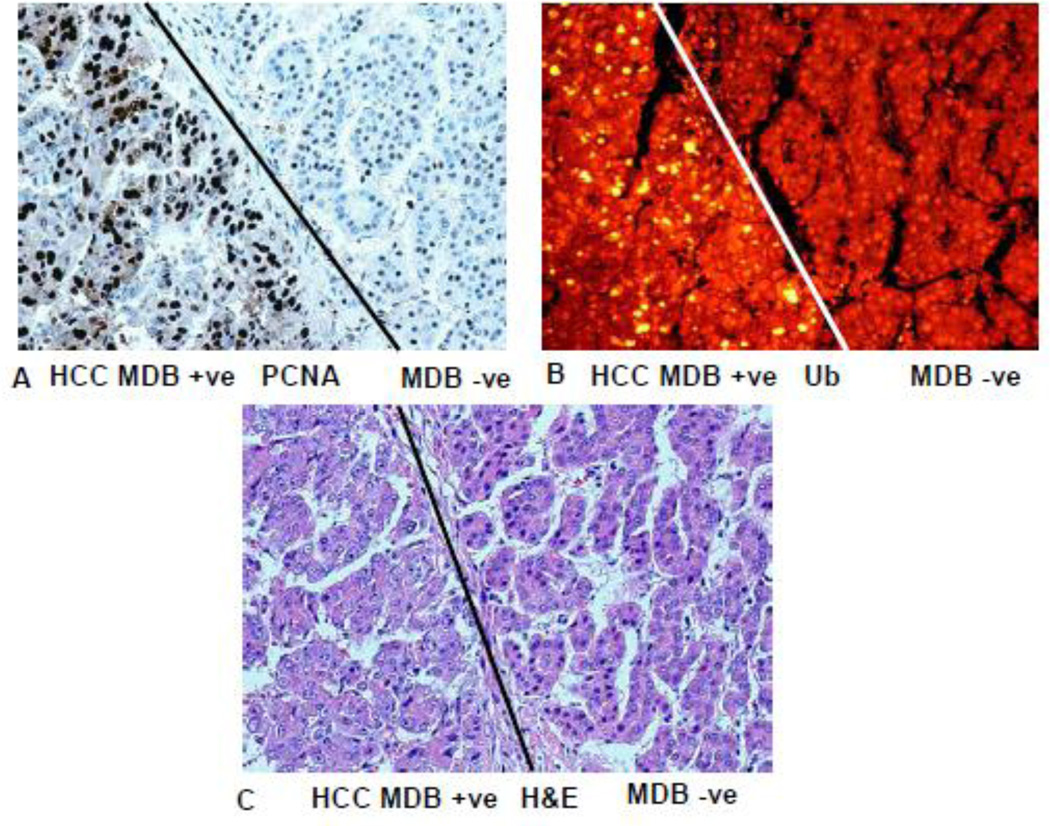
Fig 11.
Liver hepatocellular carcinoma has formed MDBs to the left of the line and not to the right of line in A, B, and C. A) PCNA stain shows that most of the nuclei in tumor cells that have formed MDBs stain positive. B) MDBs formed by the tumor cells stain positive for ubiquitin (yellow). C) Hematoxylin and eosin stain. The tumor cells that formed MDBs have vesicular nuclei compared to the tumor cells that have not formed MDBs on the right. (A×346), (B×346), (C×346).
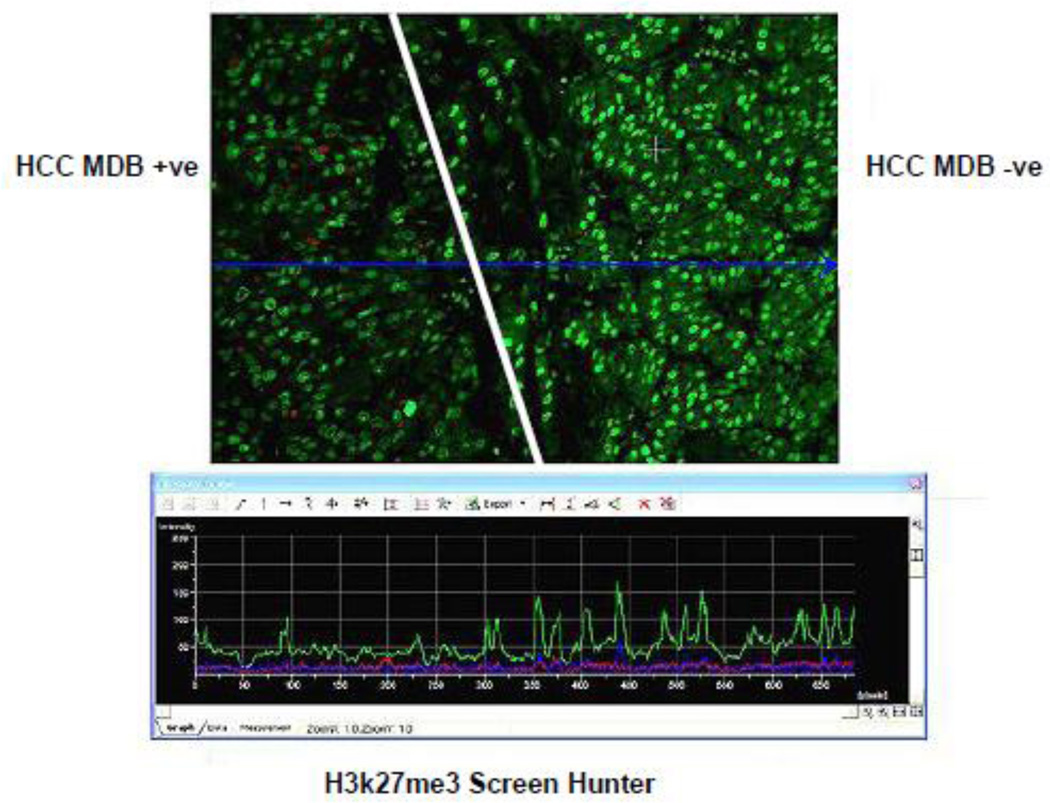
Fig 13.
Hepatocellular carcinoma, the same as shown in Fig 11 and 12, stained for H3K27me3. Note the tumor cell nuclei on the left that have formed MDBs stain with less intensity than the tumor cells on the right which have not formed MDBs. Note the green tracing of intensity below in the screen hunter (x 206).
Discussion
In both mouse liver cells that formed MDBs and human liver cells that formed MDBs in AH and HCC, the hepatocytic nuclear H3K27me3 staining was diminished, the cell proliferation was increased, and the pEZH2 was found sequestered within the MDBs. PEZH2 was increased in the nuclei of livers of the DDC refed mice compared to control mice and mice refed DDC+SAMe. Thus, the MDB expressing hepatocytes showed an increase in pEZH2 and a decrease in nuclear H3K27me3 gene silencing, leading to an increase in the number of MDB forming cells. There was a decrease in expression of EZH2 by RTPCR when SAMe was fed with DDC. This could explain why the nuclear staining intensity of 5’ methylcystosine in MDB forming mouse hepatocytes was decreased and not prevented by SAMe feeding (Oliva et al., 2008). PcG proteins like EZH2 play an essential role in regulation of methylation of DNA by H3K27me3 and the regulation of epigenetic gene silencing (Chen et al., 2010). This function of EZH2 is important for the regulation of cell renewal and cell differentiation and is involved in cancer (Varambally et al., 2002; Villasante et al., 2011).
The EZH2 gene transcript is negatively regulated by the tumor suppressor RB and the microRNA Mir-101 (Bracken et al., 2003). AKT phosphorylates EZH2 and inhibits methyltransferase activity (Cha et al., 2005) by impeding the binding of EZH2 to histone H3K27. This may explain the result in which there was a decrease in H3K27 trimethylation and a reduction of gene silencing in the DDC refed mouse model (Bardag-Gorce et al., 2010).
The number of nuclei that stained positive for PCNA, Cyclin D1, p21 and p27 was markedly increased in the liver of biopsies from patients with alcoholic hepatitis. This was in contrast to the low number of nuclei that stained positive for Ki-67. The implication is that the cell cycle progression was arrested by p21 and p27 up regulation. The increase in PCNA positive nuclei in alcoholic hepatitis was first reported by Fang et al., 1994. The increase in p21 nuclear staining and the decrease in Ki-67 in alcoholic hepatitis were first reported by Crary and Albrecht, 1998. Now we have added that p27 positive nuclei are increased in alcoholic hepatitis. P27 has tumor repressor effects as well as the ability to induce cell cycle arrest, cause genetic instability, apoptosis and potential oncogenic effects, (Abbas and Dutta, 2011); Serres et al., 2012; Serres et al., 2011. P21 is a downstream transcriptional target of p53. It functions as a mediator of G1/5 growth arrest. P21 does this both by direct inhibition of DNA synthesis by way of suppression of PCNA-dependent polymerase and indirectly by inhibiting Cyclin A and Cyclin E-cdk2 activity necessary for S-phase progression (Li et al., 2010. P21 also promotes growth arrest during the G 2 phase of the cell cycle in response to genotoxic stress due to its ability to inhibit Cdk1 complexes (Abbas and Dutta 2009; Cazzalini et al., 2010). The role of p21 and p27 increase in the cell cycle arrest induced by ethanol experimentally has recently been reported (Clemens, 2007). Koteish et al., 2002 showed that p21 and p27 were increased in animals fed ethanol for 1 month when compared to controls after a partial hepatectomy.
Fig 12.
Hepatocellular carcinoma double stained for pEZH2 and ubiquitin viewed with the tricolor filter showing co localization in MDBs (arrows). (x654).
Acknowledgement
The authors thank Adriana Flores for typing the manuscript. The study was supported by NIH/NIAAA P50-011999 Morphology Core and NIH/NIAAA 8116.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbas T, Dutta A. CRL4Cd+2; Master coordinator of cell cycle progression and genome stability. Cell cycle. 2011;10:241–249. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer intricate net works and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Dedes J, French BA, Oliva J, Li J, French SW. Mallory body formation is associated with epigenetic phenotypic change in hepatocytes in vivo. Exp Mol Pathol. 2007;83:160–168. doi: 10.1016/j.yexmp.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Lin A, Li J, French BA, French SW. SAMe prevents the up regulation of toll-like receptor signaling in Mallory-Denk body forming hepatocytes. Exp Mol Pathol. 2010;88:376–379. doi: 10.1016/j.yexmp.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Braken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha T-L, Zhou BP, Xia W, Wu Y, Yang C-C, Chen C-T, Ping B, Otto AP, Hung MC. KT-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone 3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chen L. A link between H3K27me3 mark and exon length in the gene promotes of pluripotent and differentiated cells. Discovery Note. 2010;26:855–859. doi: 10.1093/bioinformatics/btq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nature Cell Biol. 2010;12:1208–1215. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL. Effects of ethanol on hepatic cellular replication and cell cycle progression. World J Gastroenterol. 2007;13:4955–4959. doi: 10.3748/wjg.v13.i37.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crary GS, Albredht JH. Expression of cyclin-dependent kinase inhibitor p21 in human liver. Hepatology. 1988;28:738–743. doi: 10.1002/hep.510280320. [DOI] [PubMed] [Google Scholar]
- Fang JWS, Bird GLA, Nakamura T, Davis J, Lau JYN. Hepatocyte proliferation as an indicator of outome in acute alcoholic hepatitis. Lancet. 1994;33:820–828. doi: 10.1016/s0140-6736(94)92025-7. [DOI] [PubMed] [Google Scholar]
- French SW, Bardag-Gorce F, French BA, Li J, Oliva J. The role of inmate immunity in the pathogenesis of preneoplasia in drug-induced chronic hepatitis based on a mouse model. Exp Mol Pathol. 2011;91:653–659. doi: 10.1016/j.yexmp.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand S, Kortalaci PS, Farahbakhshian E, Pourfarzad F, von Lindern M, Verrijzer CP. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence, Epigenics Chromatin. 2009;2:16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteich A, Yang S, Lin H, Huang J, Diehl AM. Ethanol induces redox-sensitive cell cycle inhibitors and inhibits liver after partial hepatectomy. Alcohol Clin Exp Res. 2002;26:1710–1718. doi: 10.1097/01.ALC.0000036923.77613.59. [DOI] [PubMed] [Google Scholar]
- Lee CG, Ren J, Cheong LS, Ban KH, Ooi L, YongTan S, Kan A, Nuchprayoon I, Jin R, Lee KH, Choti M, Lee LA. Expression of the FAT10 gene is highly up regulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22:2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- Li J, Bardag-Gorce F, Dedes J, French BA, Oliva J, Amidi F, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting epigenetic memory. Hepatology. 2008;47:613–624. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jenkins CW, Nichols MA, Xiog Y. Cell cycle expression and p53 regulation of the cyclin-dependent kinase inhibitor p21. Oncogene. 1994;9:2261–2268. 1994. [PubMed] [Google Scholar]
- Nan L, Bardag-Gorce F, Wu Y, Li J, French BA, French SW. Mallory body forming cells express the preneoplastic hepatocyte phenotype. Exp Mol Path. 2006;80:109–118. doi: 10.1016/j.yexmp.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Ohta M, Marceau N, Perry G, Manetto V, Gambetti P, Antilo-Gambetti L, Metuzals J, Cadrin M, Kawahara H, French SW. Ubiquitin is present on the cytokeratin intermediate filaments and Mallory bodies of hepatocytes. Lab Invest. 1988;58:848–856. [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S, French SW. FAT10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;94:101–112. doi: 10.1016/j.yexmp.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Li J, French BA, Nguyen SK, Lu SC, French SW. Betaine prevents Mallory-Denk body formation in drug-primed mice by epigenetic mechanisms. Exp Mol Pathol. 2009;86:77–86. doi: 10.1016/j.yexmp.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Lin A, French BA, French SW. Role of cytokines in UBD promoter regulation and Mallory-Denk body-like aggresomes. Exp Mol Pathol. 2010;89:1–8. doi: 10.1016/j.yexmp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liiu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkyn SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identify and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Role of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Zapp J, Beeker H, Adani KP, Goto Y, Fell R. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrelle TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Villasante A, Piazzolla D, Li H, Gomez-Lopez G, Djabali M, Serrano M. Epigenetic regulation of Nanog expression by EZH2 in pluripotent stem cells. Cell Cycle. 2011;10:1488–1498. doi: 10.4161/cc.10.9.15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Long-Yuan L, Lang J, Su-Peng Y, Bin S, Cheng-Chieh Y, Jer-Yen, Chun-Yi L, Chen-Chen L, Mien-Chie H. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CKD1)-mediated phosphorylation of enhancer of Zeste 2 (EZH2) regulates its stability. J Biol Chem. 2011;286:28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan QX, Marceau N, French BA, Fu P, French SW. Mallory body induction in drug primed mouse liver. Hepatology. 1996;24:603–612. doi: 10.1002/hep.510240324. [DOI] [PubMed] [Google Scholar]
- Zeng XZ, Chen SA, Huang HJ. Phosphorylation of EZH2 by CDK1 and CDK2. A possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell cycle. 2011;10:579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]